Distinct Synaptic Vesicle Proteomic Signatures Associated with Pre- and Post-Natal Oxycodone-Exposure
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Oxycodone Treatment
2.3. Synaptic Vesicle Isolation
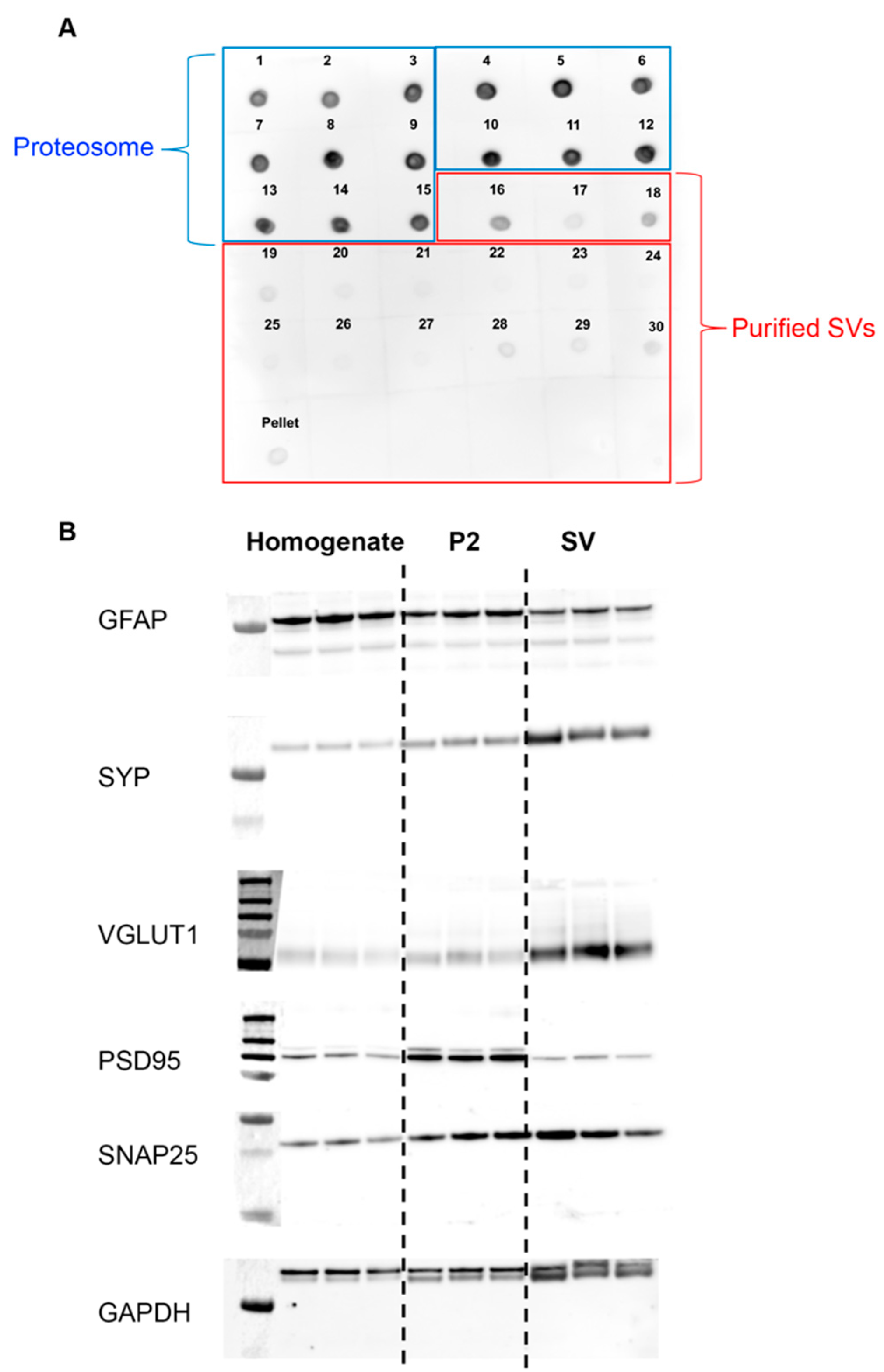
2.4. Dot Blot
2.5. Western Blot
2.6. Mass Spectrometry
2.7. Bioinformatic Analysis
2.8. Statistical Analyses
3. Results
3.1. Purity and Validation of Synaptic Vesicle Samples
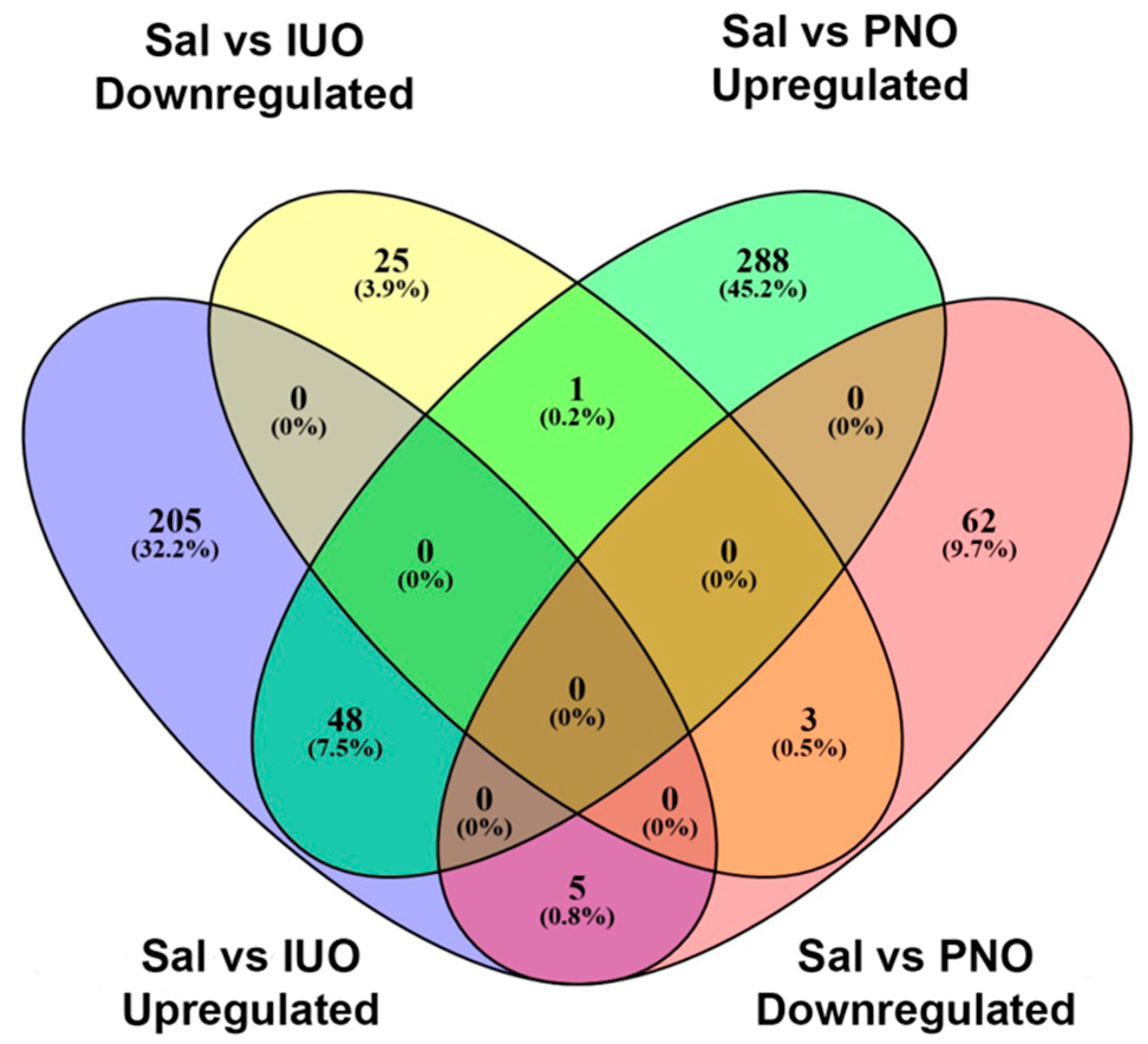
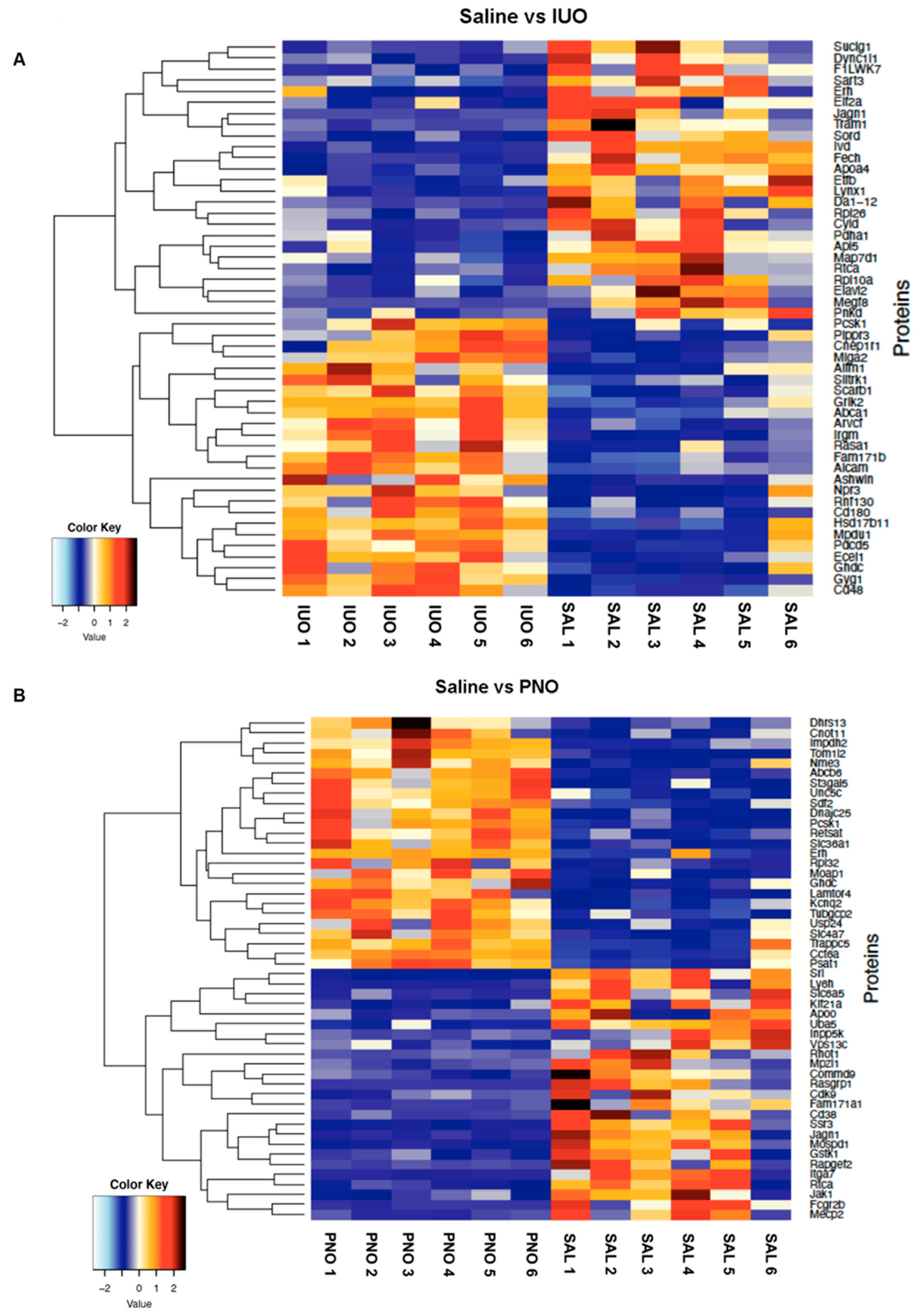
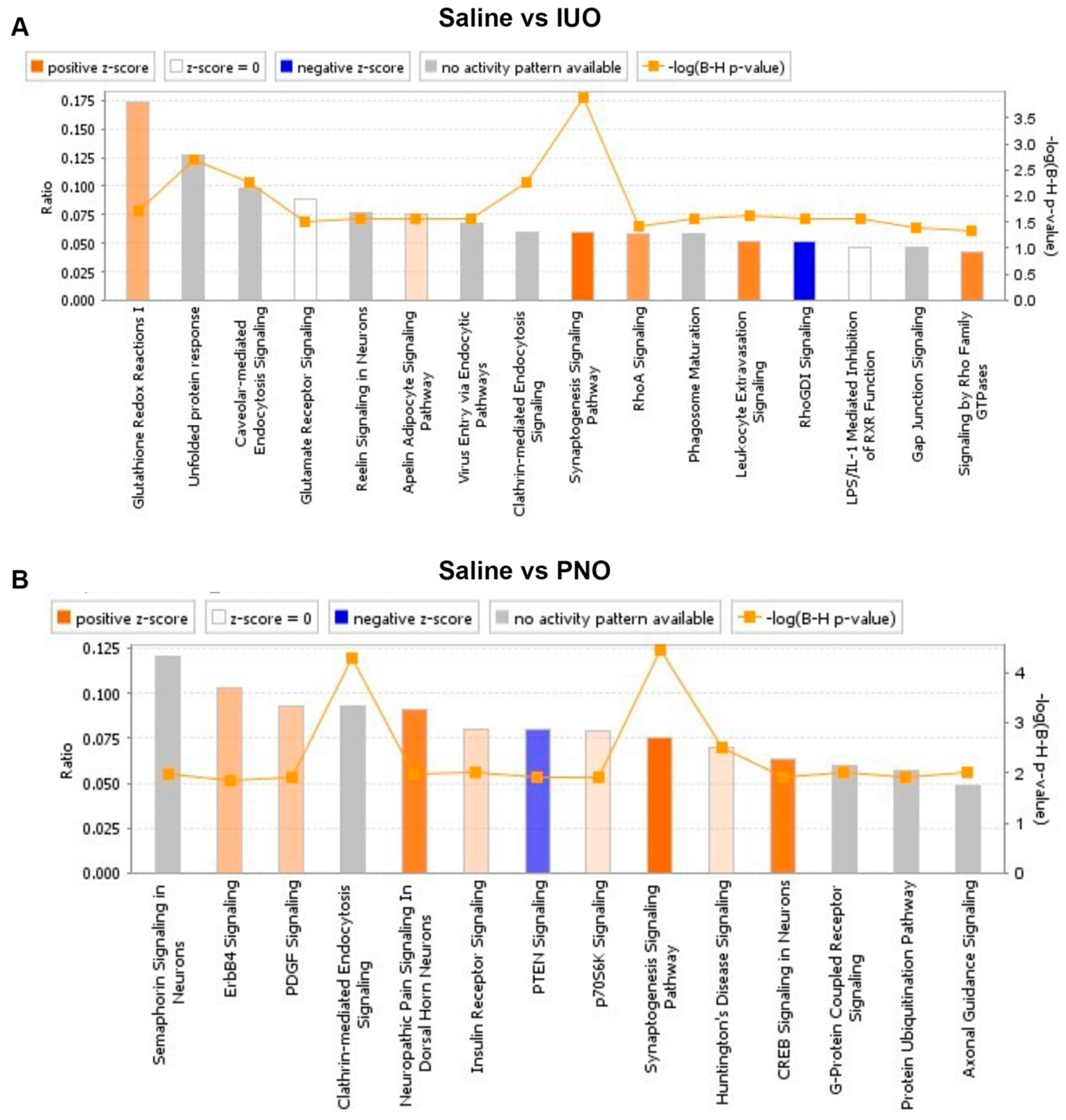
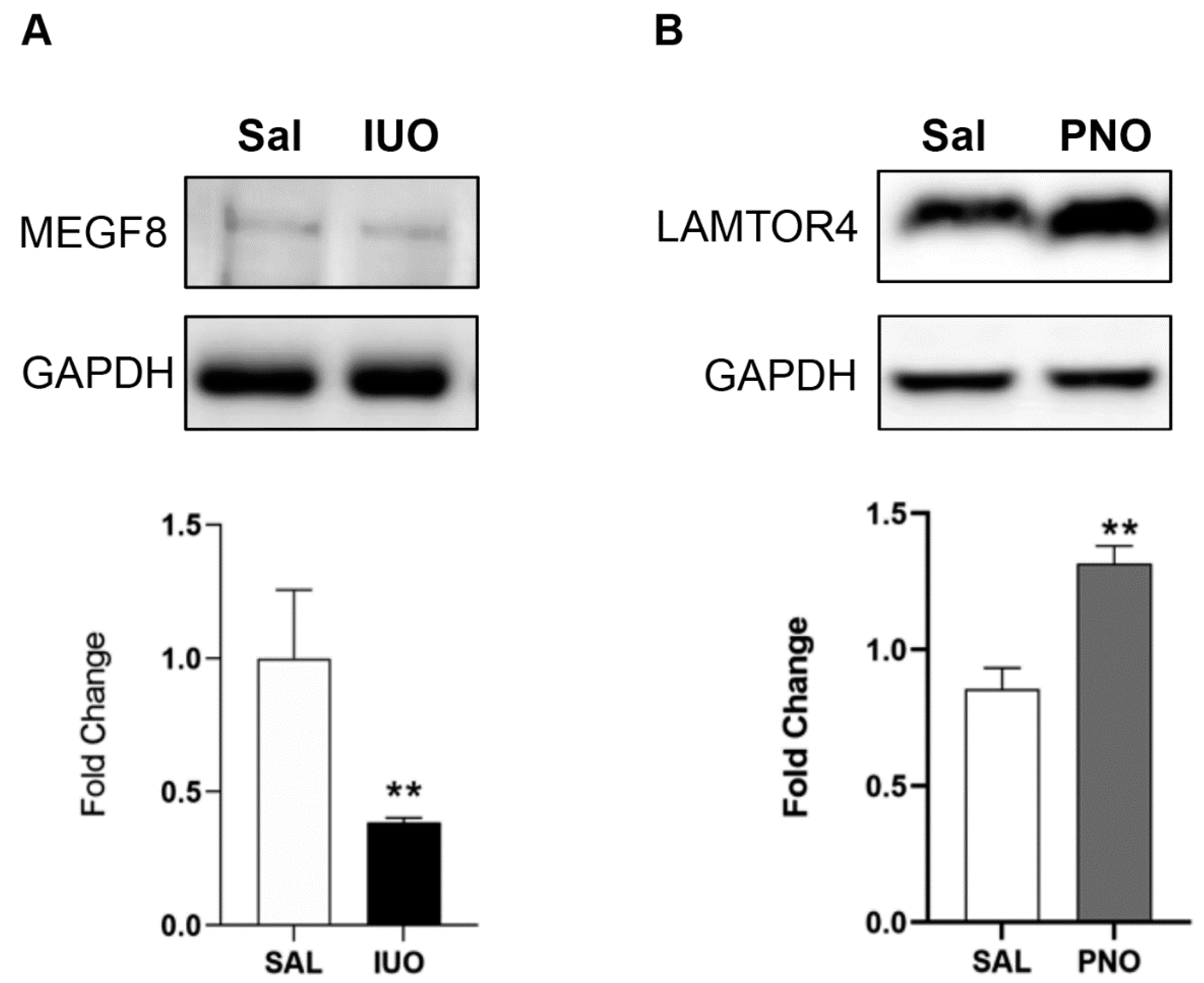
3.2. Differential Expression of Synaptic Vesicle Proteins
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Volkow, N.D.; McLellan, A.T. Opioid Abuse in Chronic Pain—Misconceptions and Mitigation Strategies. N. Engl. J. Med. 2016, 374, 1253–1263. [Google Scholar] [CrossRef] [PubMed]
- Gerdin, E.; Rane, A.; Lindberg, B. Transplacental transfer of morphine in man. J. Perinat. Med. 1990, 18, 305–312. [Google Scholar] [CrossRef] [PubMed]
- Nanovskaya, T.; Deshmukh, S.; Brooks, M.; Ahmed, M.S. Transplacental transfer and metabolism of buprenorphine. J. Pharmacol. Exp. Ther. 2002, 300, 26–33. [Google Scholar] [CrossRef] [PubMed]
- Nanovskaya, T.N.; Nekhayeva, I.A.; Hankins, G.D.; Ahmed, M.S. Transfer of methadone across the dually perfused preterm human placental lobule. Am. J. Obstet. Gynecol. 2008, 198, 126.e1–126.e4. [Google Scholar] [CrossRef]
- Kim, E.S. Oxycodone/Naloxone Prolonged Release: A Review in Severe Chronic Pain. Clin. Drug Investig. 2017, 37, 1191–1201. [Google Scholar] [CrossRef]
- Chaves, C.; Remiao, F.; Cisternino, S.; Decleves, X. Opioids and the Blood-Brain Barrier: A Dynamic Interaction with Consequences on Drug Disposition in Brain. Curr. Neuropharmacol. 2017, 15, 1156–1173. [Google Scholar] [CrossRef]
- Okura, T.; Hattori, A.; Takano, Y.; Sato, T.; Hammarlund-Udenaes, M.; Terasaki, T.; Deguchi, Y. Involvement of the pyrilamine transporter, a putative organic cation transporter, in blood-brain barrier transport of oxycodone. Drug Metab. Dispos. 2008, 36, 2005–2013. [Google Scholar] [CrossRef]
- Okura, T.; Higuchi, K.; Deguchi, Y. The blood-brain barrier transport mechanism controlling analgesic effects of opioid drugs in CNS. Yakugaku Zasshi 2015, 135, 697–702. [Google Scholar] [CrossRef]
- Byrnes, E.M.; Vassoler, F.M. Modeling prenatal opioid exposure in animals: Current findings and future directions. Front. Neuroendocrinol. 2018, 51, 1–13. [Google Scholar] [CrossRef]
- Robinson, T.E.; Kolb, B. Structural plasticity associated with exposure to drugs of abuse. Neuropharmacology 2004, 47 (Suppl. S1), 33–46. [Google Scholar] [CrossRef]
- Shahjin, F.; Guda, R.S.; Schaal, V.L.; Odegaard, K.; Clark, A.; Gowen, A.; Xiao, P.; Lisco, S.J.; Pendyala, G.; Yelamanchili, S.V. Brain-Derived Extracellular Vesicle microRNA Signatures Associated with In Utero and Postnatal Oxycodone Exposure. Cells 2019, 9, 21. [Google Scholar] [CrossRef] [PubMed]
- Odegaard, K.E. In Utero and Postnatal Oxycodone Exposure: Implications for Intergenerational Effects. Ph.D. Thesis, University of Nebraska Medical Center, Omaha, NE, USA, 2021. [Google Scholar]
- Ahmed, S.; Holt, M.; Riedel, D.; Jahn, R. Small-scale isolation of synaptic vesicles from mammalian brain. Nat. Protoc. 2013, 8, 998–1009. [Google Scholar] [CrossRef] [PubMed]
- Burré, J.; Volknandt, W. The synaptic vesicle proteome. J. Neurochem. 2007, 101, 1448–1462. [Google Scholar] [CrossRef] [PubMed]
- Abul-Husn, N.S.; Devi, L.A. Neuroproteomics of the synapse and drug addiction. J. Pharmacol. Exp. Ther. 2006, 318, 461–468. [Google Scholar] [CrossRef]
- Dieterich, D.C.; Kreutz, M.R. Proteomics of the Synapse--A Quantitative Approach to Neuronal Plasticity. Mol. Cell Proteom. 2016, 15, 368–381. [Google Scholar] [CrossRef]
- Odegaard, K.E.; Schaal, V.L.; Clark, A.R.; Koul, S.; Gowen, A.; Sankarasubramani, J.; Xiao, P.; Guda, C.; Lisco, S.J.; Yelamanchili, S.V.; et al. Characterization of the intergenerational impact of in utero and postnatal oxycodone exposure. Transl. Psychiatry 2020, 10, 329. [Google Scholar] [CrossRef]
- Odegaard, K.E.; Schaal, V.L.; Clark, A.R.; Koul, S.; Sankarasubramanian, J.; Xia, Z.; Mellon, M.; Uberti, M.; Liu, Y.; Stothert, A.; et al. A Holistic Systems Approach to Characterize the Impact of Pre- and Post-natal Oxycodone Exposure on Neurodevelopment and Behavior. Front. Cell Dev. Biol. 2020, 8, 619199. [Google Scholar] [CrossRef]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.H.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef]
- Twigg, S.R.; Lloyd, D.; Jenkins, D.; Elçioglu, N.E.; Cooper, C.D.; Al-Sannaa, N.; Annagür, A.; Gillessen-Kaesbach, G.; Hüning, I.; Knight, S.J.; et al. Mutations in multidomain protein MEGF8 identify a Carpenter syndrome subtype associated with defective lateralization. Am. J. Hum. Genet. 2012, 91, 897–905. [Google Scholar] [CrossRef]
- Yu, X.; Hu, L.; Liu, X.; Zhan, G.; Mei, M.; Wang, H.; Zhang, X.; Qiu, Z.; Zhou, W.; Yang, L. A Novel MYCN Variant Associated with Intellectual Disability Regulates Neuronal Development. Neurosci. Bull. 2018, 34, 854–858. [Google Scholar] [CrossRef]
- Shen, K.; Sidik, H.; Talbot, W.S. The Rag-Ragulator Complex Regulates Lysosome Function and Phagocytic Flux in Microglia. Cell Rep. 2016, 14, 547–559. [Google Scholar] [CrossRef] [PubMed]
- Russo, S.J.; Dietz, D.M.; Dumitriu, D.; Morrison, J.H.; Malenka, R.C.; Nestler, E.J. The addicted synapse: Mechanisms of synaptic and structural plasticity in nucleus accumbens. Trends Neurosci. 2010, 33, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Stockton, S.D.; Devi, L.A. An integrated quantitative proteomics and systems biology approach to explore synaptic protein profile changes during morphine exposure. Neuropsychopharmacology 2014, 39, 88–103. [Google Scholar] [CrossRef]
- Walhovd, K.B.; Watts, R.; Amlien, I.; Woodward, L.J. Neural tract development of infants born to methadone-maintained mothers. Pediatr. Neurol. 2012, 47, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Schweitzer, J.B.; Riggins, T.; Liang, X.; Gallen, C.; Kurup, P.K.; Ross, T.J.; Black, M.M.; Nair, P.; Salmeron, B.J. Prenatal drug exposure to illicit drugs alters working memory-related brain activity and underlying network properties in adolescence. Neurotoxicol. Teratol. 2015, 48, 69–77. [Google Scholar] [CrossRef]
- Merhar, S.L.; Parikh, N.A.; Braimah, A.; Poindexter, B.B.; Tkach, J.; Kline-Fath, B. White Matter Injury and Structural Anomalies in Infants with Prenatal Opioid Exposure. AJNR Am. J. Neuroradiol. 2019, 40, 2161–2165. [Google Scholar] [CrossRef]
- Fan, R.; Schrott, L.M.; Arnold, T.; Snelling, S.; Rao, M.; Graham, D.; Cornelius, A.; Korneeva, N.L. Chronic oxycodone induces axonal degeneration in rat brain. BMC Neurosci. 2018, 19, 15. [Google Scholar] [CrossRef]
- Prokai, L.; Zharikova, A.D.; Stevens, S.M. Effect of chronic morphine exposure on the synaptic plasma-membrane subproteome of rats: A quantitative protein profiling study based on isotope-coded affinity tags and liquid chromatography/mass spectrometry. J. Mass Spectrom. 2005, 40, 169–175. [Google Scholar] [CrossRef]
- Morón, J.A.; Abul-Husn, N.S.; Rozenfeld, R.; Dolios, G.; Wang, R.; Devi, L.A. Morphine administration alters the profile of hippocampal postsynaptic density-associated proteins: A proteomics study focusing on endocytic proteins. Mol. Cell Proteom. 2007, 6, 29–42. [Google Scholar] [CrossRef]
- Abul-Husn, N.S.; Annangudi, S.P.; Ma’ayan, A.; Ramos-Ortolaza, D.L.; Stockton, S.D.; Gomes, I.; Sweedler, J.V.; Devi, L.A. Chronic morphine alters the presynaptic protein profile: Identification of novel molecular targets using proteomics and network analysis. PLoS ONE 2011, 6, e25535. [Google Scholar] [CrossRef]
- Yang, L.; Sun, Z.S.; Zhu, Y.P. Proteomic analysis of rat prefrontal cortex in three phases of morphine-induced conditioned place preference. J. Proteome Res. 2007, 6, 2239–2247. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.N.; Liu, C.A.; Chung, M.Y.; Huang, H.C.; Yeh, G.C.; Wong, C.S.; Lin, W.W.; Yang, C.H.; Tao, P.L. Alterations of postsynaptic density proteins in the hippocampus of rat offspring from the morphine-addicted mother: Beneficial effect of dextromethorphan. Hippocampus 2006, 16, 521–530. [Google Scholar] [CrossRef] [PubMed]
- Nandhu, M.S.; Naijil, G.; Smijin, S.; Jayanarayanan, S.; Paulose, C.S. Opioid system functional regulation in neurological disease management. J. Neurosci. Res. 2010, 88, 3215–3221. [Google Scholar] [CrossRef] [PubMed]
- Nygaard, E.; Slinning, K.; Moe, V.; Fjell, A.; Walhovd, K.B. Mental health in youth prenatally exposed to opioids and poly-drugs and raised in permanent foster/adoptive homes: A prospective longitudinal study. Early Hum. Dev. 2019, 140, 104910. [Google Scholar] [CrossRef]
- Schultz, J.L.; Kamholz, J.A.; Moser, D.J.; Feely, S.M.; Paulsen, J.S.; Nopoulos, P.C. Substance abuse may hasten motor onset of Huntington disease: Evaluating the Enroll-HD database. Neurology 2017, 88, 909–915. [Google Scholar] [CrossRef]
- Mallappallil, M.; Sabu, J.; Friedman, E.A.; Salifu, M. What Do We Know about Opioids and the Kidney? Int. J. Mol. Sci. 2017, 18, 223. [Google Scholar] [CrossRef]
- Atici, S.; Cinel, I.; Cinel, L.; Doruk, N.; Eskandari, G.; Oral, U. Liver and kidney toxicity in chronic use of opioids: An experimental long term treatment model. J. Biosci. 2005, 30, 245–252. [Google Scholar] [CrossRef]
- Verna, E.C.; Schluger, A.; Brown, R.S. Opioid epidemic and liver disease. JHEP Rep. 2019, 1, 240–255. [Google Scholar] [CrossRef]
- Rahimi Darabad, B.; Vatandust, J.; Pourmousavi Khoshknab, M.M.; Hajahmadi Poorrafsanjani, M. Survey of the effect of opioid abuse on the extent of coronary artery diseases. Glob. J. Health Sci. 2014, 6, 83–91. [Google Scholar] [CrossRef]
- Ziaee, M.; Hajizadeh, R.; Khorrami, A.; Sepehrvand, N.; Momtaz, S.; Ghaffari, S. Cardiovascular Complications of Chronic Opium Consumption: A Narrative Review Article. Iran J. Public Health 2019, 48, 2154–2164. [Google Scholar] [CrossRef] [PubMed]
- Engelhard, C.; Sarsfield, S.; Merte, J.; Wang, Q.; Li, P.; Beppu, H.; Kolodkin, A.L.; Sucov, H.M.; Ginty, D.D. MEGF8 is a modifier of BMP signaling in trigeminal sensory neurons. Elife 2013, 2, e01160. [Google Scholar] [CrossRef] [PubMed]
- Lamberti, G.; De Smet, C.H.; Angelova, M.; Kremser, L.; Taub, N.; Herrmann, C.; Hess, M.W.; Rainer, J.; Tancevski, I.; Schweigreiter, R.; et al. LAMTOR/Ragulator regulates lipid metabolism in macrophages and foam cell differentiation. FEBS Lett. 2020, 594, 31–42. [Google Scholar] [CrossRef] [PubMed]
- de Araujo, M.E.G.; Naschberger, A.; Fürnrohr, B.G.; Stasyk, T.; Dunzendorfer-Matt, T.; Lechner, S.; Welti, S.; Kremser, L.; Shivalingaiah, G.; Offterdinger, M.; et al. Crystal structure of the human lysosomal mTORC1 scaffold complex and its impact on signaling. Science 2017, 358, 377–381. [Google Scholar] [CrossRef]
- Lutz, B.M.; Nia, S.; Xiong, M.; Tao, Y.-X.; Bekker, A. mTOR, a new potential target for chronic pain and opioid-induced tolerance and hyperalgesia. Mol. Pain 2015, 11, 32. [Google Scholar] [CrossRef]
- Jantzie, L.L.; Maxwell, J.R.; Newville, J.C.; Yellowhair, T.R.; Kitase, Y.; Madurai, N.; Ramachandra, S.; Bakhireva, L.N.; Northington, F.J.; Gerner, G.; et al. Prenatal opioid exposure: The next neonatal neuroinflammatory disease. Brain Behav. Immun. 2020, 84, 45–58. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Odegaard, K.E.; Gallegos, G.; Koul, S.; Schaal, V.L.; Vellichirammal, N.N.; Guda, C.; Dutoit, A.P.; Lisco, S.J.; Yelamanchili, S.V.; Pendyala, G. Distinct Synaptic Vesicle Proteomic Signatures Associated with Pre- and Post-Natal Oxycodone-Exposure. Cells 2022, 11, 1740. https://doi.org/10.3390/cells11111740
Odegaard KE, Gallegos G, Koul S, Schaal VL, Vellichirammal NN, Guda C, Dutoit AP, Lisco SJ, Yelamanchili SV, Pendyala G. Distinct Synaptic Vesicle Proteomic Signatures Associated with Pre- and Post-Natal Oxycodone-Exposure. Cells. 2022; 11(11):1740. https://doi.org/10.3390/cells11111740
Chicago/Turabian StyleOdegaard, Katherine E., Gabriel Gallegos, Sneh Koul, Victoria L. Schaal, Neetha N. Vellichirammal, Chittibabu Guda, Andrea P. Dutoit, Steven J. Lisco, Sowmya V. Yelamanchili, and Gurudutt Pendyala. 2022. "Distinct Synaptic Vesicle Proteomic Signatures Associated with Pre- and Post-Natal Oxycodone-Exposure" Cells 11, no. 11: 1740. https://doi.org/10.3390/cells11111740
APA StyleOdegaard, K. E., Gallegos, G., Koul, S., Schaal, V. L., Vellichirammal, N. N., Guda, C., Dutoit, A. P., Lisco, S. J., Yelamanchili, S. V., & Pendyala, G. (2022). Distinct Synaptic Vesicle Proteomic Signatures Associated with Pre- and Post-Natal Oxycodone-Exposure. Cells, 11(11), 1740. https://doi.org/10.3390/cells11111740




_Guda.png)


