Identification of the Transcriptional Biomarkers Panel Linked to Pathological Remodelling of the Eye Tissues in Various HD Mouse Models
Abstract
1. Introduction
2. Material and Methods
2.1. Mouse Maintenance and Genotyping
2.2. RNA Extraction and Taqman Real-Time PCR Expression Analysis
2.3. RNA Isolation for Array Analysis
2.4. Microarray Expression Study
2.5. Microarray Data Analysis
2.6. Assignment of Differentially Expressed Genes to Relevant Gene Ontology (GO) Terms
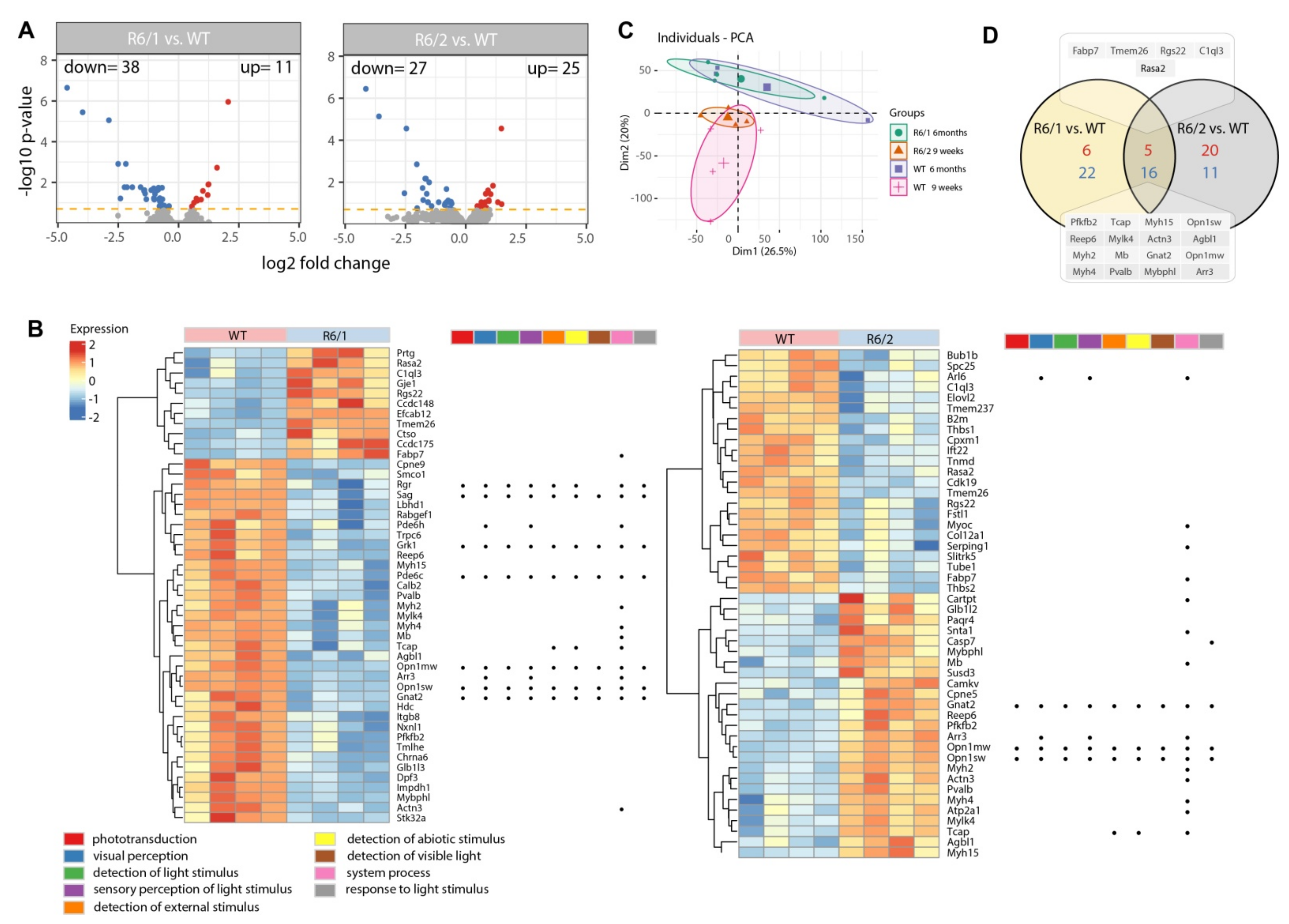
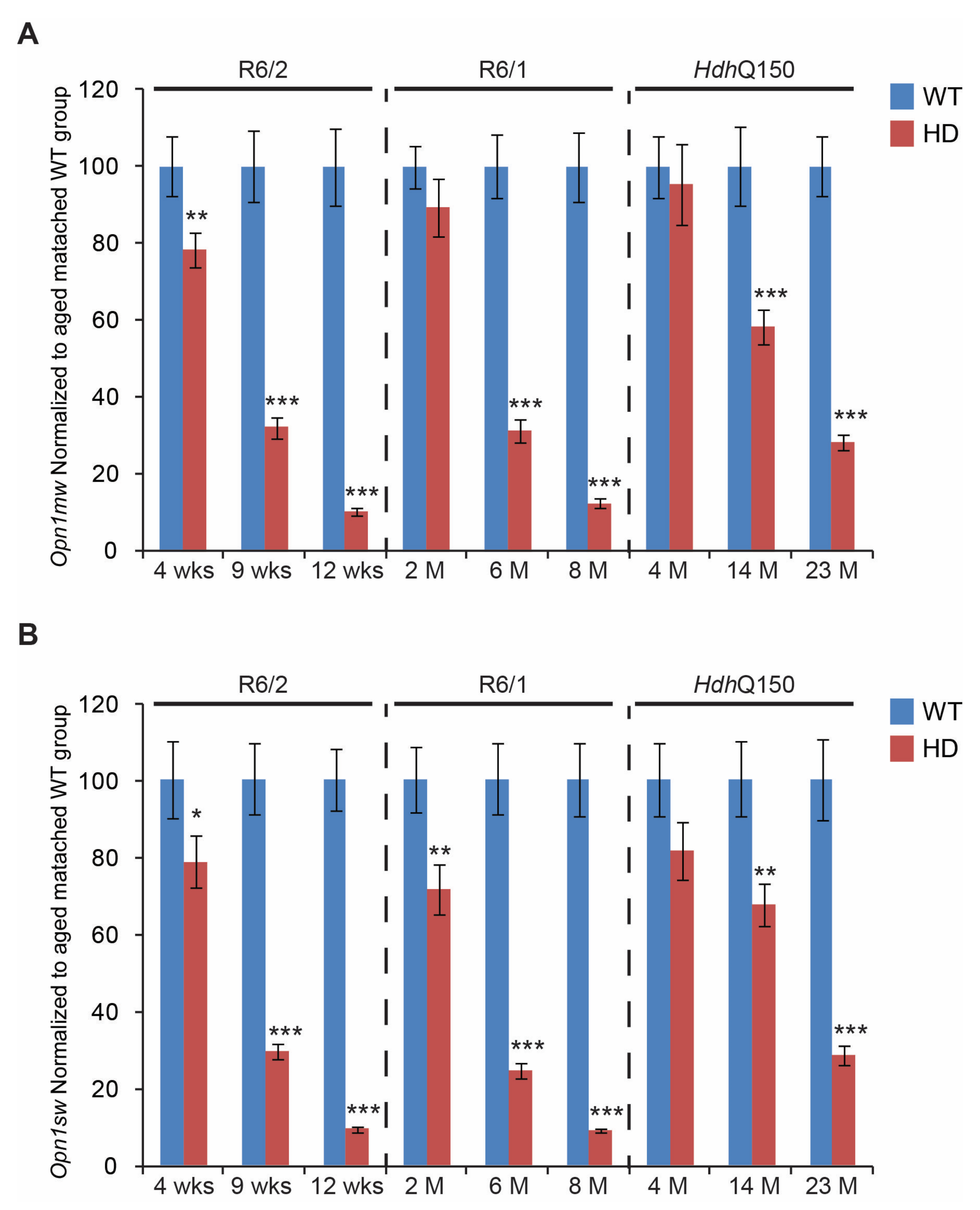
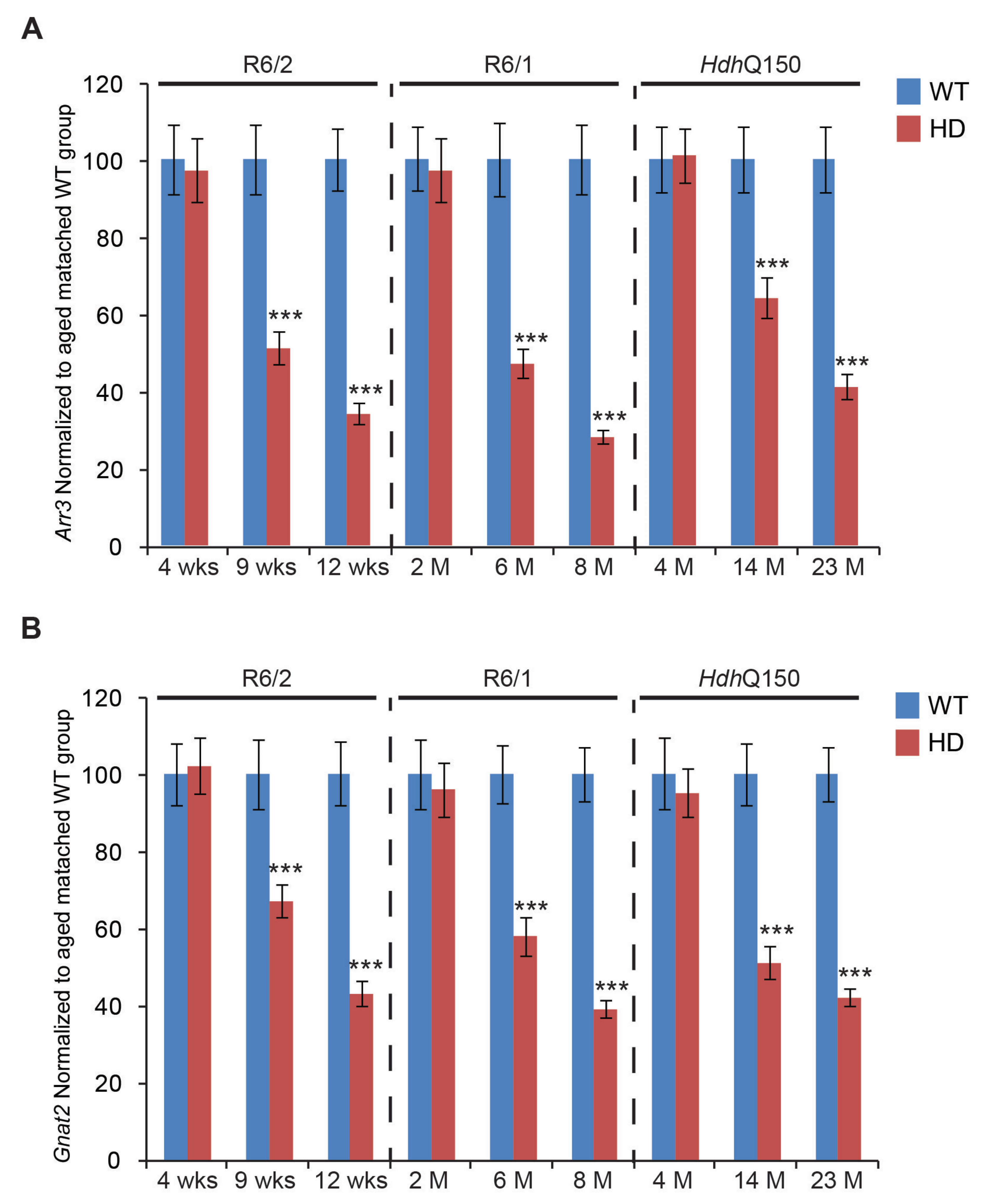
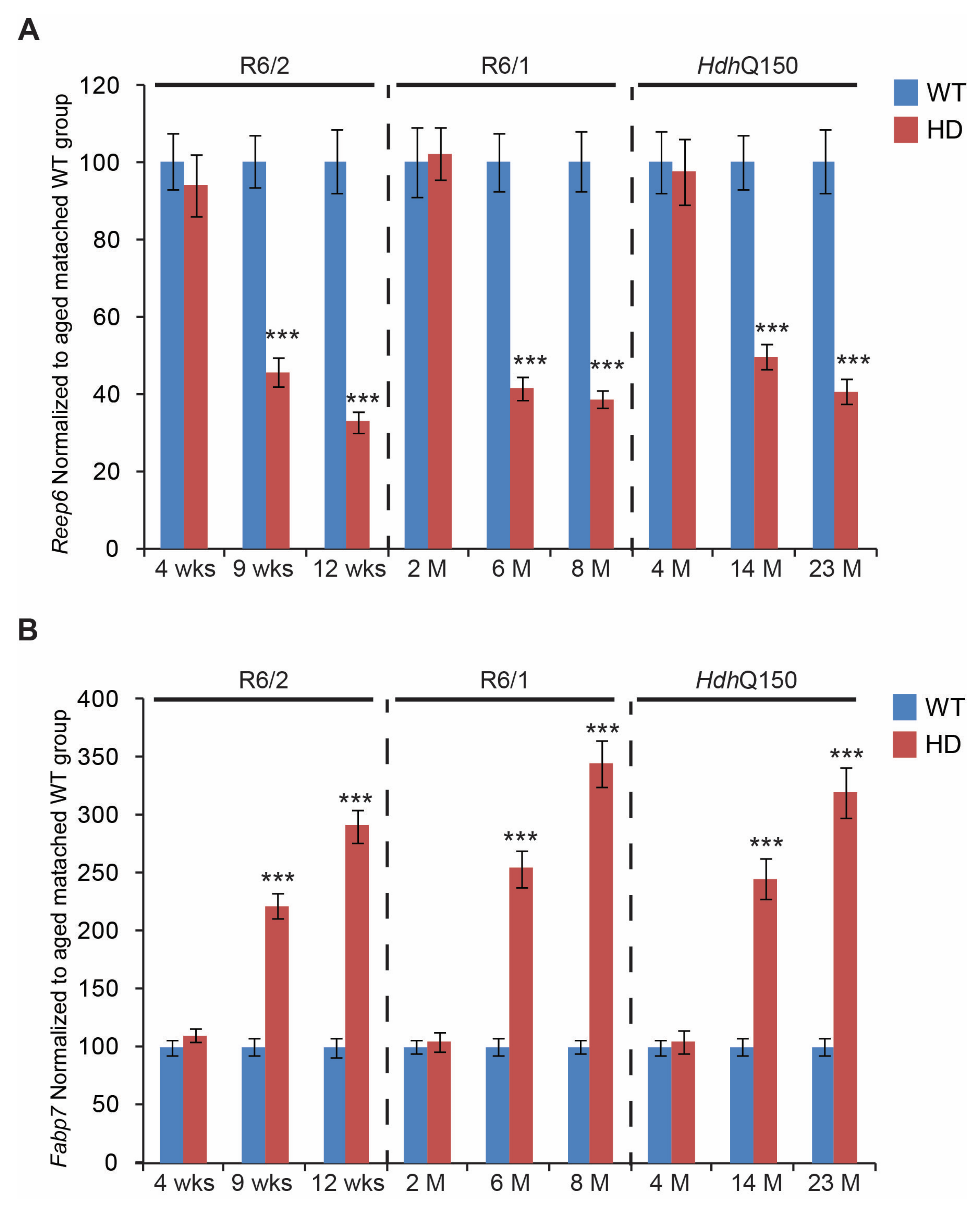
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Guidoboni, G.; Sacco, R.; Szopos, M.; Sala, L.; Verticchio Vercellin, A.C.; Siesky, B.; Harris, A. Neurodegenerative Disorders of the Eye and of the Brain: A Perspective on Their Fluid-Dynamical Connections and the Potential of Mechanism-Driven Modeling. Front. Neurosci. 2020, 14, 566428. [Google Scholar] [CrossRef] [PubMed]
- Marchesi, N.; Fahmideh, F.; Boschi, F.; Pascale, A.; Barbieri, A. Ocular Neurodegenerative Diseases: Interconnection between Retina and Cortical Areas. Cells 2021, 10, 2394. [Google Scholar] [CrossRef] [PubMed]
- Snyder, P.J.; Alber, J.; Alt, C.; Bain, L.J.; Bouma, B.E.; Bouwman, F.H.; DeBuc, D.C.; Campbell, M.C.W.; Carrillo, M.C.; Chew, E.Y.; et al. Retinal imaging in Alzheimer’s and neurodegenerative diseases. Alzheimers Dement. 2021, 17, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Mangiarini, L.; Sathasivam, K.; Seller, M.; Cozens, B.; Harper, A.; Hetherington, C.; Lawton, M.; Trottier, Y.; Lehrach, H.; Davies, S.W.; et al. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell 1996, 87, 493–506. [Google Scholar] [CrossRef]
- Schulte, J.; Littleton, J.T. The biological function of the Huntingtin protein and its relevance to Huntington’s Disease pathology. Curr. Trends Neurol. 2011, 5, 65–78. [Google Scholar] [PubMed]
- Zielonka, D.; Mielcarek, M.; Landwehrmeyer, G.B. Update on Huntington’s disease: Advances in care and emerging therapeutic options. Parkinsonism Relat. Disord 2015, 21, 169–178. [Google Scholar] [CrossRef] [PubMed]
- Zielonka, D.; Witkowski, G.; Puch, E.A.; Lesniczak, M.; Mazur-Michalek, I.; Isalan, M.; Mielcarek, M. Prevalence of Non-psychiatric Comorbidities in Pre-symptomatic and Symptomatic Huntington’s Disease Gene Carriers in Poland. Front. Med. 2020, 7, 79. [Google Scholar] [CrossRef]
- Li, S.H.; Schilling, G.; Young, W.S., III; Li, X.J.; Margolis, R.L.; Stine, O.C.; Wagster, M.V.; Abbott, M.H.; Franz, M.L.; Ranen, N.G.; et al. Huntington’s disease gene (IT15) is widely expressed in human and rat tissues. Neuron 1993, 11, 985–993. [Google Scholar] [CrossRef]
- Dragatsis, I.; Efstratiadis, A.; Zeitlin, S. Mouse mutant embryos lacking huntingtin are rescued from lethality by wild-type extraembryonic tissues. Development 1998, 125, 1529–1539. [Google Scholar] [CrossRef]
- Mielcarek, M. Huntington’s disease is a multi-system disorder. Rare Dis. 2015, 3, e1058464. [Google Scholar] [CrossRef]
- Mielcarek, M.; Isalan, M. Polyglutamine diseases: Looking beyond the neurodegenerative universe. Neural Regen. Res. 2021, 16, 1186–1187. [Google Scholar] [CrossRef] [PubMed]
- Mielcarek, M.; Inuabasi, L.; Bondulich, M.K.; Muller, T.; Osborne, G.F.; Franklin, S.A.; Smith, D.L.; Neueder, A.; Rosinski, J.; Rattray, I.; et al. Dysfunction of the CNS-heart axis in mouse models of Huntington’s disease. PLoS Genet. 2014, 10, e1004550. [Google Scholar] [CrossRef]
- Mielcarek, M.; Bondulich, M.K.; Inuabasi, L.; Franklin, S.A.; Muller, T.; Bates, G.P. The Huntington’s disease-related cardiomyopathy prevents a hypertrophic response in the R6/2 mouse model. PLoS ONE 2014, 9, e108961. [Google Scholar] [CrossRef] [PubMed]
- Toczek, M.; Zielonka, D.; Zukowska, P.; Marcinkowski, J.T.; Slominska, E.; Isalan, M.; Smolenski, R.T.; Mielcarek, M. An impaired metabolism of nucleotides underpins a novel mechanism of cardiac remodeling leading to Huntington’s disease related cardiomyopathy. Biochim. Biophys. Acta 2016, 1862, 2147–2157. [Google Scholar] [CrossRef] [PubMed]
- Critchley, B.J.; Isalan, M.; Mielcarek, M. Neuro-Cardio Mechanisms in Huntington’s Disease and Other Neurodegenerative Disorders. Front. Physiol. 2018, 9, 559. [Google Scholar] [CrossRef] [PubMed]
- Mielcarek, M.; Toczek, M.; Smeets, C.J.; Franklin, S.A.; Bondulich, M.K.; Jolinon, N.; Muller, T.; Ahmed, M.; Dick, J.R.; Piotrowska, I.; et al. HDAC4-myogenin axis as an important marker of HD-related skeletal muscle atrophy. PLoS Genet. 2015, 11, e1005021. [Google Scholar] [CrossRef]
- Mielcarek, M.; Smolenski, R.T.; Isalan, M. Transcriptional Signature of an Altered Purine Metabolism in the Skeletal Muscle of a Huntington’s Disease Mouse Model. Front. Physiol. 2017, 8, 127. [Google Scholar] [CrossRef]
- Mielcarek, M.; Isalan, M. A shared mechanism of muscle wasting in cancer and Huntington’s disease. Clin. Transl. Med. 2015, 4, 34. [Google Scholar] [CrossRef]
- Mielcarek, M.; Rattray, I.; Osborne, G.F.; Jolinon, N.; Dick, J.R.T.; Bondulich, M.K.; Franklin, S.A.; Ahmed, M.; Benjamin, A.C.; Goodwin, D.; et al. Myostatin Inhibition as a Novel Approach to Targeting Muscle Pathology in HD. J. Neurol. Neurosurg. Psychiatry 2014, 85, A97. [Google Scholar] [CrossRef]
- Helmlinger, D.; Yvert, G.; Picaud, S.; Merienne, K.; Sahel, J.; Mandel, J.L.; Devys, D. Progressive retinal degeneration and dysfunction in R6 Huntington’s disease mice. Hum. Mol. Genet. 2002, 11, 3351–3359. [Google Scholar] [CrossRef]
- Batcha, A.H.; Greferath, U.; Jobling, A.I.; Vessey, K.A.; Ward, M.M.; Nithianantharajah, J.; Hannan, A.J.; Kalloniatis, M.; Fletcher, E.L. Retinal dysfunction, photoreceptor protein dysregulation and neuronal remodelling in the R6/1 mouse model of Huntington’s disease. Neurobiol. Dis. 2012, 45, 887–896. [Google Scholar] [CrossRef] [PubMed]
- Lasker, A.G.; Zee, D.S. Ocular motor abnormalities in Huntington’s disease. Vis. Res. 1997, 37, 3639–3645. [Google Scholar] [CrossRef]
- Gulmez Sevim, D.; Unlu, M.; Gultekin, M.; Karaca, C. Retinal single-layer analysis with optical coherence tomography shows inner retinal layer thinning in Huntington’s disease as a potential biomarker. Int. Ophthalmol. 2019, 39, 611–621. [Google Scholar] [CrossRef] [PubMed]
- Mielcarek, M.; Landles, C.; Weiss, A.; Bradaia, A.; Seredenina, T.; Inuabasi, L.; Osborne, G.F.; Wadel, K.; Touller, C.; Butler, R.; et al. HDAC4 reduction: A novel therapeutic strategy to target cytoplasmic huntingtin and ameliorate neurodegeneration. PLoS Biol. 2013, 11, e1001717. [Google Scholar] [CrossRef]
- Agustin-Pavon, C.; Mielcarek, M.; Garriga-Canut, M.; Isalan, M. Deimmunization for gene therapy: Host matching of synthetic zinc finger constructs enables long-term mutant Huntingtin repression in mice. Mol. Neurodegener. 2016, 11, 64. [Google Scholar] [CrossRef]
- Mielcarek, M.; Benn, C.L.; Franklin, S.A.; Smith, D.L.; Woodman, B.; Marks, P.A.; Bates, G.P. SAHA decreases HDAC 2 and 4 levels in vivo and improves molecular phenotypes in the R6/2 mouse model of Huntington’s disease. PLoS ONE 2011, 6, e27746. [Google Scholar] [CrossRef]
- Stelcer, E.; Milecka, P.; Komarowska, H.; Jopek, K.; Tyczewska, M.; Szyszka, M.; Lesniczak, M.; Suchorska, W.; Bekova, K.; Szczepaniak, B.; et al. Adropin Stimulates Proliferation and Inhibits Adrenocortical Steroidogenesis in the Human Adrenal Carcinoma (HAC15) Cell Line. Front. Endocrinol. 2020, 11, 561370. [Google Scholar] [CrossRef]
- Szyszka, M.; Paschke, L.; Tyczewska, M.; Jopek, K.; Celichowski, P.; Milecka, P.; Sultanova, G.; Stelcer, E.; Malinska, A.; Malendowicz, L.K.; et al. Analysis of Transcriptome, Selected Intracellular Signaling Pathways, Proliferation and Apoptosis of LNCaP Cells Exposed to High Leptin Concentrations. Int. J. Mol. Sci. 2019, 20, 5412. [Google Scholar] [CrossRef]
- Jopek, K.; Tyczewska, M.; Ramanjaneya, M.; Szyszka, M.; Celichowski, P.; Milecka, P.; Malendowicz, L.K.; Rucinski, M. Effect of ACTH and hCG on the Expression of Gonadotropin-Inducible Ovarian Transcription Factor 1 (Giot1) Gene in the Rat Adrenal Gland. Int. J. Mol. Sci. 2018, 19, 2285. [Google Scholar] [CrossRef]
- Gautier, L.; Cope, L.; Bolstad, B.M.; Irizarry, R.A. Affy—Analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 2004, 20, 307–315. [Google Scholar] [CrossRef]
- MacDonald, J.W. Mogene21sttranscriptcluster.db: Affymetrix Mogene21 Annotation Data (Chip Mogene21st Transcript Cluster). R Package Version 880. 2021. Available online: https://bioconductor.org/packages/release/data/annotation/html/mogene21sttranscriptcluster.db.html (accessed on 25 March 2022).
- Gentleman, R.C.V.J.; Huber, W.; Hahne, F. Genefilter: Genefilter: Methods for Filtering Genes from High-Throughput Experiments. R Package Version 17.0. 2021. Available online: https://rdrr.io/bioc/genefilter/ (accessed on 25 March 2022).
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Kassambara, A.M.F. Factoextra: Extract and Visualize the Results of Multivariate Data Analyses. R Package Version 107. 2020. Available online: https://cran.r-project.org/web/packages/factoextra/readme/README.html (accessed on 25 March 2022).
- Dennis, G., Jr.; Sherman, B.T.; Hosack, D.A.; Yang, J.; Gao, W.; Lane, H.C.; Lempicki, R.A. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003, 4, R60. [Google Scholar] [CrossRef]
- Fresno, C.; Fernandez, E.A. RDAVIDWebService: A versatile R interface to DAVID. Bioinformatics 2013, 29, 2810–2811. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Cohen, R. Weighted false discovery rate controlling procedures for clinical trials. Biostatistics 2017, 18, 91–104. [Google Scholar] [CrossRef]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef]
- Yan, L. Ggvenn: Draw Venn Diagram by ‘ggplot2’. R Package Version 019. 2021. Available online: https://cran.r-project.org/web/packages/ggvenn/index.html (accessed on 25 March 2022).
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Lin, C.H.; Tallaksen-Greene, S.; Chien, W.M.; Cearley, J.A.; Jackson, W.S.; Crouse, A.B.; Ren, S.; Li, X.J.; Albin, R.L.; Detloff, P.J. Neurological abnormalities in a knock-in mouse model of Huntington’s disease. Hum. Mol. Genet. 2001, 10, 137–144. [Google Scholar] [CrossRef]
- Zielonka, D.; Piotrowska, I.; Marcinkowski, J.T.; Mielcarek, M. Skeletal muscle pathology in Huntington’s disease. Front. Physiol. 2014, 5, 380. [Google Scholar] [CrossRef]
- Zielonka, D.; Piotrowska, I.; Mielcarek, M. Cardiac dysfunction in Huntington’s Disease. Exp. Clin. Cardiol. 2014, 20, 2547–2554. [Google Scholar]
- Moffitt, H.; McPhail, G.D.; Woodman, B.; Hobbs, C.; Bates, G.P. Formation of polyglutamine inclusions in a wide range of non-CNS tissues in the HdhQ150 knock-in mouse model of Huntington’s disease. PLoS ONE 2009, 4, e8025. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.T.; Li, J.; Zhu, P.; Freedman, B.; Smith, W.C.; Baehr, W.; Hauswirth, W.W. Rescue of M-cone Function in Aged Opn1mw-/- Mice, a Model for Late-Stage Blue Cone Monochromacy. Investig. Ophthalmol. Vis. Sci. 2019, 60, 3644–3651. [Google Scholar] [CrossRef] [PubMed]
- Baraas, R.C.; Hagen, L.A.; Dees, E.W.; Neitz, M. Substitution of isoleucine for threonine at position 190 of S-opsin causes S-cone-function abnormalities. Vis. Res. 2012, 73, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Thapa, A.; Morris, L.M.; Michalakis, S.; Biel, M.; Frank, M.B.; Bebak, M.; Ding, X.Q. Loss of cone cyclic nucleotide-gated channel leads to alterations in light response modulating system and cellular stress response pathways: A gene expression profiling study. Hum. Mol. Genet. 2013, 22, 3906–3919. [Google Scholar] [CrossRef][Green Version]
- Pang, J.J.; Alexander, J.; Lei, B.; Deng, W.; Zhang, K.; Li, Q.; Chang, B.; Hauswirth, W.W. Achromatopsia as a potential candidate for gene therapy. Adv. Exp. Med. Biol. 2010, 664, 639–646. [Google Scholar] [CrossRef] [PubMed]
- Ronning, K.E.; Allina, G.P.; Miller, E.B.; Zawadzki, R.J.; Pugh, E.N., Jr.; Herrmann, R.; Burns, M.E. Loss of cone function without degeneration in a novel Gnat2 knock-out mouse. Exp. Eye Res. 2018, 171, 111–118. [Google Scholar] [CrossRef]
- Liang, Q.; Wu, N.; Zaneveld, S.; Liu, H.; Fu, S.; Wang, K.; Bertrand, R.; Wang, J.; Li, Y.; Chen, R. Transcript isoforms of Reep6 have distinct functions in the retina. Hum. Mol. Genet. 2021, 30, 1907–1918. [Google Scholar] [CrossRef]
- Lin, Y.; Xu, C.L.; Velez, G.; Yang, J.; Tanaka, A.J.; Breazzano, M.P.; Mahajan, V.B.; Sparrow, J.R.; Tsang, S.H. Novel REEP6 gene mutation associated with autosomal recessive retinitis pigmentosa. Doc. Ophthalmol. 2020, 140, 67–75. [Google Scholar] [CrossRef]
- Zaneveld, S.A.; Eblimit, A.; Liang, Q.; Bertrand, R.; Wu, N.; Liu, H.; Nguyen, Q.; Zaneveld, J.; Wang, K.; Li, Y.; et al. Gene Therapy Rescues Retinal Degeneration in Receptor Expression-Enhancing Protein 6 Mutant Mice. Hum. Gene Ther. 2019, 30, 302–315. [Google Scholar] [CrossRef]
- Su, X.; Tan, Q.S.; Parikh, B.H.; Tan, A.; Mehta, M.N.; Sia Wey, Y.; Tun, S.B.; Li, L.J.; Han, X.Y.; Wong, T.Y.; et al. Characterization of Fatty Acid Binding Protein 7 (FABP7) in the Murine Retina. Vis. Sci. 2016, 57, 3397–3408. [Google Scholar] [CrossRef][Green Version]
- Killoy, K.M.; Harlan, B.A.; Pehar, M.; Vargas, M.R. FABP7 upregulation induces a neurotoxic phenotype in astrocytes. Glia 2020, 68, 2693–2704. [Google Scholar] [CrossRef] [PubMed]
- Rider, M.H.; Bertrand, L.; Vertommen, D.; Michels, P.A.; Rousseau, G.G.; Hue, L. 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase: Head-to-head with a bifunctional enzyme that controls glycolysis. Biochem. J. 2004, 381, 561–579. [Google Scholar] [CrossRef] [PubMed]
- Tang, N.; Zhang, J.; Fu, X.; Xie, W.; Qiu, Y. PP2Acalpha inhibits PFKFB2-induced glycolysis to promote termination of liver regeneration. Biochem. Biophys. Res. Commun. 2020, 526, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Moon, J.S.; Jin, W.J.; Kwak, J.H.; Kim, H.J.; Yun, M.J.; Kim, J.W.; Park, S.W.; Kim, K.S. Androgen stimulates glycolysis for de novo lipid synthesis by increasing the activities of hexokinase 2 and 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 2 in prostate cancer cells. Biochem. J. 2011, 433, 225–233. [Google Scholar] [CrossRef]
- Byerly, M.S.; Swanson, R.; Wei, Z.; Seldin, M.M.; McCulloh, P.S.; Wong, G.W. A central role for C1q/TNF-related protein 13 (CTRP13) in modulating food intake and body weight. PLoS ONE 2013, 8, e62862. [Google Scholar] [CrossRef]
- Gupta, R.; Nguyen, D.C.; Schaid, M.D.; Lei, X.; Balamurugan, A.N.; Wong, G.W.; Kim, J.A.; Koltes, J.E.; Kimple, M.E.; Bhatnagar, S. Complement 1q-like-3 protein inhibits insulin secretion from pancreatic beta-cells via the cell adhesion G protein-coupled receptor BAI3. J. Biol. Chem. 2018, 293, 18086–18098. [Google Scholar] [CrossRef]
- Wei, Z.; Peterson, J.M.; Wong, G.W. Metabolic regulation by C1q/TNF-related protein-13 (CTRP13): Activation OF AMP-activated protein kinase and suppression of fatty acid-induced JNK signaling. J. Biol. Chem. 2011, 286, 15652–15665. [Google Scholar] [CrossRef]
- O’Bryhim, B.E.; Apte, R.S.; Kung, N.; Coble, D.; Van Stavern, G.P. Association of Preclinical Alzheimer Disease With Optical Coherence Tomographic Angiography Findings. JAMA Ophthalmol. 2018, 136, 1242–1248. [Google Scholar] [CrossRef]
- Huang, L.; Zhang, D.; Ji, J.; Wang, Y.; Zhang, R. Central retina changes in Parkinson’s disease: A systematic review and meta-analysis. J. Neurol. 2021, 268, 4646–4654. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mazur-Michałek, I.; Ruciński, M.; Sowiński, M.; Pietras, P.; Leśniczak-Staszak, M.; Szaflarski, W.; Isalan, M.; Mielcarek, M. Identification of the Transcriptional Biomarkers Panel Linked to Pathological Remodelling of the Eye Tissues in Various HD Mouse Models. Cells 2022, 11, 1675. https://doi.org/10.3390/cells11101675
Mazur-Michałek I, Ruciński M, Sowiński M, Pietras P, Leśniczak-Staszak M, Szaflarski W, Isalan M, Mielcarek M. Identification of the Transcriptional Biomarkers Panel Linked to Pathological Remodelling of the Eye Tissues in Various HD Mouse Models. Cells. 2022; 11(10):1675. https://doi.org/10.3390/cells11101675
Chicago/Turabian StyleMazur-Michałek, Iwona, Marcin Ruciński, Mateusz Sowiński, Paulina Pietras, Marta Leśniczak-Staszak, Witold Szaflarski, Mark Isalan, and Michal Mielcarek. 2022. "Identification of the Transcriptional Biomarkers Panel Linked to Pathological Remodelling of the Eye Tissues in Various HD Mouse Models" Cells 11, no. 10: 1675. https://doi.org/10.3390/cells11101675
APA StyleMazur-Michałek, I., Ruciński, M., Sowiński, M., Pietras, P., Leśniczak-Staszak, M., Szaflarski, W., Isalan, M., & Mielcarek, M. (2022). Identification of the Transcriptional Biomarkers Panel Linked to Pathological Remodelling of the Eye Tissues in Various HD Mouse Models. Cells, 11(10), 1675. https://doi.org/10.3390/cells11101675






