Discovery of a Novel Leaf Rust (Puccinia recondita) Resistance Gene in Rye (Secale cereale L.) Using Association Genomics
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and DNA Extraction
2.2. Molecular Marker Resource and SNP Genotyping
2.3. Collection of Puccinia Recondite f. sp. secalis Populations
2.4. Field Trial
2.5. Field Inoculation
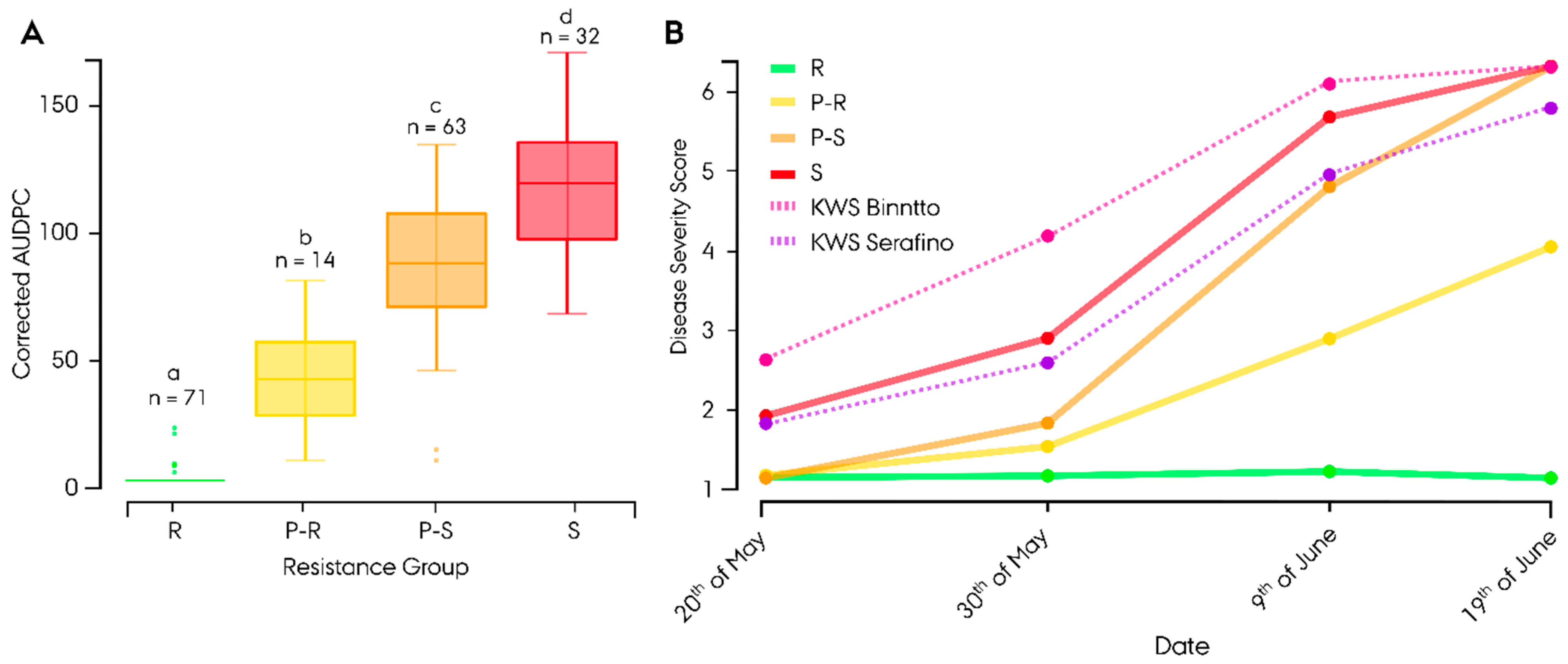
2.6. Disease Scoring
2.7. Analysis of Disease Scoring Data
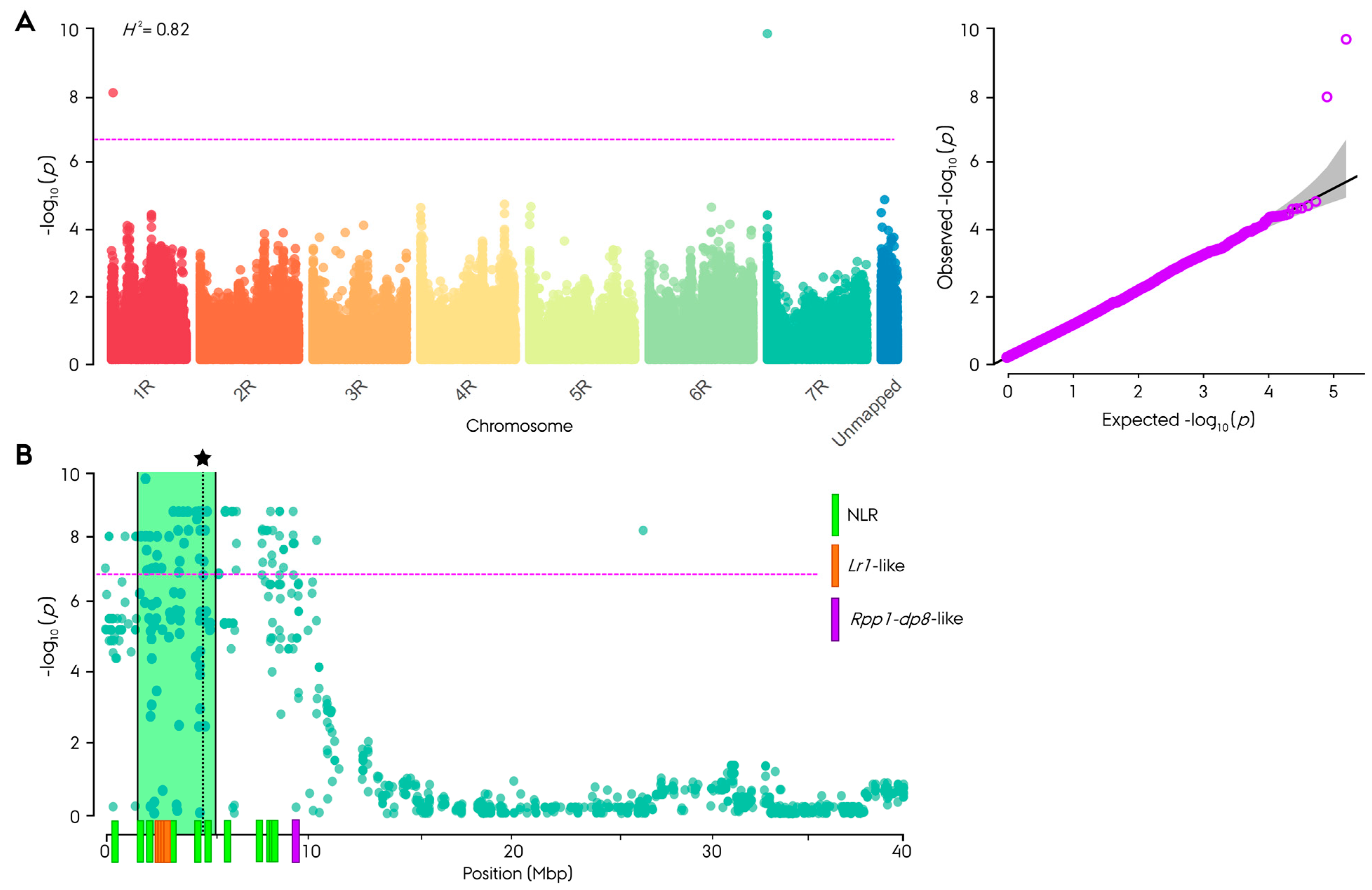
2.8. Genome-Wide Association Study
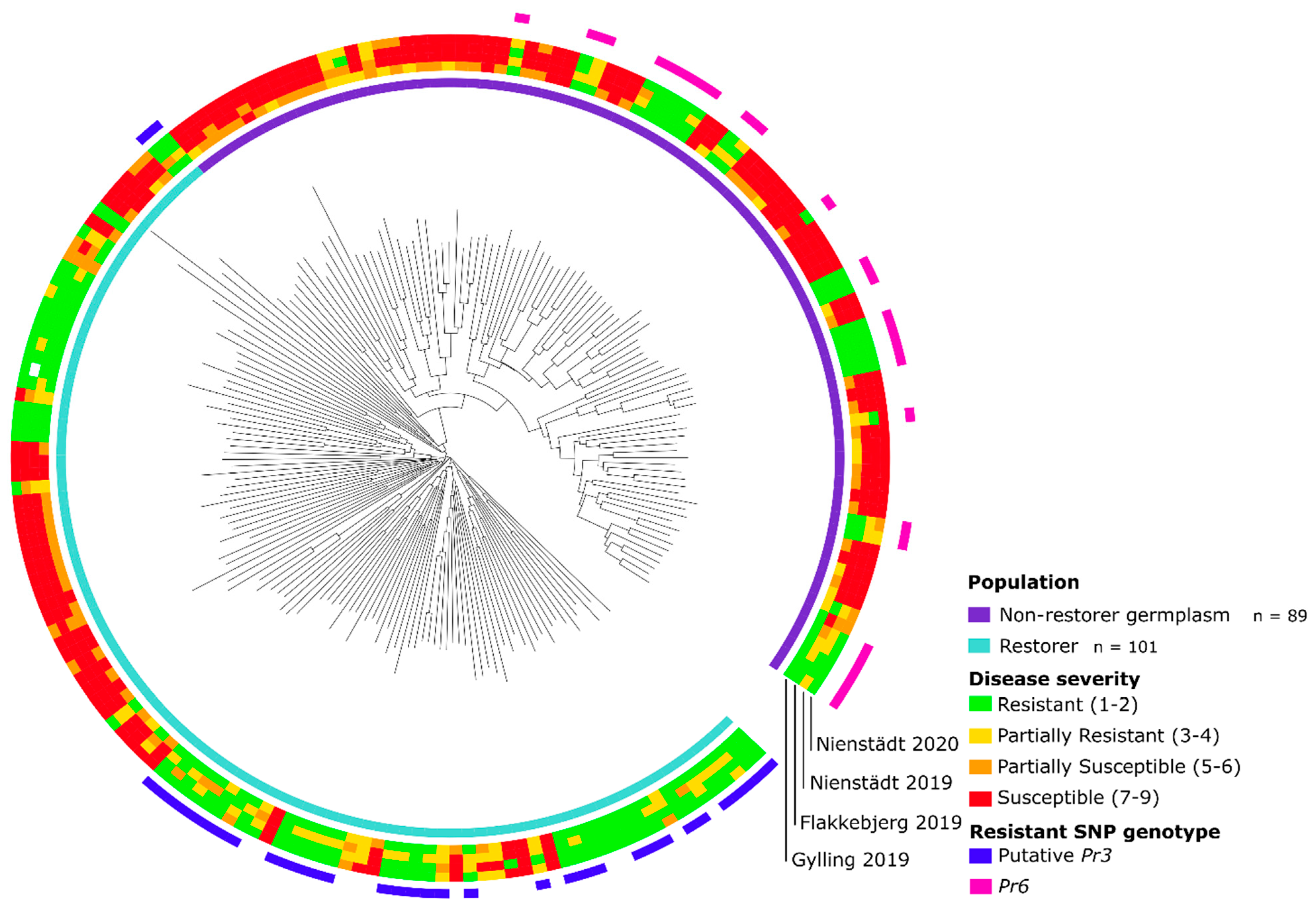
2.9. Phylogenetic Analysis of Lines
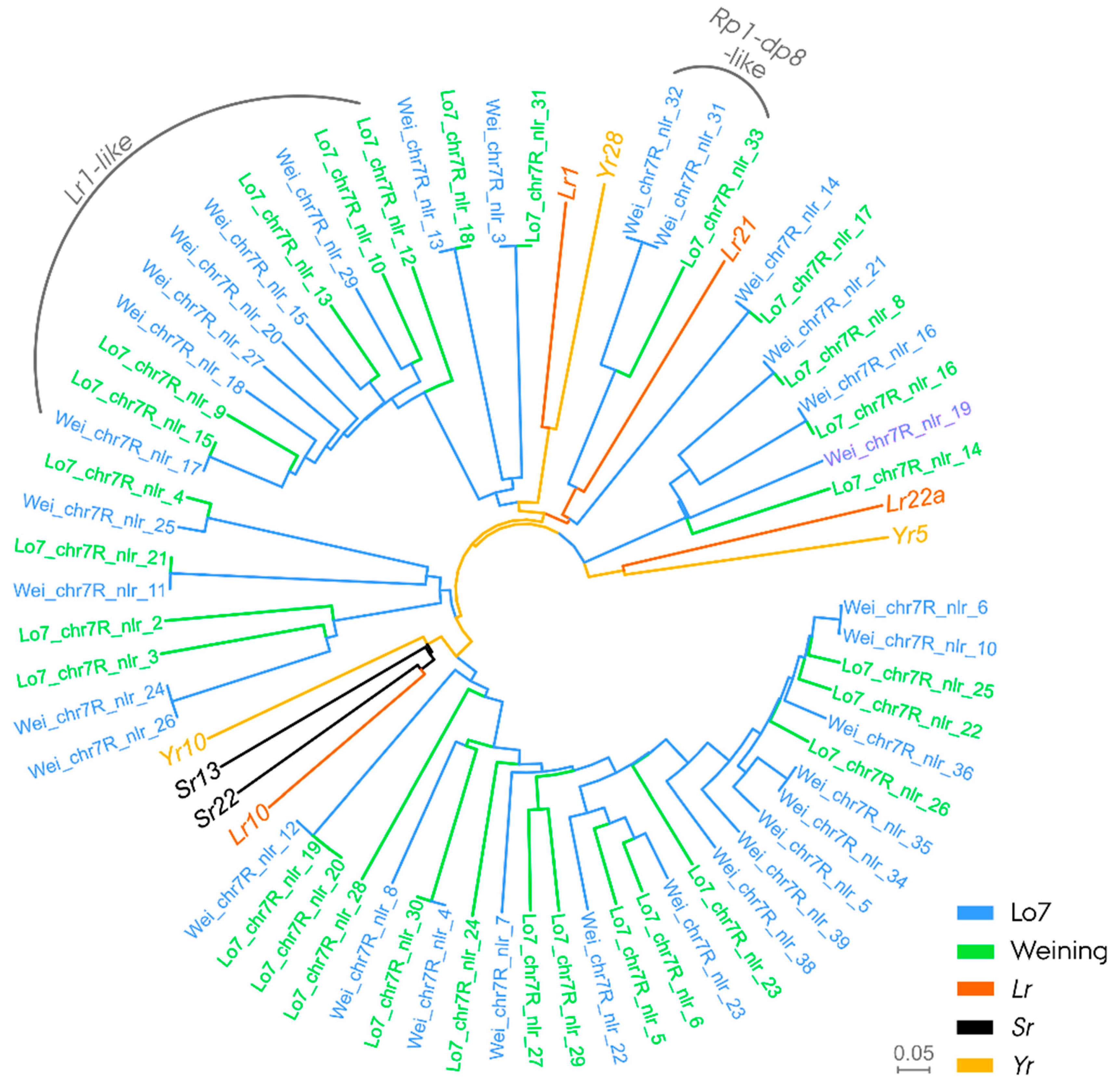
2.10. Phylogenetic Analysis and In Silico Characterization of Nucleotide-Binding Leucine-Rich Repeat Genes in Leaf Rust Resistance-Associated Regions
2.11. Graphical Editing
3. Results
3.1. 600 K SNP Genotyping of Panel
3.2. Phenotyping of Leaf Rust Resistance
3.3. Genome-Wide Association Study
3.4. Phylogenetic Analysis and In Silico Characterization of Nucleotide-Binding Leucine-Rich Repeat Genes in Leaf Rust Resistance-Associated Block on Chromosome Arm 7RS
4. Discussion
4.1. Evidence of Quantitative Resistance in the Germplasm
4.2. Discovery of a Novel Major Pr Gene on Rye Chromosome Arm 7RS in the Non-Restorer Germplasm Population
4.3. Discovery of a Putative Pr3 Gene in the Restorer Population
4.4. Pr6, a Potential Ortholog to Wheat Leaf Rust Resistance Gene Lr1 on Rye Chromosome Arm 7RS
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data availability statement
Acknowledgments
Conflicts of Interest
References
- FAOSTAT. Food and Agriculture Data. Available online: https://fao.org (accessed on 20 August 2021).
- Miedaner, T.; Mirdita, V.; Rodemann, B.; Drobeck, T.; Rentel, D. Genetic variation of winter rye cultivars for their ergot (Claviceps purpurea) reaction tested in a field design with minimized interplot interference. Plant Breed. 2010, 129, 58–62. [Google Scholar] [CrossRef]
- Mielke, H. Investigations on the control of ergot. Nachr. Dtsch. Pflanzenschutzd. 1993, 45, 97–102. [Google Scholar]
- Miedaner, T.; Kodisch, A.; Raditschnig, A.; Eifler, J. Ergot alkaloid contents in hybrid rye are reduced by breeding. Agriculture 2021, 11, 526. [Google Scholar] [CrossRef]
- Miedaner, T.; Geiger, H.H. Biology, genetics, and management of ergot (Claviceps spp.) in rye, sorghum, and pearl millet. Toxins 2015, 7, 659–678. [Google Scholar] [CrossRef] [PubMed]
- Miedaner, T.; Gey, A.-K.M.; Sperling, U.; Geiger, H.H. Quantitative-genetic analysis of leaf-rust resistance in seedling and adult-plant stages of inbred lines and their testcrosses in winter rye. Plant Breed. 2002, 121, 475–479. [Google Scholar] [CrossRef]
- Sortsinfo. The Danish Official Trial Records. Available online: https://sortinfo.dk/ (accessed on 10 February 2021).
- Bundessortenamt. Bescribende Sortenlisten—Getreide, Mais Öl- und Faserpflanzen, Leguminosen, Rüben, Zwischenfrüchte. Available online: https://www.bundessortenamt.de/bsa/sorten/beschreibende-sortenlisten/download-bsl-im-pdf-format (accessed on 1 October 2021).
- Miedaner, T.; Sperling, U. Effect of leaf rust and yield components of winter rye hybrids and assessment of quantitative resistance. J. Phytopathol. 1995, 143, 725–730. [Google Scholar] [CrossRef]
- Rodriguez-Algaba, J.; Walter, S.; Sørensen, C.K.; Hovmøller, M.S.; Justesen, A.F. Sexual structures and recombination of the wheat rust fungus Puccinia striiformis on Berberis vulgaris. Fungal Genet. Biol. 2014, 70, 77–85. [Google Scholar] [CrossRef]
- Jin, Y.; Szabo, L.J.; Carson, M. Century-old mystery of Puccinia striiformis life history solved with the identification of Berberis as an alternate host. Phytopathology 2010, 100, 432–435. [Google Scholar] [CrossRef] [PubMed]
- Anikster, Y.; Bushnell, W.; Roelfs, A.; Eilam, T.; Manisterski, J. Puccinia recondita causing leaf rust on cultivated wheats, wild wheats, and rye. Can. J. Bot. 1997, 75, 2082–2096. [Google Scholar] [CrossRef]
- Roelfs, A. Epidemiology of the cereal rusts in North America. Can. J. Plant Pathol. 1989, 11, 86–90. [Google Scholar] [CrossRef]
- Zadoks, J. Plant disease epidemiology in the twentieth century: A picture by means of selected controversies. Plant Dis. 2001, 85, 808–816. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhao, J.; Wang, M.; Chen, X.; Kang, Z. Role of alternate hosts in epidemiology and pathogen variation of cereal rusts. Annu. Rev. Phytopathol. 2016, 54, 207–228. [Google Scholar] [CrossRef] [PubMed]
- Roelf, A.P. Development and impact of regional cereal rust epidemics. Plant Dis. Epidemiol. 1986, 1, 129–150. [Google Scholar]
- Kolmer, J.A.; Ordonez, M.E.; German, S.; Morgounov, A.; Pretorius, Z.; Visser, B.; Goyeau, H.; Anikster, Y.; Acevedo, M. Multilocus genotypes of the wheat leaf rust fungus puccinia triticina in worldwide regions indicate past and current long-distance migration. Phytopathology 2019, 109, 1453–1463. [Google Scholar] [CrossRef]
- Hovmoller, M.S.; Yahyaoui, A.H.; Milus, E.A.; Justesen, A.F. Rapid global spread of two aggressive strains of a wheat rust fungus. Mol. Ecol. 2008, 17, 3818–3826. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.P.; Hodson, D.P.; Jin, Y.; Lagudah, E.S.; Ayliffe, M.A.; Bhavani, S.; Rouse, M.N.; Pretorius, Z.A.; Szabo, L.J.; Huerta-Espino, J.; et al. Emergence and spread of new races of wheat stem rust fungus: Continued threat to food security and prospects of genetic control. Phytopathology 2015, 105, 872–884. [Google Scholar] [CrossRef] [PubMed]
- Hermansen, J.E.; Torp, U.; Prahm, L.P. Studies of transport of live spores of cereal mildew and rust fungi across the North Sea. Grana 1978, 17, 41–46. [Google Scholar] [CrossRef]
- Hovmøller, M.; Justesen, A.; Brown, J. Clonality and long-distance migration of Puccinia striiformis f. sp. tritici in north-west Europe. Plant Pathol. 2002, 51, 24–32. [Google Scholar] [CrossRef]
- Hovmøller, M.S.; Walter, S.; Bayles, R.A.; Hubbard, A.; Flath, K.; Sommerfeldt, N.; Leconte, M.; Czembor, P.; Rodriguez-Algaba, J.; Thach, T.; et al. Replacement of the European wheat yellow rust population by new races from the centre of diversity in the near-Himalayan region. Plant Pathol. 2016, 65, 402–411. [Google Scholar] [CrossRef]
- Figueroa, M.; Dodds, P.N.; Henningsen, E.C. Evolution of virulence in rust fungi—Multiple solutions to one problem. Curr. Opin. Plant Biol. 2020, 56, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Goyeau, H.; Berder, J.; Czerepak, C.; Gautier, A.; Lanen, C.; Lannou, C. Low diversity and fast evolution in the population of Puccinia triticina causing durum wheat leaf rust in France from 1999 to 2009, as revealed by an adapted differential set. Plant Pathol. 2012, 61, 761–772. [Google Scholar] [CrossRef]
- Kolmer, J. Collections of Puccinia triticina in different provinces of China are highly related for virulence and molecular genotype. Phytopathology 2015, 105, 700–706. [Google Scholar] [CrossRef][Green Version]
- Ali, S.; Rodriguez-Algaba, J.; Thach, T.; Sorensen, C.K.; Hansen, J.G.; Lassen, P.; Nazari, K.; Hodson, D.P.; Justesen, A.F.; Hovmoller, M.S. Yellow rust epidemics worldwide were caused by pathogen races from divergent genetic lineages. Front. Plant Sci. 2017, 8, 1057. [Google Scholar] [CrossRef] [PubMed]
- Thach, T.; Ali, S.; Justesen, A.; Rodriguez-Algaba, J.; Hovmøller, M. Recovery and virulence phenotyping of the historic ‘Stubbs collection’of the yellow rust fungus Puccinia striiformis from wheat. Ann. Appl. Biol. 2015, 167, 314–326. [Google Scholar] [CrossRef]
- Li, F.; Upadhyaya, N.M.; Sperschneider, J.; Matny, O.; Nguyen-Phuc, H.; Mago, R.; Raley, C.; Miller, M.E.; Silverstein, K.A.T.; Henningsen, E.; et al. Emergence of the Ug99 lineage of the wheat stem rust pathogen through somatic hybridisation. Nat. Commun. 2019, 10, 5068. [Google Scholar] [CrossRef]
- Kim, W.; Shang, H.; Samborski, D. Electrophoretic analysis of detergent-soluble polypeptides of Puccinia recondita f. sp. tritici, P. recondita f. sp. secalis, P. hordei, and P. coronata. Can. J. Plant Pathol. 1985, 7, 287–293. [Google Scholar] [CrossRef]
- Hanzlik, K.; Gerowitt, B. Occurrence and distribution of important weed species in German winter oilseed rape fields. J. Plant Dis. Prot. 2012, 119, 107–120. [Google Scholar] [CrossRef]
- De Mol, F.; Von Redwitz, C.; Gerowitt, B. Weed species composition of maize fields in Germany is influenced by site and crop sequence. Weed Res. 2015, 55, 574–585. [Google Scholar] [CrossRef]
- Andreasen, C.; Stryhn, H. Increasing weed flora in Danish beet, pea and winter barley fields. Crop. Prot. 2012, 36, 11–17. [Google Scholar] [CrossRef]
- Miedaner, T.; Klocke, B.; Flath, K.; Geiger, H.H.; Weber, W.E. Diversity, spatial variation, and temporal dynamics of virulences in the German leaf rust (Puccinia recondita f. sp. secalis) population in winter rye. Eur. J. Plant Pathol. 2011, 132, 23–35. [Google Scholar] [CrossRef]
- Wehling, P.; Linz, A.; Hackauf, B.; Roux, S.R.; Ruge, B.; Klocke, B. Leaf-rust resistance in rye (Secale cereale L.). 1. Genetic analysis and mapping of resistance genes Pr1 and Pr2. Theor. Appl. Genet. 2003, 107, 432–438. [Google Scholar] [CrossRef]
- Roux, S.R.; Hackauf, B.; Linz, A.; Ruge, B.; Klocke, B.; Wehling, P. Leaf-rust resistance in rye (Secale cereale L.). 2. Genetic analysis and mapping of resistance genes Pr3, Pr4, and Pr5. Theor. Appl. Genet. 2004, 110, 192–201. [Google Scholar] [CrossRef] [PubMed]
- McIntosh, R.A.; Frieb, B.; Jiang, J.; Gill, B.S. Cytogenetical studies in wheat XVI. Chromosomal location of a new gene for resistance to leaf rust in a Japanese wheat-rye translocation line. Euphytica 1995, 82, 141–147. [Google Scholar] [CrossRef]
- Friebe, B.; Jiang, J.; Raupp, W.J.; McIntosh, R.A.; Gill, B.S. Characterization of wheat-alien translocations conferring resistance to diseases and pests: Current status. Euphytica 1996, 91, 59–87. [Google Scholar] [CrossRef]
- Kourelis, J.; Van Der Hoorn, R.A. Defended to the nines: 25 years of resistance gene cloning identifies nine mechanisms for R protein function. Plant Cell 2018, 30, 285–299. [Google Scholar] [CrossRef] [PubMed]
- Rabanus-Wallace, M.T.; Hackauf, B.; Mascher, M.; Lux, T.; Wicker, T.; Gundlach, H.; Báez, M.; Houben, A.; Mayer, K.F.X.; Guo, L.; et al. Chromosome-scale genome assembly provides insights into rye biology, evolution and agronomic potential. Nat. Genet. 2021, 53, 564–573. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Wang, L.; Yang, J.; He, H.; Jin, H.; Li, X.; Ren, T.; Ren, Z.; Li, F.; Han, X.; et al. A high-quality genome assembly highlights rye genomic characteristics and agronomically important genes. Nat. Genet. 2021, 53, 574–584. [Google Scholar] [CrossRef]
- Krasileva, K.V.; Dahlbeck, D.; Staskawicz, B.J. Activation of an Arabidopsis resistance protein is specified by the in planta association of its leucine-rich repeat domain with the cognate oomycete effector. Plant Cell 2010, 22, 2444–2458. [Google Scholar] [CrossRef]
- Takken, F.L.; Goverse, A. How to build a pathogen detector: Structural basis of NB-LRR function. Curr. Opin. Plant Biol. 2012, 15, 375–384. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Han, M.; Liu, Y. Diversity, structure and function of the coiled-coil domains of plant NLR immune receptors. J. Integr. Plant Biol. 2021, 63, 283–296. [Google Scholar] [CrossRef]
- Miedaner, T.; Korzun, V. Marker-assisted selection for disease resistance in wheat and barley breeding. Phytopathology 2012, 102, 560–566. [Google Scholar] [CrossRef] [PubMed]
- Mundt, C.C. Pyramiding for resistance durability: Theory and practice. Phytopathology 2018, 108, 792–802. [Google Scholar] [CrossRef]
- Koller, T.; Brunner, S.; Herren, G.; Hurni, S.; Keller, B. Pyramiding of transgenic Pm3 alleles in wheat results in improved powdery mildew resistance in the field. Theor. Appl. Genet. 2018, 131, 861–871. [Google Scholar] [CrossRef]
- Bauer, E.; Schmutzer, T.; Barilar, I.; Mascher, M.; Gundlach, H.; Martis, M.M.; Twardziok, S.O.; Hackauf, B.; Gordillo, A.; Wilde, P.; et al. Towards a whole-genome sequence for rye (Secale cereale L.). Plant J. 2017, 89, 853–869. [Google Scholar] [CrossRef] [PubMed]
- Vendelbo, N.M.; Mahmood, K.; Sarup, P.; Kristensen, P.S.; Orabi, J.; Jahoor, A. Genomic Scan of Male Fertility Restoration Genes in a ‘Gülzow’Type Hybrid Breeding System of Rye (Secale cereale L.). Int. J. Mol. Sci. 2021, 22, 9277. [Google Scholar] [CrossRef] [PubMed]
- Miedaner, T.; Glass, C.; Dreyer, F.; Wilde, P.; Wortmann, H.; Geiger, H.H. Mapping of genes for male-fertility restoration in ‘Pampa’ CMS winter rye (Secale cereale L.). Theor. Appl. Genet. 2000, 101, 1226–1233. [Google Scholar] [CrossRef]
- Vendelbo, N.M.; Sarup, P.; Orabi, J.; Kristensen, P.S.; Jahoor, A. Genetic structure of a germplasm for hybrid breeding in rye (Secale cereale L.). PLoS ONE 2020, 15, e0239541. [Google Scholar] [CrossRef]
- USDA. United States Department of Agriculture: Wheat and Barley DNA Extraction Protocol (96-Well Plate Format). Available online: https://www.ars.usda.gov/ARSUserFiles/60701500/SmallGrainsGenotypingLaboratory/Protocols/wheat%20and%20barleyDNA%20extraction_original.pdf (accessed on 7 July 2021).
- Pallotta, M.A.; Warner, P.; Fox, R.L.; Kuchel, H.; Jefferies, S.J.; Langridge, P. Marker assisted wheat breeding in the southern region of Australia. Proc. Tenth Int. Wheat Genet. Symp. 2003, 789–791. [Google Scholar]
- Vendelbo, N.; Mahmood, K.; Sarup, P.; Kristensen, P.; Orabi, J.; Jahoor, A. Discovery of a novel powdery mildew (Blumeria graminis) resistance locus in rye (Secale cereale L.). Sci. Rep. 2021, 11, 23057. [Google Scholar] [CrossRef] [PubMed]
- Patterson, H.D.; Hunter, E.A. The efficiency of incomplete block designs in national list and recommend list cereal variety trials. J. Agric. Sci. 1983, 101, 427–433. [Google Scholar] [CrossRef]
- Arias, N.; Lafarga, A.; Virto, C. RustWatch: Recommendations for Coordinated and Harmonized Wheat Rust Surveillance in Europe Based on Input from Stakeholders. Available online: https://ec.europa.eu/research/participants/documents/downloadPublic?documentIds=080166e5c8e0c491&appId=PPGMS (accessed on 6 June 2021).
- De Mendiburu, F.; Reinhard, S. Agrocolae-Ten years of open source statistical tool for experiments in breeding, agriculture and biology. PeerJ PrePrints 2015, 3, e1748. [Google Scholar]
- Lipka, A.E.; Tian, F.; Wang, Q.; Peiffer, J.; Li, M.; Bradbury, P.J.; Gore, M.A.; Buckler, E.S.; Zhang, Z. GAPIT: Genome association and prediction integrated tool. Bioinformatics 2012, 28, 2397–2399. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. ggplot2. Wiley Interdiscip. Rev. Comput. Stat. 2011, 3, 180–185. [Google Scholar] [CrossRef]
- Huang, M.; Liu, X.; Zhou, Y.; Summers, R.M.; Zhang, Z. BLINK: A package for the next level of genome-wide association studies with both individuals and markers in the millions. Gigascience 2019, 8, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Mago, R.; Miah, H.; Lawrence, G.J.; Wellings, C.R.; Spielmeyer, W.; Bariana, H.S.; McIntosh, R.A.; Pryor, A.J.; Ellis, J.G. High-resolution mapping and mutation analysis separate the rust resistance genes Sr31, Lr26 and Yr9 on the short arm of rye chromosome 1. Theor. Appl. Genet. 2005, 112, 41–50. [Google Scholar] [CrossRef]
- NCBI. National Center for Biotechnology Information. Available online: https://www.ncbi.nlm.nih.gov (accessed on 13 June 2021).
- Paradis, E.; Schliep, K. ape 5.0: An environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 2019, 35, 526–528. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, 256–259. [Google Scholar] [CrossRef] [PubMed]
- Stanke, M.; Steinkamp, R.; Waack, S.; Morgenstern, B. AUGUSTUS: A web server for gene finding in eukaryotes. Nucleic Acids Res. 2004, 32, W309–W312. [Google Scholar] [CrossRef]
- The UniProt Consortium. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- Toparslan, E.; Karabag, K.; Bilge, U. A workflow with R: Phylogenetic analyses and visualizations using mitochondrial cytochrome b gene sequences. PLoS ONE 2020, 15, e0243927. [Google Scholar] [CrossRef] [PubMed]
- Bodenhofer, U.; Bonatesta, E.; Horejs-Kainrath, C.; Hochreiter, S. msa: An R package for multiple sequence alignment. Bioinformatics 2015, 31, 3997–3999. [Google Scholar] [CrossRef]
- Charif, D.; Lobry, J.R. SeqinR 1.0-2: A contributed package to the R project for statistical computing devoted to biological sequences retrieval and analysis. In Structural Approaches to Sequence Evolution; Springer: Berlin/Heidelberg, Germany, 2007; pp. 207–232. [Google Scholar]
- Yu, G. Using ggtree to visualize data on tree-like structures. Curr. Protoc. Bioinform. 2020, 69, e96. [Google Scholar] [CrossRef]
- Lolle, S.; Stevens, D.; Coaker, G. Plant NLR-triggered immunity: From receptor activation to downstream signaling. Curr. Opin. Immunol. 2020, 62, 99–105. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, L.; Wu, W.; He, L.; Yang, X.; Pan, Q. Function and evolution of Magnaporthe oryzae avirulence gene AvrPib responding to the rice blast resistance gene Pib. Sci. Rep. 2015, 5, 11642. [Google Scholar] [CrossRef]
- Chen, J.; Upadhyaya, N.M.; Ortiz, D.; Sperschneider, J.; Li, F.; Bouton, C.; Breen, S.; Dong, C.; Xu, B.; Zhang, X. Loss of AvrSr50 by somatic exchange in stem rust leads to virulence for Sr50 resistance in wheat. Science 2017, 358, 1607–1610. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Bakkeren, G.; McCallum, B. Virulence and molecular polymorphisms of the wheat leaf rust fungus Puccinia triticina in Canada from 1997 to 2007. Botany 2010, 88, 575–589. [Google Scholar] [CrossRef]
- McCallum, B.D.; Hiebert, C.W.; Cloutier, S.; Bakkeren, G.; Rosa, S.B.; Humphreys, D.G.; Marais, G.F.; McCartney, C.A.; Panwar, V.; Rampitsch, C.; et al. A review of wheat leaf rust research and the development of resistant cultivars in Canada. Can. J. Plant Pathol. 2016, 38, 1–18. [Google Scholar] [CrossRef]
- Cowger, C.; Mehra, L.; Arellano, C.; Meyers, E.; Murphy, J.P. Virulence differences in Blumeria graminis f. sp. tritici from the Central and Eastern United States. Phytopathology 2018, 108, 402–411. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.P.; Huerta-Espino, J.; William, H.M. Genetics and breeding for durable resistance to leaf and stripe rusts in wheat. Turk. J. Agric. For. 2005, 29, 121–127. [Google Scholar]
- Fukuoka, S.; Saka, N.; Mizukami, Y.; Koga, H.; Yamanouchi, U.; Yoshioka, Y.; Hayashi, N.; Ebana, K.; Mizobuchi, R.; Yano, M. Gene pyramiding enhances durable blast disease resistance in rice. Sci. Rep. 2015, 5, 7773. [Google Scholar] [CrossRef] [PubMed]
- Sucher, J.; Boni, R.; Yang, P.; Rogowsky, P.; Buchner, H.; Kastner, C.; Kumlehn, J.; Krattinger, S.G.; Keller, B. The durable wheat disease resistance gene Lr34 confers common rust and northern corn leaf blight resistance in maize. Plant Biotechnol. J. 2017, 15, 489–496. [Google Scholar] [CrossRef] [PubMed]
- Rinaldo, A.; Gilbert, B.; Boni, R.; Krattinger, S.G.; Singh, D.; Park, R.F.; Lagudah, E.; Ayliffe, M. The Lr34 adult plant rust resistance gene provides seedling resistance in durum wheat without senescence. Plant Biotechnol. J. 2017, 15, 894–905. [Google Scholar] [CrossRef]
- Pinto da Silva, G.B.; Zanella, C.M.; Martinelli, J.A.; Chaves, M.S.; Hiebert, C.W.; McCallum, B.D.; Boyd, L.A. Quantitative trait loci conferring leaf rust resistance in hexaploid wheat. Phytopathology 2018, 108, 1344–1354. [Google Scholar] [CrossRef]
- Das, M.K.; Raaram, S.; Mundt, C.C.; Kronstad, W.E. Inheritance of slow-rusting resistance to leaf rust in wheat. Crop. Sci. 1992, 32, 1452–1456. [Google Scholar] [CrossRef]
- Pilet-Nayel, M.L.; Moury, B.; Caffier, V.; Montarry, J.; Kerlan, M.C.; Fournet, S.; Durel, C.E.; Delourme, R. Quantitative resistance to plant pathogens in pyramiding strategies for durable crop protection. Front. Plant Sci. 2017, 8, 1838. [Google Scholar] [CrossRef]
- Huerta-Espino, J.; Singh, R.; Crespo-Herrera, L.A.; Villasenor-Mir, H.E.; Rodriguez-Garcia, M.F.; Dreisigacker, S.; Barcenas-Santana, D.; Lagudah, E. Adult plant slow rusting genes confer high levels of resistance to rusts in bread wheat cultivars from Mexico. Front. Plant Sci. 2020, 11, 1–15. [Google Scholar] [CrossRef]
- Cobb, J.N.; Biswas, P.S.; Platten, J.D. Back to the future: Revisiting MAS as a tool for modern plant breeding. Theor. Appl. Genet. 2019, 132, 647–667. [Google Scholar] [CrossRef] [PubMed]
- Beukert, U.; Thorwarth, P.; Zhao, Y.; Longin, C.F.H.; Serfling, A.; Ordon, F.; Reif, J.C. Comparing the potential of marker-assisted selection and genomic prediction for improving rust resistance in hybrid wheat. Front. Plant Sci. 2020, 11, 594113. [Google Scholar] [CrossRef] [PubMed]
- Reichel, A. Untersuchungen zur Horizontalen Resistenz von Roggen und Weizen Gegenüber Braunrost. Ph.D. Thesis, University of Halle, Halle, Germany, 1981. [Google Scholar]
- Solodukhina, O.V. Genetic characterization of rye accessions with regard to leaf rust resistance. Russ. J. Genet. 2002, 38, 497–506. [Google Scholar] [CrossRef]
- Korte, A.; Farlow, A. The advantages and limitations of trait analysis with GWAS: A review. Plant Methods 2013, 9, 29. [Google Scholar] [CrossRef]
- Coriton, O.; Jahier, J.; Leconte, M.; Huteau, V.; Trotoux, G.; Dedryver, F.; de-Vallavieille-Pope, C. Double dose efficiency of the yellow rust resistance gene Yr17 in bread wheat lines. Plant Breed. 2020, 139, 263–271. [Google Scholar] [CrossRef]
- Ellis, J.G.; Lawrence, G.J.; Luck, J.E.; Dodds, P.N. Identification of regions in alleles of the flax rust resistance gene L that determine differences in gene-for-gene specificity. Plant Cell 1999, 11, 495–506. [Google Scholar] [CrossRef]
- Liu, H.J.; Yan, J. Crop genome-wide association study: A harvest of biological relevance. Plant J. 2019, 97, 8–18. [Google Scholar] [CrossRef] [PubMed]
- Alqudah, A.M.; Sallam, A.; Stephen Baenziger, P.; Borner, A. GWAS: Fast-forwarding gene identification and characterization in temperate cereals: Lessons from barley—A review. J. Adv. Res. 2020, 22, 119–135. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Challa, G.S.; Zhu, H.; Wei, W. Recurrence of chromosome rearrangements and reuse of dna breakpoints in the evolution of the triticeae genomes. G3 Genes Genomes Genet. 2016, 6, 3837–3847. [Google Scholar] [CrossRef]
- Devos, K.M. Updating the ‘crop circle’. Curr. Opin. Plant Biol. 2005, 8, 155–162. [Google Scholar] [CrossRef] [PubMed]
- Dracatos, P.M.; Khatkar, M.S.; Singh, D.; Park, R.F. Genetic mapping of a new race specific resistance allele effective to Puccinia hordei at the Rph9/Rph12 locus on chromosome 5HL in barley. BMC Plant Biol. 2014, 14, 1598. [Google Scholar] [CrossRef] [PubMed]
- Borovkova, I.; Jin, Y.; Steffenson, B. Chromosomal location and genetic relationship of leaf rust resistance genes Rph9 and Rph12 in barley. Phytopathology 1998, 88, 76–80. [Google Scholar] [CrossRef][Green Version]
- Martis, M.M.; Zhou, R.; Haseneyer, G.; Schmutzer, T.; Vrana, J.; Kubalakova, M.; Konig, S.; Kugler, K.G.; Scholz, U.; Hackauf, B.; et al. Reticulate evolution of the rye genome. Plant Cell 2013, 25, 3685–3698. [Google Scholar] [CrossRef]
- McIntosh, R.A.; Ducovsky, W.J.; Appels, R.; Xia, X.C. Catalogue of Gene Symbols for Wheat: 2017 Supplement. KOMUGI Integrated Wheat Science Database. Available online: https://shigen.nig.ac.jp/wheat/komugi/about/about.jsp (accessed on 6 July 2021).
- Chen, N.W.G.; Thareau, V.; Ribeiro, T.; Magdelenat, G.; Ashfield, T.; Innes, R.W.; Pedrosa-Harand, A.; Geffroy, V. Common bean subtelomeres are hot spots of recombination and favor resistance gene evolution. Front. Plant Sci. 2018, 9, 1185. [Google Scholar] [CrossRef] [PubMed]
- Flint-Garcia, S.A.; Thornsberry, J.M.; Buckler, E.S., IV. Structure of linkage disequilibrium in plants. Annu. Rev. Plant Biol. 2003, 54, 357–374. [Google Scholar] [CrossRef] [PubMed]
- Nieri, D.; Di Donato, A.; Ercolano, M.R. Analysis of tomato meiotic recombination profile reveals preferential chromosome positions for NB-LRR genes. Euphytica 2017, 213, 206. [Google Scholar] [CrossRef]
- Mondragon-Palomino, M.; Meyers, B.C.; Michelmore, R.W.; Gaut, B.S. Patterns of positive selection in the complete NBS-LRR gene family of Arabidopsis thaliana. Genome Res. 2002, 12, 1305–1315. [Google Scholar] [CrossRef] [PubMed]
- Florence, J.; Vernaldi, S.; Maekawa, T. Evolution and conservation of plant NLR functions. Front. Immunol. 2013, 4, 297. [Google Scholar] [CrossRef]
- Feuillet, C.; Messmer, M.; Schachermayr, G.; Keller, B. Genetic and physical characterization of the Lr1 leaf rust resistance locus in wheat (Triticum aestivum L.). Mol. Gen. Genet. MGG 1995, 248, 553–562. [Google Scholar] [CrossRef] [PubMed]
- Ling, H.Q.; Zhu, Y.; Keller, B. High-resolution mapping of the leaf rust disease resistance gene Lr1 in wheat and characterization of BAC clones from the Lr1 locus. Theor. Appl. Genet. 2003, 106, 875–882. [Google Scholar] [CrossRef] [PubMed]
- Cloutier, S.; McCallum, B.D.; Loutre, C.; Banks, T.W.; Wicker, T.; Feuillet, C.; Keller, B.; Jordan, M.C. Leaf rust resistance gene Lr1, isolated from bread wheat (Triticum aestivum L.) is a member of the large psr567 gene family. Plant Mol. Biol. 2007, 65, 93–106. [Google Scholar] [CrossRef]
- Khlestkina, E.K.; Than, M.H.; Pestsova, E.G.; Roder, M.S.; Malyshev, S.V.; Korzun, V.; Borner, A. Mapping of 99 new microsatellite-derived loci in rye (Secale cereale L.) including 39 expressed sequence tags. Theor. Appl. Genet. 2004, 109, 725–732. [Google Scholar] [CrossRef]
- Hurni, S.; Brunner, S.; Buchmann, G.; Herren, G.; Jordan, T.; Krukowski, P.; Wicker, T.; Yahiaoui, N.; Mago, R.; Keller, B. Rye Pm8 and wheat Pm3 are orthologous genes and show evolutionary conservation of resistance function against powdery mildew. Plant J. 2013, 76, 957–969. [Google Scholar] [CrossRef]
- Arora, S.; Steuernagel, B.; Gaurav, K.; Chandramohan, S.; Long, Y.; Matny, O.; Johnson, R.; Enk, J.; Periyannan, S.; Singh, N.; et al. Resistance gene cloning from a wild crop relative by sequence capture and association genetics. Nat. Biotechnol. 2019, 37, 139–143. [Google Scholar] [CrossRef]
- Rollar, S.; Serfling, A.; Geyer, M.; Hartl, L.; Mohler, V.; Ordon, F. QTL mapping of adult plant and seedling resistance to leaf rust (Puccinia triticina Eriks.) in a multiparent advanced generation intercross (MAGIC) wheat population. Theor. Appl. Genet. 2021, 134, 37–51. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Hou, W.; Lan, C.; Basnet, B.R.; Singh, R.P.; Zhu, W.; Cheng, X.; Cui, D.; Chen, F. QTL analysis and nested association mapping for adult plant resistance to powdery mildew in two bread wheat populations. Front. Plant Sci. 2017, 8, 1212. [Google Scholar] [CrossRef] [PubMed]
- Crespo-Herrera, L.A.; Garkava-Gustavsson, L.; Åhman, I. A systematic review of rye (Secale cereale L.) as a source of resistance to pathogens and pests in wheat (Triticum aestivum L.). Hereditas 2017, 154, 14. [Google Scholar] [CrossRef] [PubMed]
| Reference Genome | Coding Sequence (aa) | BlastP | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| NLR ID | Position (Mbp) | Gene Length (bp) | Predicted Protein Sequence Length (aa) | Hit | Species | Alignment Length (aa) | Identity (%) | Gaps (%) | ||
| Lo7 | Lo7_chr7R_nlr_9 | 2.37 | 3294 | 1098 | 1408 | Lr1 disease protein | Triticum aestivum | 1183 | 84.60 | 4.18 |
| Lo7_chr7R_nlr_10 | 2.41 | 3141 | 1047 | 1326 | Lr1 disease protein | Triticum aestivum | 1270 | 80.48 | 6.31 | |
| Lo7_chr7R_nlr_12 | 2.80 | 3258 | 1086 | 1387 | Lr1 disease protein | Triticum aestivum | 1341 | 80.67 | 5.44 | |
| Lo7_chr7R_nlr_13 | 2.81 | 3246 | 1082 | 1429 | Lr1 disease protein | Triticum aestivum | 1195 | 81.99 | 4.00 | |
| Lo7_chr7R_nlr_15 | 2.87 | 3207 | 1068 | 1438 | Lr1 disease protein | Triticum aestivum | 1195 | 84.69 | 3.75 | |
| Lo7_chr7R_nlr_33 | 9.57 | 2294 | 526 | - | Putative rust resistance protein Rp1-dp8 | Brachypodium distachyon | 350 | 62.90 | 17.74 | |
| Weining | Wei_chr7R_nlr_15 | 12.21 | 3687 | 1075 | - | Lr1 disease protein | Triticum aestivum | 1064 | 83.72 | 4.29 |
| Wei_chr7R_nlr_17 | 12.35 | 3207 | 1069 | - | Lr1 disease protein | Triticum aestivum | 1054 | 85.41 | 3.88 | |
| Wei_chr7R_nlr_18 | 12.46 | 6438 | 1730 | - | Lr1 disease protein | Triticum aestivum | 1113 | 86.03 | 3.87 | |
| Wei_chr7R_nlr_20 | 12.53 | 3294 | 1098 | - | Lr1 disease protein | Triticum aestivum | 1078 | 84.32 | 3.87 | |
| Wei_chr7R_nlr_27 | 14.41 | 1392 | 464 | - | Lr1 disease protein | Triticum aestivum | 464 | 81.66 | 3.41 | |
| Wei_chr7R_nlr_29 | 14.52 | 11,906 | 1044 | - | Lr1 disease protein | Triticum aestivum | 915 | 82.43 | 5.75 | |
| Wei_chr7R_nlr_31 | 18.91 | 2054 | 500 | - | Putative rust resistance protein Rp1-dp8 | Brachypodium distachyon | 534 | 54.68 | 25.72 | |
| Wei_chr7R_nlr_32 | 19.09 | 2053 | 440 | - | Putative rust resistance protein Rp1-dp8 | Brachypodium distachyon | 366 | 59.95 | 18.11 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vendelbo, N.M.; Mahmood, K.; Sarup, P.; Hovmøller, M.S.; Justesen, A.F.; Kristensen, P.S.; Orabi, J.; Jahoor, A. Discovery of a Novel Leaf Rust (Puccinia recondita) Resistance Gene in Rye (Secale cereale L.) Using Association Genomics. Cells 2022, 11, 64. https://doi.org/10.3390/cells11010064
Vendelbo NM, Mahmood K, Sarup P, Hovmøller MS, Justesen AF, Kristensen PS, Orabi J, Jahoor A. Discovery of a Novel Leaf Rust (Puccinia recondita) Resistance Gene in Rye (Secale cereale L.) Using Association Genomics. Cells. 2022; 11(1):64. https://doi.org/10.3390/cells11010064
Chicago/Turabian StyleVendelbo, Nikolaj Meisner, Khalid Mahmood, Pernille Sarup, Mogens S. Hovmøller, Annemarie Fejer Justesen, Peter Skov Kristensen, Jihad Orabi, and Ahmed Jahoor. 2022. "Discovery of a Novel Leaf Rust (Puccinia recondita) Resistance Gene in Rye (Secale cereale L.) Using Association Genomics" Cells 11, no. 1: 64. https://doi.org/10.3390/cells11010064
APA StyleVendelbo, N. M., Mahmood, K., Sarup, P., Hovmøller, M. S., Justesen, A. F., Kristensen, P. S., Orabi, J., & Jahoor, A. (2022). Discovery of a Novel Leaf Rust (Puccinia recondita) Resistance Gene in Rye (Secale cereale L.) Using Association Genomics. Cells, 11(1), 64. https://doi.org/10.3390/cells11010064






