A Novel Hyperactive Nud1 Mitotic Exit Network Scaffold Causes Spindle Position Checkpoint Bypass in Budding Yeast
Abstract
1. Introduction
2. Materials and Methods
2.1. Yeast Strains and Growth Conditions
2.2. Fixed-Cell Imaging and Analysis
2.3. Live-Cell Imaging and Analysis
2.4. Protein Extraction, Immunoprecipitation, and Western Blot Analysis
3. Results
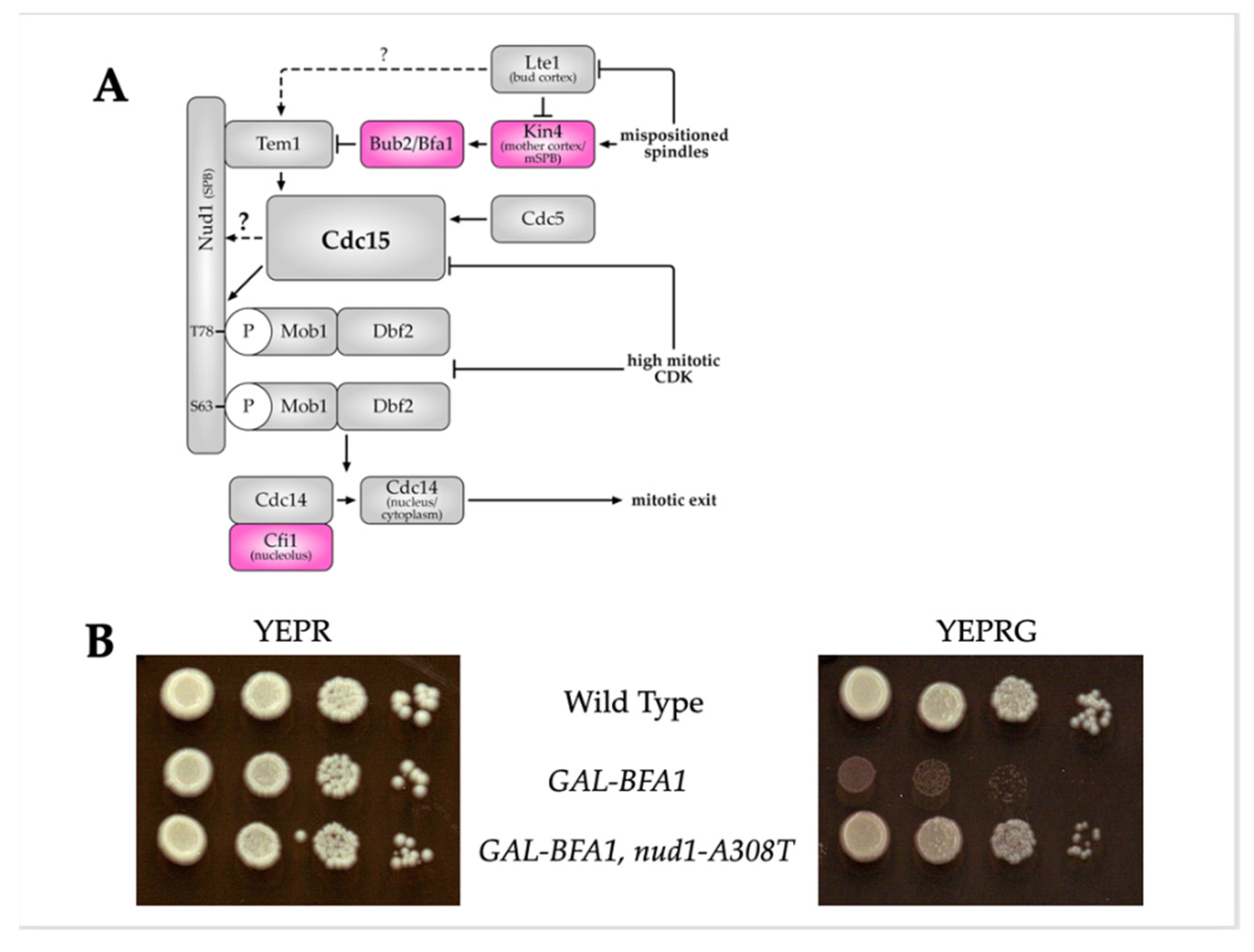
3.1. Nud1-A308T Partially Suppresses the Cell Cycle Defects of GAL-BFA1
3.2. Nud1-A308T Exhibits Weak Bypass of the Spindle Assembly Checkpoint
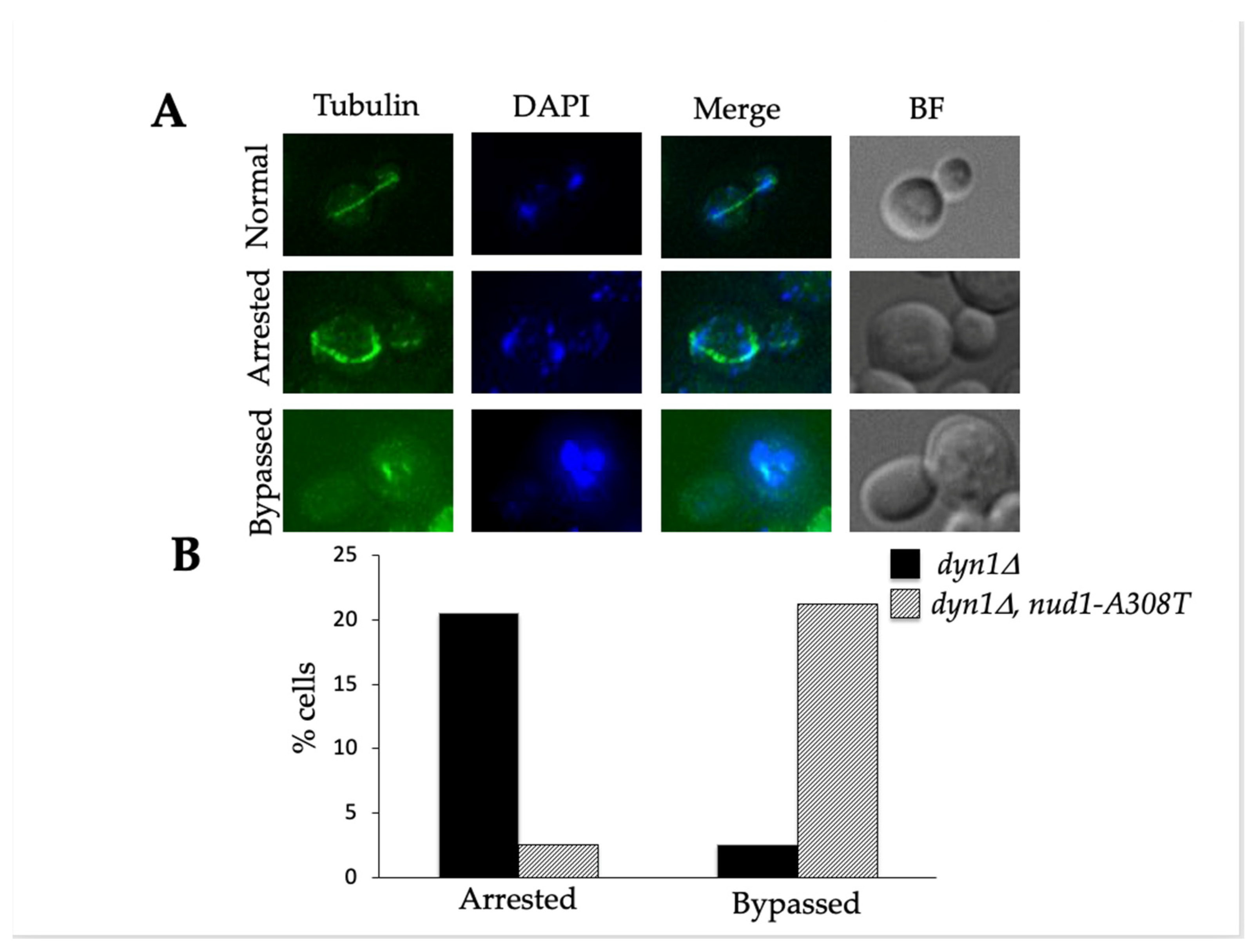
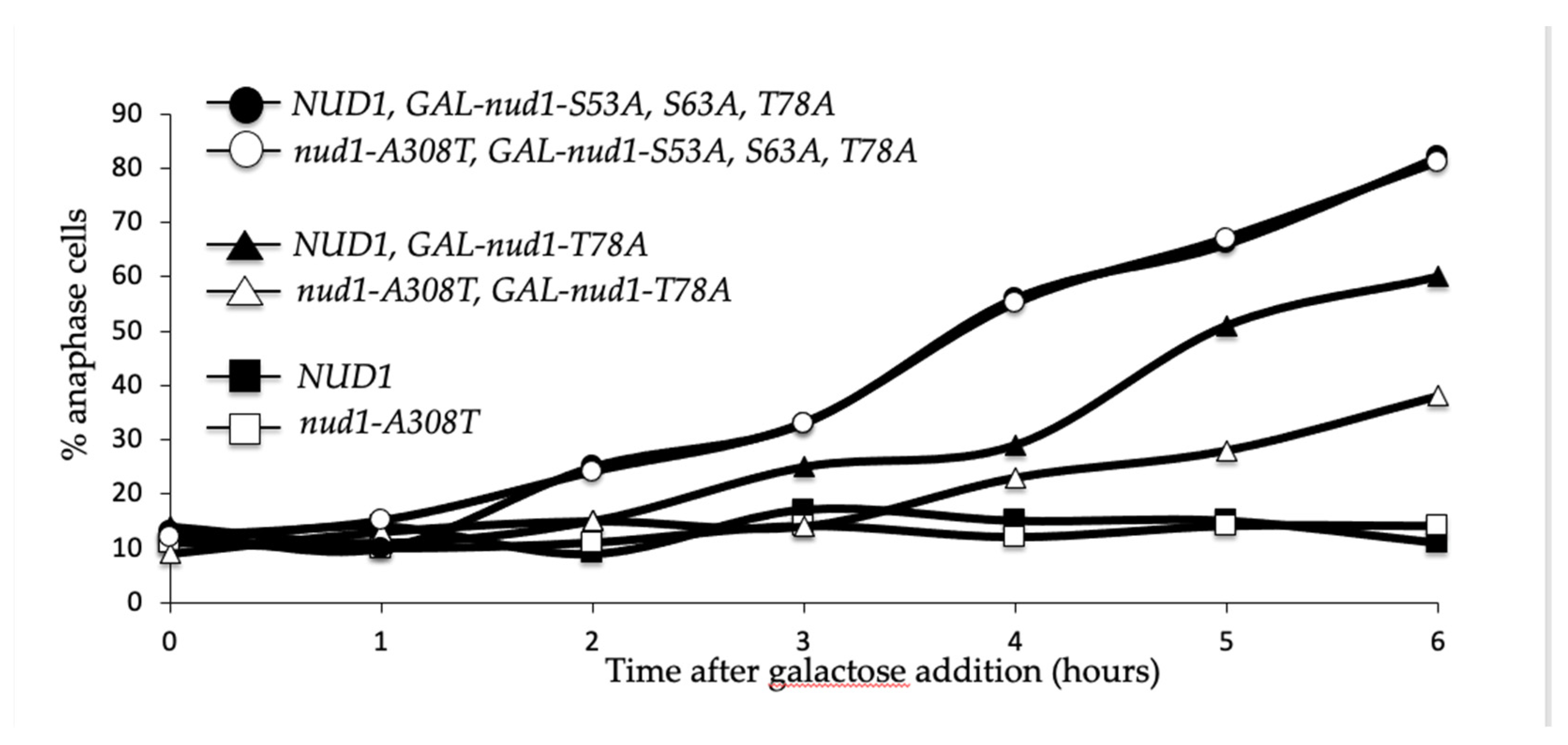
3.3. Nud1-A308T Suppresses the Spindle Position Checkpoint
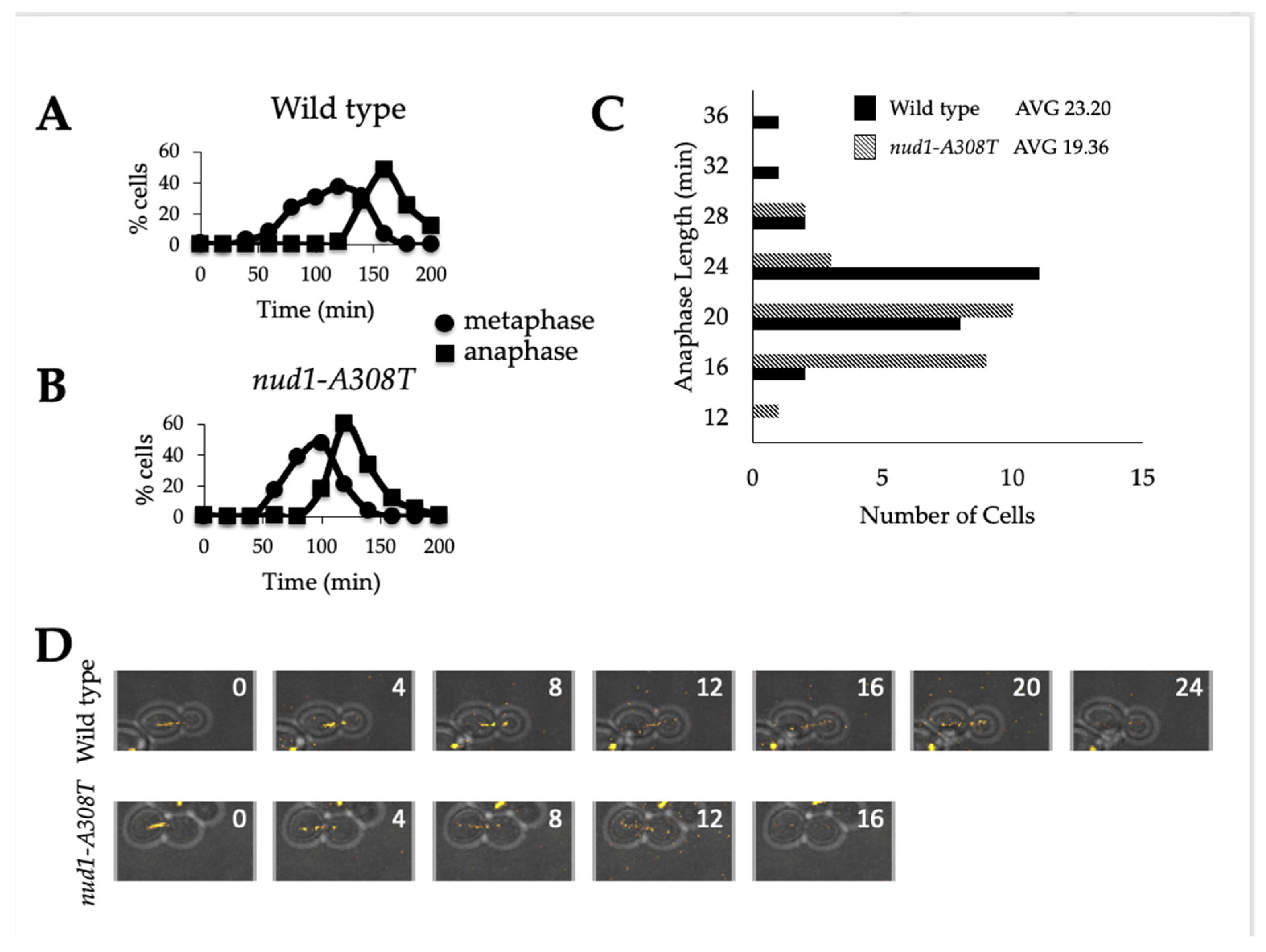
3.4. Nud1-A308T Cells Have a Shortened Anaphase
3.5. Nud1-A308T Does Not Promote Association of Tem1 to SPBs
3.6. Cdc15 Is Recruited to SPBs in All Cell Cycle Phases in the Majority of Nud1-A308T Cells
3.7. Nud1-A308T Prematurely Recruits Dbf2 to the Mother SPB in Metaphase
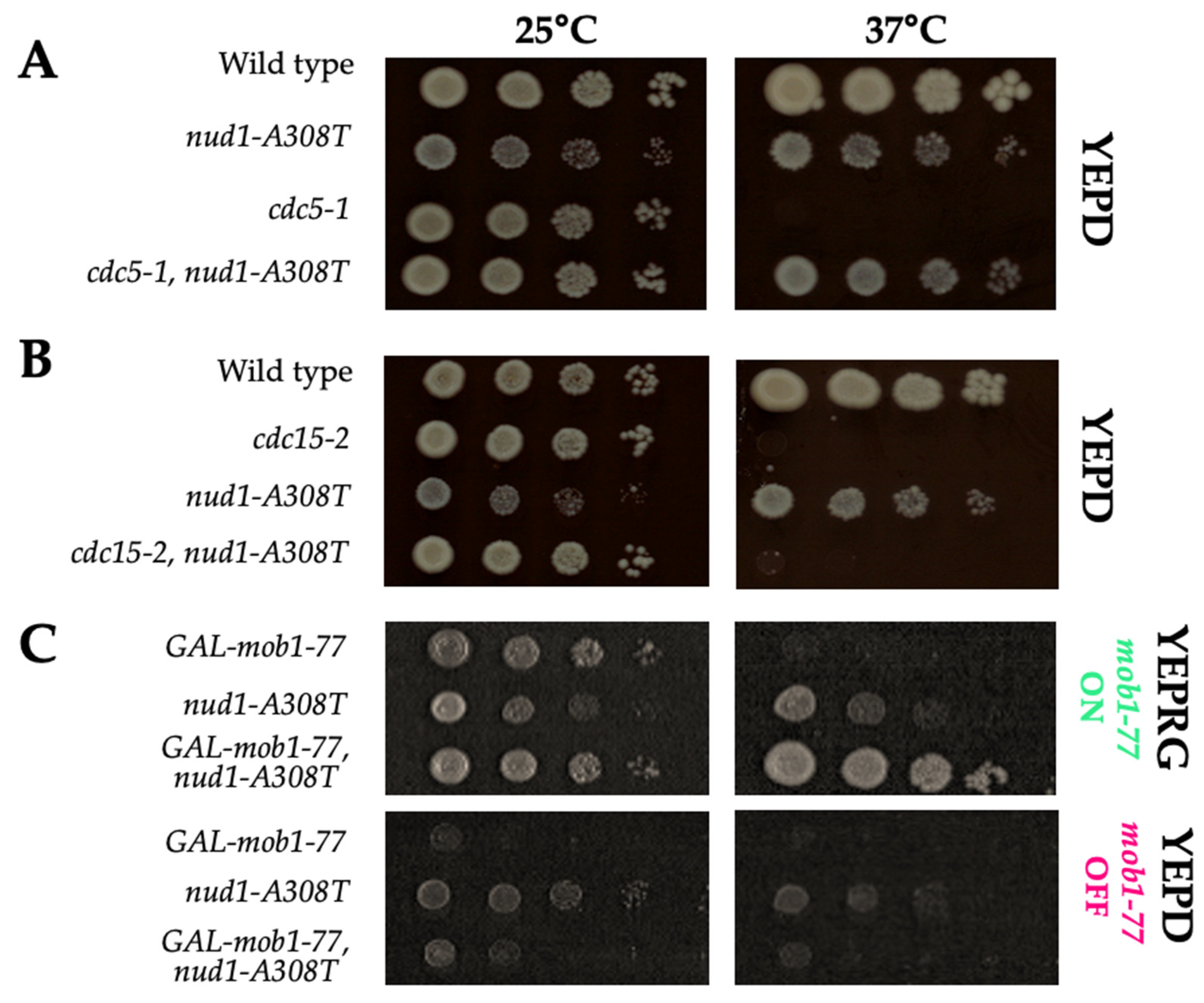
3.8. Nud1-A308T Function Requires CDC15 and MOB1, but Not CDC5
3.9. Nud1-A308T Recruits Mob1 in Metaphase and Suppresses the Dominant GAL-nud1-T78A
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Murray, A.W.; Kirschner, M.W. Cyclin synthesis drives the early embryonic cell cycle. Nat. Cell Biol. 1989, 339, 275–280. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, M.; Morgan, D. Finishing mitosis, one step at a time. Nat. Rev. Mol. Cell Biol. 2007, 8, 894–903. [Google Scholar] [CrossRef]
- Irniger, S.; Piatti, S.; Michaelis, C.; Nasmyth, K. Genes involved in sister chromatid separation are needed for b-type cyclin proteolysis in budding yeast. Cell 1995, 81, 269–277. [Google Scholar] [CrossRef]
- King, R.W.; Peters, J.-M.; Tugendreich, S.; Rolfe, M.; Hieter, P.; Kirschner, M.W. A 20s complex containing CDC27 and CDC16 catalyzes the mitosis-specific conjugation of ubiquitin to cyclin B. Cell 1995, 81, 279–288. [Google Scholar] [CrossRef]
- Sudakin, V.; Ganoth, D.; Dahan, A.; Heller, H.; Hershko, J.; Luca, F.C.; Ruderman, J.V.; Hershko, A. The cyclosome, a large complex containing cyclin-selective ubiquitin ligase activity, targets cyclins for destruction at the end of mitosis. Mol. Biol. Cell 1995, 6, 185–197. [Google Scholar] [CrossRef]
- Tugendreich, S.; Tomkiel, J.; Earnshaw, W.; Hieter, P. CDC27Hs colocalizes with CDC16Hs to the centrosome and mitotic spindle and is essential for the metaphase to anaphase transition. Cell 1995, 81, 261–268. [Google Scholar] [CrossRef]
- Jaspersen, S.L.; Charles, J.; Tinker-Kulberg, R.L.; Morgan, D.O. A Late Mitotic Regulatory Network Controlling Cyclin Destruction in Saccharomyces cerevisiae. Mol. Biol. Cell 1998, 9, 2803–2817. [Google Scholar] [CrossRef] [PubMed]
- Visintin, R.; Craig, K.; Hwang, E.S.; Prinz, S.; Tyers, M.; Amon, A. The Phosphatase Cdc14 Triggers Mitotic Exit by Reversal of Cdk-Dependent Phosphorylation. Mol. Cell 1998, 2, 709–718. [Google Scholar] [CrossRef]
- Zachariae, W.; Schwab, M.; Nasmyth, K.; Seufert, W. Control of Cyclin Ubiquitination by CDK-Regulated Binding of Hct1 to the Anaphase Promoting Complex. Science 1998, 282, 1721–1724. [Google Scholar] [CrossRef]
- Musacchio, A.; Salmon, E.D. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 2007, 8, 379–393. [Google Scholar] [CrossRef]
- Yeh, E.; Skibbens, R.V.; Cheng, J.W.; Salmon, E.D.; Bloom, K. Spindle dynamics and cell cycle regulation of dynein in the budding yeast, Saccharomyces cerevisiae. J. Cell Biol. 1995, 130, 687–700. [Google Scholar] [CrossRef]
- Caydasi, A.K.; Ibrahim, B.; Pereira, G. Monitoring spindle orientation: Spindle position checkpoint in charge. Cell Div. 2010, 5, 28. [Google Scholar] [CrossRef] [PubMed]
- Stegmeier, F.; Amon, A. Closing Mitosis: The Functions of the Cdc14 Phosphatase and Its Regulation. Annu. Rev. Genet. 2004, 38, 203–232. [Google Scholar] [CrossRef] [PubMed]
- Bardin, A.J.; Visintin, R.; Amon, A. A Mechanism for Coupling Exit from Mitosis to Partitioning of the Nucleus. Cell 2000, 102, 21–31. [Google Scholar] [CrossRef]
- Pereira, G.; Höfken, T.; Grindlay, J.; Manson, C.; Schiebel, E. The Bub2p Spindle Checkpoint Links Nuclear Migration with Mitotic Exit. Mol. Cell 2000, 6, 1–10. [Google Scholar] [CrossRef]
- Molk, J.N.; Schuyler, S.C.; Liu, J.Y.; Evans, J.G.; Salmon, E.D.; Pellman, D.; Bloom, K. The Differential Roles of Budding Yeast Tem1p, Cdc15p, and Bub2p Protein Dynamics in Mitotic Exit. Mol. Biol. Cell 2004, 15, 1519–1532. [Google Scholar] [CrossRef][Green Version]
- Campbell, I.W.; Zhou, X.; Amon, A. Spindle pole bodies function as signal amplifiers in the Mitotic Exit Network. Mol. Biol. Cell 2020, 31, 906–916. [Google Scholar] [CrossRef]
- D’Aquino, K.E.; Monje-Casas, F.; Paulson, J.; Reiser, V.; Charles, G.M.; Lai, L.; Shokat, K.M.; Amon, A. The Protein Kinase Kin4 Inhibits Exit from Mitosis in Response to Spindle Position Defects. Mol. Cell 2005, 19, 223–234. [Google Scholar] [CrossRef]
- Pereira, G.; Schiebel, E. Kin4 Kinase Delays Mitotic Exit in Response to Spindle Alignment Defects. Mol. Cell 2005, 19, 209–221. [Google Scholar] [CrossRef] [PubMed]
- Alexandru, G.; Zachariae, W.; Schleiffer, A.; Nasmyth, K. Sister chromatid separation and chromosome re-duplication are regulated by different mechanisms in response to spindle damage. EMBO J. 1999, 18, 2707–2721. [Google Scholar] [CrossRef]
- Fesquet, D.; Fitzpatrick, P.J.; Johnson, A.L.; Kramer, K.M.; Toyn, J.H.; Johnston, L.H. A Bub2p-dependent spindle checkpoint pathway regulates the Dbf2p kinase in budding yeast. EMBO J. 1999, 18, 2424–2434. [Google Scholar] [CrossRef]
- Fraschini, R.; Formenti, E.; Lucchini, G.; Piatti, S. Budding Yeast Bub2 Is Localized at Spindle Pole Bodies and Activates the Mitotic Checkpoint via a Different Pathway from Mad2. J. Cell Biol. 1999, 145, 979–991. [Google Scholar] [CrossRef]
- Maekawa, H.; Priest, C.; Lechner, J.; Pereira, G.; Schiebel, E. The yeast centrosome translates the positional information of the anaphase spindle into a cell cycle signal. J. Cell Biol. 2007, 179, 423–436. [Google Scholar] [CrossRef] [PubMed]
- Bertazzi, D.T.; Kurtulmus, B.; Pereira, G. The cortical protein Lte1 promotes mitotic exit by inhibiting the spindle position checkpoint kinase Kin4. J. Cell Biol. 2011, 193, 1033–1048. [Google Scholar] [CrossRef]
- Falk, J.; Chan, L.; Amon, A. Lte1 promotes mitotic exit by controlling the localization of the spindle position checkpoint kinase Kin4. Proc. Natl. Acad. Sci. USA 2011, 108, 12584–12590. [Google Scholar] [CrossRef] [PubMed]
- Falk, J.E.; Campbell, I.W.; Joyce, K.; Whalen, J.; Seshan, A.; Amon, A. LTE1 promotes exit from mitosis by multiple mech-anisms. Mol. Biol. Cell 2016, 27, 3991–4001. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Jang, S.S.; Song, K. Different Levels of Bfa1/Bub2 GAP Activity Are Required to Prevent Mitotic Exit of Budding Yeast Depending on the Type of Perturbations. Mol. Biol. Cell 2008, 19, 4328–4340. [Google Scholar] [CrossRef] [PubMed]
- Valerio-Santiago, M.; Monje-Casas, F. Tem1 localization to the spindle pole bodies is essential for mitotic exit and impairs spindle checkpoint function. J. Cell Biol. 2011, 192, 599–614. [Google Scholar] [CrossRef]
- Chan, L.; Amon, A. Spindle Position Is Coordinated with Cell-Cycle Progression through Establishment of Mitotic Exit-Activating and -Inhibitory Zones. Mol. Cell 2010, 39, 444–454. [Google Scholar] [CrossRef]
- Falk, J.E.; Tsuchiya, D.; Verdaasdonk, J.; Lacefield, S.; Bloom, K.; Amon, A. Spatial signals link exit from mitosis to spindle position. eLife 2016, 5, e14036. [Google Scholar] [CrossRef] [PubMed]
- Scarfone, I.; Venturetti, M.; Hotz, M.; Lengefeld, J.; Barral, Y.; Piatti, S. Asymmetry of the Budding Yeast Tem1 GTPase at Spindle Poles Is Required for Spindle Positioning But Not for Mitotic Exit. PLoS Genet. 2015, 11, e1004938. [Google Scholar] [CrossRef]
- Whalen, J.; Sniffen, C.; Gartland, S.; Vannini, M.; Seshan, A. Budding Yeast BFA1 Has Multiple Positive Roles in Directing Late Mitotic Events. G3 Genes Genomes Genet. 2018, 8, 3397–3410. [Google Scholar] [CrossRef] [PubMed]
- Asakawa, K.; Yoshida, S.; Otake, F.; Toh-E, A. A Novel Functional Domain of Cdc15 Kinase Is Required for Its Interaction with Tem1 GTPase in Saccharomyces cerevisiae. Genetics 2001, 157, 1437–1450. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.E.; Frenz, L.M.; Wells, N.J.; Johnson, A.L.; Johnston, L.H. Order of function of the budding-yeast mitotic exit-network proteins Tem1, Cdc15, Mob1, Dbf2, and Cdc5. Curr. Biol. 2001, 11, 784–788. [Google Scholar] [CrossRef][Green Version]
- Visintin, R.; Amon, A. Regulation of the Mitotic Exit Protein Kinases Cdc15 and Dbf2. Mol. Biol. Cell 2001, 12, 2961–2974. [Google Scholar] [CrossRef]
- Rock, J.M.; Amon, A. Cdc15 integrates Tem1 GTPase-mediated spatial signals with Polo kinase-mediated temporal cues to activate mitotic exit. Genes Dev. 2011, 25, 1943–1954. [Google Scholar] [CrossRef] [PubMed]
- Mah, A.S.; Jang, J.; Deshaies, R.J. Protein kinase Cdc15 activates the Dbf2-Mob1 kinase complex. Proc. Natl. Acad. Sci. USA 2001, 98, 7325–7330. [Google Scholar] [CrossRef]
- Rock, J.M.; Lim, D.; Stach, L.; Ogrodowicz, R.W.; Keck, J.M.; Jones, M.H.; Wong, C.C.L.; Yates, J.R.; Winey, M.; Smerdon, S.J.; et al. Activation of the Yeast Hippo Pathway by Phosphorylation-Dependent Assembly of Signaling Complexes. Science 2013, 340, 871–875. [Google Scholar] [CrossRef]
- Mohl, D.A.; Huddleston, M.J.; Collingwood, T.S.; Annan, R.S.; Deshaies, R.J. Dbf2–Mob1 drives relocalization of protein phosphatase Cdc14 to the cytoplasm during exit from mitosis. J. Cell Biol. 2009, 184, 527–539. [Google Scholar] [CrossRef]
- Manzoni, R.; Montani, F.; Visintin, C.; Caudron, F.; Ciliberto, A.; Visintin, R. Oscillations in Cdc14 release and sequestration reveal a circuit underlying mitotic exit. J. Cell Biol. 2010, 190, 209–222. [Google Scholar] [CrossRef]
- Zhou, X.; Li, W.; Liu, Y.; Amon, A. Cross-compartment signal propagation in the mitotic exit network. eLife 2021, 10, 63645. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Hunke, L.; Hardy, C.F.J. Cell Cycle Regulation of the Saccharomyces cerevisiae Polo-Like Kinase Cdc5p. Mol. Cell. Biol. 1998, 18, 7360–7370. [Google Scholar] [CrossRef] [PubMed]
- Park, J.E.; Park, C.J.; Sakchaisri, K.; Karpova, T.; Asano, S.; McNally, J.; Lee, K.S. Novel functional dissection of the lo-calization-specific roles of budding yeast polo kinase Cdc5p. Mol. Cell. Biol. 2004, 24, 9873–9886. [Google Scholar] [CrossRef] [PubMed]
- Botchkarev, V.V.; Rossio, V.; Yoshida, S. The budding yeast Polo-like kinase Cdc5 is released from the nucleus during anaphase for timely mitotic exit. Cell Cycle 2014, 13, 3260–3270. [Google Scholar] [CrossRef]
- Campbell, I.W.; Zhou, X.; Amon, A. The Mitotic Exit Network integrates temporal and spatial signals by distributing regulation across multiple components. eLife 2019, 8, 41139. [Google Scholar] [CrossRef]
- Jaspersen, S.L.; Morgan, D. Cdc14 activates Cdc15 to promote mitotic exit in budding yeast. Curr. Biol. 2000, 10, 615–618. [Google Scholar] [CrossRef]
- König, C.; Maekawa, H.; Schiebel, E. Mutual regulation of cyclin-dependent kinase and the mitotic exit network. J. Cell Biol. 2010, 188, 351–368. [Google Scholar] [CrossRef][Green Version]
- Shirayama, M.; Tóth, A.; Gálová, M.; Nasmyth, K. APCCdc20 promotes exit from mitosis by destroying the anaphase inhibitor Pds1 and cyclin Clb5. Nat. Cell Biol. 1999, 402, 203–207. [Google Scholar] [CrossRef]
- Bardin, A.J.; Boselli, M.G.; Amon, A. Mitotic Exit Regulation through Distinct Domains within the Protein Kinase Cdc15. Mol. Cell. Biol. 2003, 23, 5018–5030. [Google Scholar] [CrossRef][Green Version]
- Visintin, R.; Hwang, E.S.; Amon, A. Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus. Nat. Cell Biol. 1999, 398, 818–823. [Google Scholar] [CrossRef]
- Ro, H.-S.; Song, S.; Lee, K.S. Bfa1 can regulate Tem1 function independently of Bub2 in the mitotic exit network of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 2002, 99, 5436–5441. [Google Scholar] [CrossRef] [PubMed]
- Chan, L.Y.; Amon, A. The protein phosphatase 2A functions in the spindle position checkpoint by regulating the checkpoint kinase Kin4. Genes Dev. 2009, 23, 1639–1649. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gruneberg, U.; Campbell, K.; Simpson, C.; Grindlay, J.; Schiebel, E. Nud1p links astral microtubule organization and the control of exit from mitosis. EMBO J. 2000, 19, 6475–6488. [Google Scholar] [CrossRef]
- Luca, F.C.; Winey, M. MOB1, an Essential Yeast Gene Required for Completion of Mitosis and Maintenance of Ploidy. Mol. Biol. Cell 1998, 9, 29–46. [Google Scholar] [CrossRef][Green Version]
- Pereira, G.; Tanaka, T.; Nasmyth, K.; Schiebel, E. Modes of spindle pole body inheritance and segregation of the Bfa1p-Bub2p checkpoint protein complex. EMBO J. 2001, 20, 6359–6370. [Google Scholar] [CrossRef]
- Hotz, M.; Lengefeld, J.; Barral, Y. The MEN mediates the effects of the spindle assembly checkpoint on Kar9-dependent spindle pole body inheritance in budding yeast. Cell Cycle 2012, 11, 3109–3116. [Google Scholar] [CrossRef]
- Lengefeld, J.; Hotz, M.; Rollins, M.; Baetz, K.; Barral, Y. Budding yeast Wee1 distinguishes spindle pole bodies to guide their pattern of age-dependent segregation. Nat. Cell Biol. 2017, 19, 941–951. [Google Scholar] [CrossRef]
- Lengfeld, J.; Barral, Y. Asymmetric Segregation of Aged Spindle Pole Bodies During Cell Division: Mechanisms and Relevance Beyond Budding Yeast? BioEssays 2018, 40, e1800038. [Google Scholar] [CrossRef]
- Manzano-Lopez, J.; Matellán, L.; Álvarez-Llamas, A.; Blanco-Mira, J.C.; Monje-Casas, F. Asymmetric inheritance of spindle microtubule-organizing centres preserves replicative lifespan. Nat. Cell Biol. 2019, 21, 952–965. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Sanawar, R.; Li, X.; Li, F. Structure, biochemistry, and biology of PAK kinases. Gene 2017, 605, 20–31. [Google Scholar] [CrossRef] [PubMed]
- Park, C.J.; Park, J.-E.; Karpova, T.S.; Soung, N.-K.; Yu, L.-R.; Song, S.; Lee, K.H.; Xia, X.; Kang, E.; Dabanoglu, I.; et al. Requirement for the Budding Yeast Polo Kinase Cdc5 in Proper Microtubule Growth and Dynamics. Eukaryot. Cell 2008, 7, 444–453. [Google Scholar] [CrossRef] [PubMed]
- Tamborrini, D.; Juanes, M.A.; Ibanes, S.; Rancati, G.; Piatti, S. Recruitment of the mitotic exit network to yeast centrosomes couples septin displacement to actomyosin constriction. Nat. Commun. 2018, 9, 4308. [Google Scholar] [CrossRef]
- Gromley, A.; Jurczyk, A.; Sillibourne, J.; Halilovic, E.; Mogensen, M.; Groisman, S.; Blomberg, M.; Doxsey, S. A novel human protein of the maternal centriole is required for the final stages of cytokinesis and entry into S phase. J. Cell Biol. 2003, 161, 535–545. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-T.; Hehnly, H.; Yu, Q.; Farkas, D.; Zheng, G.; Redick, S.D.; Hung, H.-F.; Samtani, R.; Jurczyk, A.; Akbarian, S.; et al. A Unique Set of Centrosome Proteins Requires Pericentrin for Spindle-Pole Localization and Spindle Orientation. Curr. Biol. 2014, 24, 2327–2334. [Google Scholar] [CrossRef] [PubMed]





| Number | Genotype |
|---|---|
| 3 | MAT a ade2-1, leu2-3, ura3, trp1-1, his3-1115, can1-100, GAL, [phi+] (W303) |
| 5 | MAT a GAL-GFP-BFA1::HIS3MX6 |
| 15 | MAT a TEM1-GFP:HIS3MX6 |
| 53 | MAT a nud1-A308T |
| 79 | MAT a TEM1-GFP:HIS3MX6, HIS3MX6::GAL-BFA1 |
| 112 | MAT a bub2::HIS3MX6 |
| 138 | MAT a CDC14-3HA |
| 228 | MAT a cdc14-3, MOB1-6HA:HIS3MX6, NUD1-3V5::kanMX6 |
| 296 | MAT a TEM1-GFP:HIS3MX6, HIS3MX6::GAL-BFA1, nud1(A308T)-3V5::kanMX6 |
| 303 | MAT a nud1(A308T)-3V5::kanMX6, CDC14-3HA |
| 326 | MAT a NUD1-3V5::kanMX6 |
| 383 | MAT a NUD1-3V5::kanMX6, trp1::YIP204-HIS3MX6::GAL-nud1(S53A,T78A)-3V5::TRP1 |
| 384 | MAT a NUD1-3V5::kanMX6, trp1::YIP204-HIS3MX6::GAL-nud1(T78A)-3V5::TRP1 |
| 385 | MAT a NUD1-3V5::kanMX6, trp1:: YIP204-HIS3MX6::GAL-nud1(S63A,T78A)-3V5::TRP1 |
| 387 | MAT a nud1(A308T)-3V5::kanMX6 |
| 388 | MAT a NUD1-3V5::kanMX6, trp1::YIP204-HIS3MX6::GAL-nud1(S53A,S63A,T78A)-3V5::TRP1 |
| 390 | MAT a nud1(A308T)-3V5::kanMX6, trp1::YIP204-HIS3MX6::GAL-nud1(S53A,S63A,T78A)-3V5::TRP1 |
| 392 | MAT a nud1(A308T)-3V5::kanMX6, trp1::YIP204-HIS3MX6::GAL-nud1(S63A,T78A)-3V5::TRP1 |
| 395 | MAT a nud1(A308T)-3V5::kanMX6, trp1::YIP204-HIS3MX6::GAL-nud1(T78A)-3V5::TRP1 |
| 397 | MAT a nud1(A308T)-3V5::kanMX6, trp1::YIP204-HIS3MX6::GAL-nud1(S53A,T78A)-3V5::TRP1 |
| 402 | MAT a kanMX6::GAL-mob1-77 |
| 407 | MAT a cdc14-3, MOB1-6HA:HIS3MX6 |
| 417 | MAT a dyn1::HIS3MX6,ura3::pAFS125-TUB1p-GFP-TUB1::URA3 |
| 439 | MAT a cdc14-3, MOB1-6HA:HIS3MX6, nud1(A308T)-3V5::kanMX6 |
| 447 | MAT a cdc15-2, nud1(A308T)-3V5::kanMX6 |
| 474 | MAT a dyn1::HIS3MX6,ura3::pAFS125-TUB1p-GFP-TUB1::URA3, nud1(A308T)-3V5::kanMX6 |
| 476 | MAT a DBF2-eGFP::HIS3MX6, NUD1-3V5::kanMX6,ura3::pRS306-TUB1-mCherry-tub1::URA3 |
| 477 | MAT a DBF2-eGFP::HIS3MX6, nud1(A308T)-3V5::kanMX6,ura3::pRS306-TUB1-mCherry-tub1::URA3 |
| 478 | MAT a nud1(A308T)-3V5::kanMX6, kanMX6::GAL-mob1-77 |
| 513 | MAT a CDC15-eGFP::kanMX6, NUD1-3V5::kanMX6,ura3::pRS306-TUB1-mCherry-tub1::URA3 |
| 517 | MAT a CDC15-eGFP::kanMX6, nud1(A308T)-3V5::kanMX6,ura3::pRS306-TUB1-mCherry-tub1::URA3 |
| 564 | MAT a cdc5-1 |
| 570 | MAT a cdc5-1, nud1(A308T)-3V5::kanMX6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vannini, M.; Mingione, V.R.; Meyer, A.; Sniffen, C.; Whalen, J.; Seshan, A. A Novel Hyperactive Nud1 Mitotic Exit Network Scaffold Causes Spindle Position Checkpoint Bypass in Budding Yeast. Cells 2022, 11, 46. https://doi.org/10.3390/cells11010046
Vannini M, Mingione VR, Meyer A, Sniffen C, Whalen J, Seshan A. A Novel Hyperactive Nud1 Mitotic Exit Network Scaffold Causes Spindle Position Checkpoint Bypass in Budding Yeast. Cells. 2022; 11(1):46. https://doi.org/10.3390/cells11010046
Chicago/Turabian StyleVannini, Michael, Victoria R. Mingione, Ashleigh Meyer, Courtney Sniffen, Jenna Whalen, and Anupama Seshan. 2022. "A Novel Hyperactive Nud1 Mitotic Exit Network Scaffold Causes Spindle Position Checkpoint Bypass in Budding Yeast" Cells 11, no. 1: 46. https://doi.org/10.3390/cells11010046
APA StyleVannini, M., Mingione, V. R., Meyer, A., Sniffen, C., Whalen, J., & Seshan, A. (2022). A Novel Hyperactive Nud1 Mitotic Exit Network Scaffold Causes Spindle Position Checkpoint Bypass in Budding Yeast. Cells, 11(1), 46. https://doi.org/10.3390/cells11010046






