Functional Characterization of Human Pluripotent Stem Cell-Derived Models of the Brain with Microelectrode Arrays
Abstract
1. Introduction
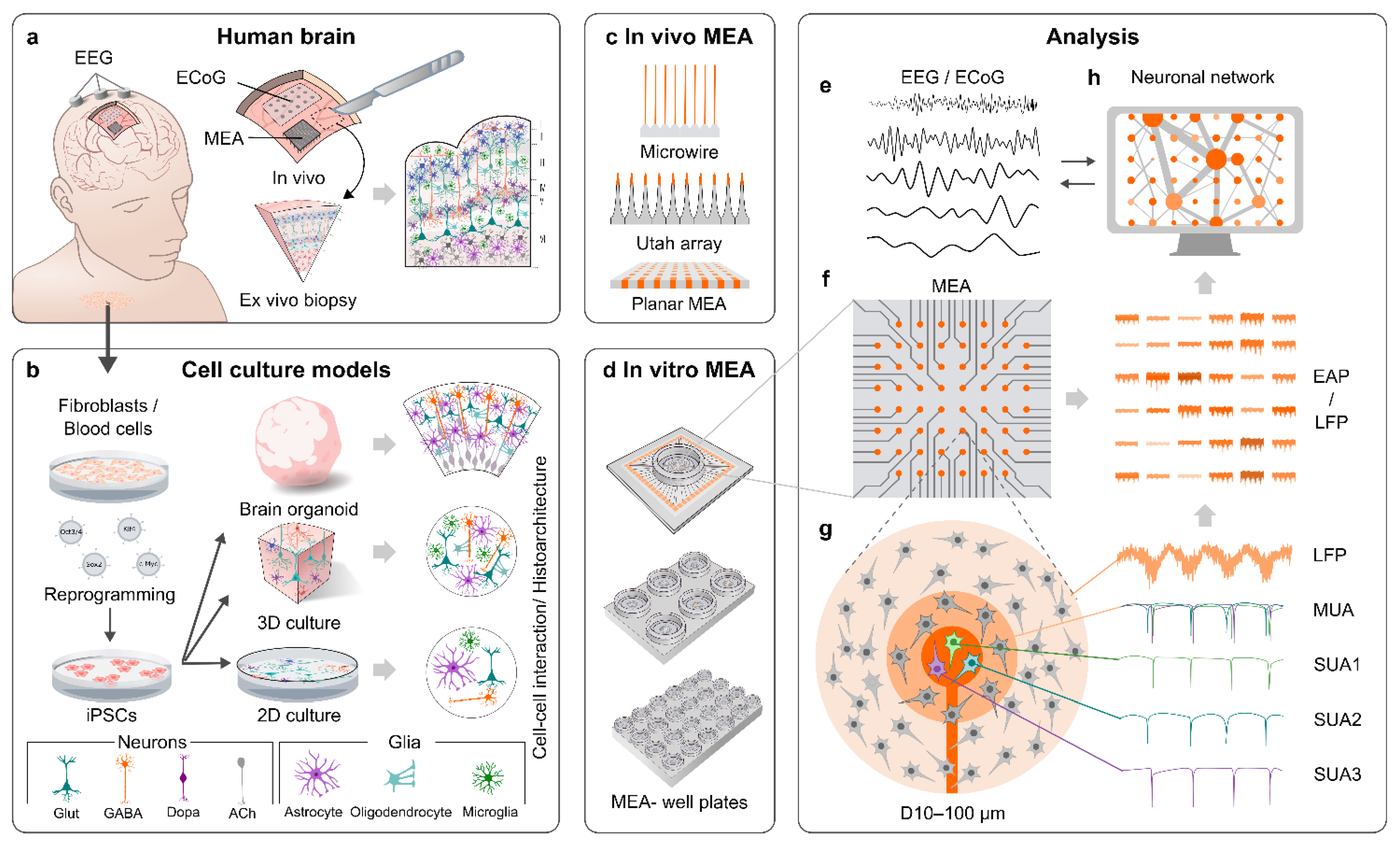
2. MEAs
2.1. General Properties of MEAs
2.2. MEAs for In Vivo Applications
2.3. MEAs for In Vitro Applications
3. MEA Recordings of the Human Brain
3.1. Requirements for MEA Recordings Set by In Vivo Neuronal Properties
3.2. Challenges Set by the 3D Structure of the Brain to MEA Recordings
3.3. Non-Neuronal Cells Affect Neuronal Activity
3.4. Neuronal Firing in the Human Brain in MEA Recordings In Vivo
3.5. Neural Oscillations of the Human Brain in MEA Recordings In Vivo
3.6. Functional Connectivity of the Human Brain in MEA Recordings In Vivo
3.7. Detecting Epileptic and Other Pathological Activity in the Human Brain with MEAs In Vivo
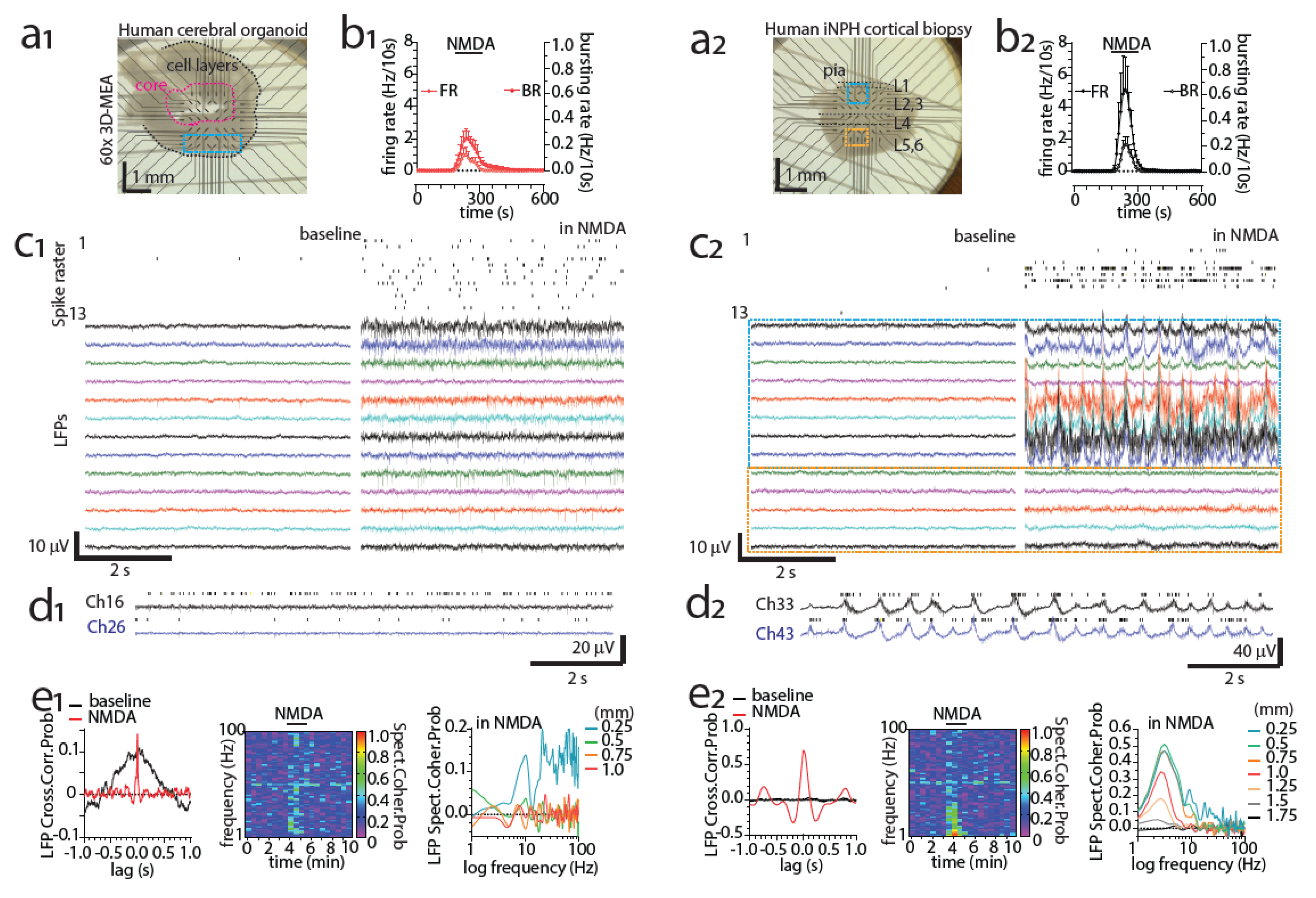
3.8. Detecting Activity of the Human Brain with MEAs Ex Vivo
4. hPSC-Derived 2D Brain Models of MEAs
4.1. General Properties of 2D Cultures on MEAs and Early hPSC-Derived Brain Models
4.2. Human Neurons Produced with Dual-SMAD Inhibition Differentiation on MEAs
4.3. Cell-Type Specific Differentiation Methods for Producing 2D Brain Models of MEAs
4.4. Main Limitations of 2D hPSC-Derived Neuronal Cultures and Overcoming Them
4.5. Guiding the Orientation of 2D Cultures
4.6. hPSC-Derived 2D Neuronal Cultures on MEAs as Models of Physiology
4.7. hPSC-Derived 2D Neuronal Cultures on MEAs as Models of Pathology
5. hPSC-Derived 3D Neuronal Cultures and Organoids on MEAs
5.1. Properties and Scaffolds of hPSC-Derived 3D Brain Models
5.2. Properties and Differentiation of hPSC-Derived Brain Organoids
5.3. Adaptation of 3D Brain Models to In Vitro MEAs and Vice Versa
5.4. Modeling Neuronal Development and Physiology with Organoids on MEAs
5.5. Modeling Neuronal Pathology with Organoids on MEAs
5.6. Overcoming Limitations of Organoids and Other 3D Brain Models
6. Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Thomson, J.A.; Itskovitz-Eldor, J.; Shapiro, S.S.; Waknitz, M.A.; Swiergiel, J.J.; Marshall, V.S.; Jones, J.M. Embryonic Stem Cell Lines Derived from Human Blastocysts. Science 1998, 282, 1145–1147. [Google Scholar] [CrossRef]
- Carpenter, M.K.; Inokuma, M.S.; Denham, J.; Mujtaba, T.; Chiu, C.P.; Rao, M.S. Enrichment of neurons and neural precursors from human embryonic stem cells. Exp. Neurol. 2001, 172, 383–397. [Google Scholar] [CrossRef]
- Charitos, I.A.; Ballini, A.; Cantore, S.; Boccellino, M.; Di Domenico, M.; Borsani, E.; Nocini, R.; Di Cosola, M.; Santacroce, L.; Bottalico, L. Stem Cells: A Historical Review about Biological, Religious, and Ethical Issues. Stem Cells Int. 2021, 2021, 9978837. [Google Scholar] [CrossRef]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef]
- Dimos, J.T.; Rodolfa, K.T.; Niakan, K.K.; Weisenthal, L.M.; Mitsumoto, H.; Chung, W.; Croft, G.F.; Saphier, G.; Leibel, R.; Goland, R.; et al. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science 2008, 321, 1218–1221. [Google Scholar] [CrossRef]
- Lancaster, M.A.; Knoblich, J.A. Generation of cerebral organoids from human pluripotent stem cells. Nat. Protoc. 2014, 9, 2329–2340. [Google Scholar] [CrossRef] [PubMed]
- Mossink, B.; Verboven, A.H.A.; van Hugte, E.J.H.; Klein Gunnewiek, T.M.; Parodi, G.; Linda, K.; Schoenmaker, C.; Kleefstra, T.; Kozicz, T.; van Bokhoven, H.; et al. Human neuronal networks on micro-electrode arrays are a highly robust tool to study disease-specific genotype-phenotype correlations in vitro. Stem Cell Rep. 2021, 16, 2182–2196. [Google Scholar] [CrossRef] [PubMed]
- Cash, S.S.; Hochberg, L.R. The emergence of single neurons in clinical neurology. Neuron 2015, 86, 79–91. [Google Scholar] [CrossRef] [PubMed]
- Fried, I.; Wilson, C.L.; Maidment, N.T.; Engel, J.; Behnke, E.; Fields, T.A.; Macdonald, K.A.; Morrow, J.W.; Ackerson, L. Cerebral microdialysis combined with single-neuron and electroencephalographic recording in neurosurgical patients: Technical note. J. Neurosurg. 1999, 91, 697–705. [Google Scholar] [CrossRef] [PubMed]
- Marg, E.; Adams, J.E. Indwelling multiple micro-electrodes in the brain. Electroencephalogr. Clin. Neurophysiol. 1967, 23, 277–280. [Google Scholar] [CrossRef]
- Hochberg, L.R.; Serruya, M.D.; Friehs, G.M.; Mukand, J.A.; Saleh, M.; Caplan, A.H.; Branner, A.; Chen, D.; Penn, R.D.; Donoghue, J.P. Neuronal ensemble control of prosthetic devices by a human with tetraplegia. Nature 2006, 442, 164–171. [Google Scholar] [CrossRef]
- Ulbert, I.; Heit, G.; Madsen, J.; Karmos, G.; Halgren, E. Laminar Analysis of Human Neocortical Interictal Spike Generation and Propagation: Current Source Density and Multiunit Analysis In Vivo. Epilepsia 2004, 45, 48–56. [Google Scholar] [CrossRef]
- Trujillo, C.A.; Gao, R.; Negraes, P.D.; Gu, J.; Buchanan, J.; Preissl, S.; Wang, A.; Wu, W.; Haddad, G.G.; Chaim, I.A.; et al. Complex Oscillatory Waves Emerging from Cortical Organoids Model Early Human Brain Network Development. Cell Stem Cell 2019, 25, 558–569.e7. [Google Scholar] [CrossRef] [PubMed]
- Shcheglovitov, A.; Peterson, R.T. Screening Platforms for Genetic Epilepsies—Zebrafish, iPSC-Derived Neurons, and Organoids. Neurotherapeutics 2021, 18, 1478–1489. [Google Scholar] [CrossRef] [PubMed]
- Samarasinghe, R.A.; Miranda, O.A.; Buth, J.E.; Mitchell, S.; Ferando, I.; Watanabe, M.; Allison, T.F.; Kurdian, A.; Fotion, N.N.; Gandal, M.J.; et al. Identification of neural oscillations and epileptiform changes in human brain organoids. Natl. Neurosci. 2021. [Google Scholar] [CrossRef] [PubMed]
- Odawara, A.; Katoh, H.; Matsuda, N.; Suzuki, I. Physiological maturation and drug responses of human induced pluripotent stem cell-derived cortical neuronal networks in long-term culture. Sci. Rep. 2016, 6, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Shimba, K.; Sakai, K.; Iida, S.; Kotani, K.; Jimbo, Y. Long-term developmental process of the human cortex revealed in vitro by axon-targeted recording using a microtunnel-augmented microelectrode array. IEEE Trans. Biomed. Eng. 2019, 66, 2538–2545. [Google Scholar] [CrossRef] [PubMed]
- Colachis, S.C.; Dunlap, C.F.; Annetta, N.V.; Tamrakar, S.M.; Bockbrader, M.A.; Friedenberg, D.A. Long-term intracortical microelectrode array performance in a human: A 5 year retrospective analysis. J. Neural Eng. 2021, 18, 0460d7. [Google Scholar] [CrossRef]
- Szymanski, L.J.; Kellis, S.; Liu, C.Y.; Jones, K.T.; Andersen, R.A.; Commins, D.; Lee, B.; McCreery, D.B.; Miller, C.A. Neuropathological effects of chronically implanted, intracortical microelectrodes in a tetraplegic patient. J. Neural Eng. 2021, 18. [Google Scholar] [CrossRef] [PubMed]
- Kamioka, H.; Maeda, E.; Jimbo, Y.; Robinson, H.P.C.; Kawana, A. Spontaneous periodic synchronized bursting during formation of mature patterns of connections in cortical cultures. Neurosci. Lett. 1996, 206, 109–112. [Google Scholar] [CrossRef]
- Peyrache, A.; Dehghani, N.; Eskandar, E.N.; Madsen, J.R.; Anderson, W.S.; Donoghue, J.A.; Hochberg, L.R.; Halgren, E.; Cash, S.S.; Destexhe, A. Spatiotemporal dynamics of neocortical excitation and inhibition during human sleep. Proc. Natl. Acad. Sci. USA 2012, 109, 1731–1736. [Google Scholar] [CrossRef]
- Hodge, R.D.; Bakken, T.E.; Miller, J.A.; Smith, K.A.; Barkan, E.R.; Graybuck, L.T.; Close, J.L.; Long, B.; Johansen, N.; Penn, O.; et al. Conserved cell types with divergent features in human versus mouse cortex. Nature 2019, 573, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Beaulieu-Laroche, L.; Toloza, E.H.S.; van der Goes, M.S.; Lafourcade, M.; Barnagian, D.; Williams, Z.M.; Eskandar, E.N.; Frosch, M.P.; Cash, S.S.; Harnett, M.T. Enhanced Dendritic Compartmentalization in Human Cortical Neurons. Cell 2018, 175, 643–651.e14. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, J.; Kahana, M.J.; Ekstrom, A.D.; Fried, I. Brain oscillations control timing of single-neuron activity in humans. J. Neurosci. 2007, 27, 3839–3844. [Google Scholar] [CrossRef]
- Oberheim, N.A.; Wang, X.; Goldman, S.; Nedergaard, M. Astrocytic complexity distinguishes the human brain. Trends Neurosci. 2006, 29, 547–553. [Google Scholar] [CrossRef] [PubMed]
- Susaki, E.A.; Shimizu, C.; Kuno, A.; Tainaka, K.; Li, X.; Nishi, K.; Morishima, K.; Ono, H.; Ode, K.L.; Saeki, Y.; et al. Versatile whole-organ/body staining and imaging based on electrolyte-gel properties of biological tissues. Nat. Commun. 2020, 11, 1–22. [Google Scholar] [CrossRef]
- Li, J.; Pan, L.; Pembroke, W.G.; Rexach, J.E.; Godoy, M.I.; Condro, M.C.; Alvarado, A.G.; Harteni, M.; Chen, Y.-W.; Stiles, L.; et al. Conservation and divergence of vulnerability and responses to stressors between human and mouse astrocytes. Nat. Commun. 2021, 12, 3958. [Google Scholar] [CrossRef]
- Zhou, Y.; Song, W.M.; Andhey, P.S.; Swain, A.; Levy, T.; Miller, K.R.; Poliani, P.L.; Cominelli, M.; Grover, S.; Gilfillan, S.; et al. Human and mouse single-nucleus transcriptomics reveal TREM2-dependent and TREM2-independent cellular responses in Alzheimer’s disease. Nat. Med. 2020, 26, 131–142. [Google Scholar] [CrossRef]
- Edler, M.K.; Mhatre-Winters, I.; Richardson, J.R. Microglia in Aging and Alzheimer’s Disease: A Comparative Species Review. Cells 2021, 10, 1138. [Google Scholar] [CrossRef]
- Cavanaugh, S.E.; Pippin, J.J.; Barnard, N.D. Animal models of Alzheimer disease: Historical pitfalls and a path forward. ALTEX 2014, 31, 279–302. [Google Scholar] [CrossRef]
- Srinivas, N.; Maffuid, K.; Kashuba, A.D.M. Clinical Pharmacokinetics and Pharmacodynamics of Drugs in the Central Nervous System. Clin. Pharmacokinet. 2018, 57, 1059–1074. [Google Scholar] [CrossRef]
- Borkholder, D.A.; Bao, J.; Maluf, N.I.; Perl, E.R.; Kovacs, G.T.A. Microelectrode arrays for stimulation of neural slice preparations. J. Neurosci. Methods 1997, 77, 61–66. [Google Scholar] [CrossRef]
- Swan, B.D.; Gasperson, L.B.; Krucoff, M.O.; Grill, W.M.; Turner, D.A. Sensory percepts induced by microwire array and DBS microstimulation in human sensory thalamus. Brain Stimul. 2018, 11, 416–422. [Google Scholar] [CrossRef]
- Herreras, O. Local field potentials: Myths and misunderstandings. Front. Neural Circuits 2016, 10, 1–16. [Google Scholar] [CrossRef]
- Buzsáki, G.; Anastassiou, C.A.; Koch, C. The origin of extracellular fields and currents-EEG, ECoG, LFP and spikes. Nat. Rev. Neurosci. 2012, 13, 407–420. [Google Scholar] [CrossRef] [PubMed]
- Strumwasser, F. Long-term recording from single neurons in brain of unrestrained mammals. Science 1958, 127, 469–470. [Google Scholar] [CrossRef]
- Csicsvari, J.; Hirase, H.; Czurkó, A.; Mamiya, A.; Buzsáki, G. Oscillatory coupling of hippocampal pyramidal cells and interneurons in the behaving rat. J. Neurosci. 1999, 19, 274–287. [Google Scholar] [CrossRef] [PubMed]
- Henze, D.A.; Borhegyi, Z.; Csicsvari, J.; Mamiya, A.; Harris, K.D.; Buzsáki, G. Intracellular features predicted by extracellular recordings in the hippocampus in vivo. J. Neurophysiol. 2000, 84, 390–400. [Google Scholar] [CrossRef]
- Buzsáki, G. Large-scale recording of neuronal ensembles. Nat. Neurosci. 2004, 7, 446–451. [Google Scholar] [CrossRef] [PubMed]
- Wise, K.D.; Angell, J.B.; Starr, A. An Integrated-Circuit Approach to Extracellular Microelectrodes. IEEE Trans. Biomed. Eng. 1970, BME-17, 238–247. [Google Scholar] [CrossRef] [PubMed]
- Wise, K.D.; Angell, J.B. A Low-Capacitance Multielectrode Probe for Use in Extracellular Neurophysiology. IEEE Trans. Biomed. Eng. 1975, BME-22, 212–219. [Google Scholar] [CrossRef] [PubMed]
- Bai, Q.; Wise, K.D. Single-unit neural recording with active microelectrode arrays. IEEE Trans. Biomed. Eng. 2001, 48, 911–920. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wise, K.D.; Hetke, J.F.; Bledsoe, S.C. A multichannel neural probe for selective chemical delivery at the cellular level. IEEE Trans. Biomed. Eng. 1997, 44, 760–769. [Google Scholar] [CrossRef] [PubMed]
- Csicsvari, J.; Henze, D.A.; Jamieson, B.; Harris, K.D.; Sirota, A.; Barthó, P.; Wise, K.D.; Buzsáki, G. Massively parallel recording of unit and local field potentials with silicon-based electrodes. J. Neurophysiol. 2003, 90, 1314–1323. [Google Scholar] [CrossRef] [PubMed]
- Jun, J.J.; Steinmetz, N.A.; Siegle, J.H.; Denman, D.J.; Bauza, M.; Barbarits, B.; Lee, A.K.; Anastassiou, C.A.; Andrei, A.; Aydin, Ç.; et al. Fully integrated silicon probes for high-density recording of neural activity. Nature 2017, 551, 232–236. [Google Scholar] [CrossRef]
- Campbell, P.K.; Jones, K.E.; Huber, R.J.; Horch, K.W.; Normann, R.A. A Silicon-Based, Three-Dimensional Neural Interface: Manufacturing Processes for an Intracortical Electrode Array. IEEE Trans. Biomed. Eng. 1991, 38, 758–768. [Google Scholar] [CrossRef]
- Hansen, B.J.; Dragoi, V. Adaptation-induced synchronization in laminar cortical circuits. Proc. Natl. Acad. Sci. USA 2011, 108, 10720–10725. [Google Scholar] [CrossRef]
- Burmeister, J.J.; Moxon, K.; Gerhardt, G.A. Ceramic-based multisite microelectrodes for electrochemical recordings. Anal. Chem. 2000, 72, 187–192. [Google Scholar] [CrossRef]
- Pomerleau, F.; Day, B.; Huettl, P.; Burmeister, J.J.; Gerhardt, G.A. Real Time in Vivo Measures of l-Glutamate in the Rat Central Nervous System Using Ceramic-Based Multisite Microelectrode Arrays. Ann. N. Y. Acad. Sci. 2003, 1003, 454–457. [Google Scholar] [CrossRef]
- Stieglitz, T.; Beutel, H.; Meyer, J.U. A flexible, light-weight multichannel sieve electrode with integrated cables for interfacing regenerating peripheral nerves. Sens. Act. A Phys. 1997, 60, 240–243. [Google Scholar] [CrossRef]
- Cheung, K.C. Implantable microscale neural interfaces. Biomed. Microdev. 2007, 9, 923–938. [Google Scholar] [CrossRef] [PubMed]
- Howard, M.A.; Volkov, I.O.; Granner, M.A.; Damasio, H.M.; Ollendieck, M.C.; Bakken, H.E. A hybrid clinical-research depth electrode for acute and chronic in vivo microelectrode recording of human brain neurons. J. Neurosurg. 1996, 84, 129–132. [Google Scholar] [CrossRef]
- Paulk, A.C.; Yang, J.C.; Cleary, D.R.; Soper, D.J.; Halgren, M.; O’Donnell, A.R.; Lee, S.H.; Ganji, M.; Ro, Y.G.; Oh, H.; et al. Microscale Physiological Events on the Human Cortical Surface. Cereb. Cortex 2021, 31, 3678–3700. [Google Scholar] [CrossRef]
- Kang, Y.N.; Chou, N.; Jang, J.W.; Choe, H.K.; Kim, S. A 3D flexible neural interface based on a microfluidic interconnection cable capable of chemical delivery. Microsyst. Nanoeng. 2021, 7, 1–11. [Google Scholar] [CrossRef]
- Yuk, H.; Lu, B.; Lin, S.; Qu, K.; Xu, J.; Luo, J.; Zhao, X. 3D printing of conducting polymers. Nat. Commun. 2020, 11, 4–11. [Google Scholar] [CrossRef]
- Willett, F.R.; Avansino, D.T.; Hochberg, L.R.; Henderson, J.M.; Shenoy, K.V. High-performance brain-to-text communication via handwriting. Nature 2021, 593, 249–254. [Google Scholar] [CrossRef]
- Tóth, E.; Fabó, D.; Entz, L.; Ulbert, I.; Eross, L. Intracranial neuronal ensemble recordings and analysis in epilepsy. J. Neurosci. Methods 2016, 260, 261–269. [Google Scholar] [CrossRef]
- Thomas, C.A.; Springer, P.A.; Loeb, G.E.; Berwald-Netter, Y.; Okun, L.M. A miniature microelectrode array to monitor the bioelectric activity of cultured cells. Exp. Cell Res. 1972, 74, 61–66. [Google Scholar] [CrossRef]
- Nam, Y.; Wheeler, B.C. In vitro microelectrode array technology and neural recordings. Crit. Rev. Biomed. Eng. 2011, 39, 45–61. [Google Scholar]
- Ryynänen, T.; Pelkonen, A.; Grigoras, K.; Ylivaara, O.M.E.; Hyvärinen, T.; Ahopelto, J.; Prunnila, M.; Narkilahti, S.; Lekkala, J. Microelectrode Array with Transparent ALD TiN Electrodes. Front. Neurosci. 2019, 13, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Ryynänen, T.; Mzezewa, R.; Meriläinen, E.; Hyvärinen, T.; Lekkala, J.; Narkilahti, S.; Kallio, P. Transparent microelectrode arrays fabricated by ion beam assisted deposition for neuronal cell in vitro recordings. Micromachines 2020, 11, 497. [Google Scholar] [CrossRef] [PubMed]
- Hierlemann, A.; Frey, U.; Hafizovic, S.; Heer, F. Growing cells atop microelectronic chips: Interfacing electrogenic cells in vitro with CMOS-based microelectrode arrays. Proc. IEEE 2011, 99, 249–251. [Google Scholar] [CrossRef]
- Müller, J.; Ballini, M.; Livi, P.; Chen, Y.; Radivojevic, M.; Shadmani, A.; Viswam, V.; Jones, I.L.; Fiscella, M.; Diggelmann, R.; et al. High-resolution CMOS MEA platform to study neurons at subcellular, cellular, and network levels. Lab Chip 2015, 15, 2767–2780. [Google Scholar] [CrossRef] [PubMed]
- McConnell, E.R.; McClain, M.A.; Ross, J.; LeFew, W.R.; Shafer, T.J. Evaluation of multi-well microelectrode arrays for neurotoxicity screening using a chemical training set. Neurotoxicology 2012, 33, 1048–1057. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Guo, C.; Lim, L.; Cheong, S.; Zhang, Q.; Tang, K.; Reboud, J. Compact microelectrode array system: Tool for in situ monitoring of drug effects on neurotransmitter release from neural cells. Anal. Chem. 2008, 80, 1133–1140. [Google Scholar] [CrossRef] [PubMed]
- van de Wijdeven, R.; Ramstad, O.H.; Valderhaug, V.D.; Köllensperger, P.; Sandvig, A.; Sandvig, I.; Halaas, Ø. A novel lab-on-chip platform enabling axotomy and neuromodulation in a multi-nodal network. Biosens. Bioelectron. 2019, 140. [Google Scholar] [CrossRef] [PubMed]
- Moutaux, E.; Charlot, B.; Genoux, A.; Saudou, F.; Cazorla, M. An integrated microfluidic/microelectrode array for the study of activity-dependent intracellular dynamics in neuronal networks. Lab Chip 2018, 18, 3425–3435. [Google Scholar] [CrossRef]
- Dworak, B.J.; Wheeler, B.C. Novel MEA platform with PDMS microtunnels enables the detection of action potential propagation from isolated axons in culture. Lab Chip 2009, 9, 404–410. [Google Scholar] [CrossRef]
- Kleber, C.K.; Martina, M.; Burkhardt, C.J.; Guenther, E.; Kraushaar, U. A Novel 3D Microelectrode Array for Extracellular Signal Recording of Acute Brain Slices. In Proceedings of the 9th Int. Meeting on Substrate-Integrated Microelectrode Arrays, Reutlingen, Germany, 1–4 July 2014; pp. 280–281. [Google Scholar]
- Soscia, D.A.; Lam, D.; Tooker, A.C.; Enright, H.A.; Triplett, M.; Karande, P.; Peters, S.K.G.; Sales, A.P.; Wheeler, E.K.; Fischer, N.O. A flexible 3-dimensional microelectrode array for in vitro brain models. Lab Chip 2020, 20, 901–911. [Google Scholar] [CrossRef] [PubMed]
- Obien, M.E.J.; Deligkaris, K.; Bullmann, T.; Bakkum, D.J.; Frey, U. Revealing neuronal function through microelectrode array recordings. Front. Neurosci. 2015, 9, 423. [Google Scholar] [CrossRef] [PubMed]
- Stetson, D.S.; Albers, J.W.; Barbara, A.; Wolfe, R.A. Effects of Age, Sex, and Anthropometric Factors on Nerve Conduction Measures. Muscle Nerve 1992, 15, 1095–1104. [Google Scholar] [CrossRef]
- Mansour, A.A.; Gonçalves, J.T.; Bloyd, C.W.; Li, H.; Fernandes, S.; Quang, D.; Johnston, S.; Parylak, S.L.; Jin, X.; Gage, F.H. An in vivo model of functional and vascularized human brain organoids. Nat. Biotechnol. 2018, 36, 432–441. [Google Scholar] [CrossRef]
- Quiroga, R.Q.; Nadasdy, Z.; Ben-Shaul, Y. Unsupervised spike detection and sorting with wavelets and superparamagnetic clustering. Neural Comput. 2004, 16, 1661–1687. [Google Scholar] [CrossRef] [PubMed]
- Mayer, M.; Arrizabalaga, O.; Lieb, F.; Ciba, M.; Ritter, S.; Thielemann, C. Electrophysiological investigation of human embryonic stem cell derived neurospheres using a novel spike detection algorithm. Biosens. Bioelectron. 2018, 100, 462–468. [Google Scholar] [CrossRef]
- Viskontas, I.V.; Ekstrom, A.D.; Wilson, C.L.; Fried, I. Characterizing interneuron and pyramidal cells in the human medial temporal lobe in vivo using extracellular recordings. Hippocampus 2007, 17, 49–57. [Google Scholar] [CrossRef]
- Pakkenberg, B.; Gundersen, H.J.G. Neocortical neuron number in humans: Effect of sex and age. J. Comp. Neurol. 1997, 384, 312–320. [Google Scholar] [CrossRef]
- Carlson, D.; Carin, L. Continuing progress of spike sorting in the era of big data. Curr. Opin. Neurobiol. 2019, 55, 90–96. [Google Scholar] [CrossRef]
- Hagler, D.J.; Ulbert, I.; Wittner, L.; Erőss, L.; Madsen, J.R.; Devinsky, O.; Doyle, W.; Fabó, D.; Cash, S.S.; Halgren, E. Heterogeneous origins of human sleep spindles in different cortical layers. J. Neurosci. 2018, 38, 3013–3025. [Google Scholar] [CrossRef]
- Halgren, M.; Fabó, D.; Ulbert, I.; Madsen, J.R.; Eross, L.; Doyle, W.K.; Devinsky, O.; Schomer, D.; Cash, S.S.; Halgren, E. Superficial Slow Rhythms Integrate Cortical Processing in Humans. Sci. Rep. 2018, 8, 1–12. [Google Scholar] [CrossRef]
- Markram, H.; Toledo-Rodriguez, M.; Wang, Y.; Gupta, A.; Silberberg, G.; Wu, C. Interneurons of the neocortical inhibitory system. Nat. Rev. Neurosci. 2004, 5, 793–807. [Google Scholar] [CrossRef]
- Köhling, R.; Qü, M.; Zilles, K.; Speckmann, E.J. Current-source-density profiles associated with sharp waves in human epileptic neocortical tissue. Neuroscience 1999, 94, 1039–1050. [Google Scholar] [CrossRef]
- Vargas-Irwin, C.E.; Feldman, J.M.; King, B.; Simeral, J.D.; Sorice, B.L.; Oakley, E.M.; Cash, S.S.; Eskandar, E.N.; Friehs, G.M.; Hochberg, L.R.; et al. Watch, imagine, attempt: Motor cortex single-unit activity reveals context-dependent movement encoding in humans with tetraplegia. Front. Hum. Neurosci. 2018, 12, 450. [Google Scholar] [CrossRef]
- Truccolo, W.; Ahmed, O.J.; Harrison, M.T.; Eskandar, E.N.; Rees Cosgrove, G.; Madsen, J.R.; Blum, A.S.; Stevenson Potter, N.; Hochberg, L.R.; Cash, S.S. Neuronal ensemble synchrony during human focal seizures. J. Neurosci. 2014, 34, 9927–9944. [Google Scholar] [CrossRef]
- Truccolo, W.; Donoghue, J.A.; Hochberg, L.R.; Eskandar, E.N.; Madsen, J.R.; Anderson, W.S.; Brown, E.N.; Halgren, E.; Cash, S.S. Single-neuron dynamics in human focal epilepsy. Nat. Neurosci. 2011, 14, 635–643. [Google Scholar] [CrossRef] [PubMed]
- Gelbard-Sagiv, H.; Mukamel, R.; Harel, M.; Malach, R.; Fried, I. Internally generated reactivation of single neurons in human hippocampus during free recall. Science 2008, 322, 96–101. [Google Scholar] [CrossRef] [PubMed]
- Le Van Quyen, M.; Staba, R.; Bragin, A.; Dickson, C.; Valderrama, M.; Fried, I.; Engel, J. Large-scale microelectrode recordings of high-frequency gamma oscillations in human cortex during sleep. J. Neurosci. 2010, 30, 7770–7782. [Google Scholar] [CrossRef] [PubMed]
- Lang, E.J.; Rosenbluth, J. Role of myelination in the development of a uniform olivocerebellar conduction time. J. Neurophysiol. 2003, 89, 2259–2270. [Google Scholar] [CrossRef]
- Chever, O.; Dossi, E.; Pannasch, U.; Derangeon, M.; Rouach, N. Astroglial networks promote neuronal coordination. Sci. Signal. 2016, 9, 1–9. [Google Scholar] [CrossRef]
- Sardinha, V.M.; Guerra-Gomes, S.; Caetano, I.; Tavares, G.; Martins, M.; Reis, J.S.; Correia, J.S.; Teixeira-Castro, A.; Pinto, L.; Sousa, N.; et al. Astrocytic signaling supports hippocampal–prefrontal theta synchronization and cognitive function. Glia 2017, 65, 1944–1960. [Google Scholar] [CrossRef]
- Chen, Z.; Jalabi, W.; Hu, W.; Park, H.J.; Gale, J.T.; Kidd, G.J.; Bernatowicz, R.; Gossman, Z.C.; Chen, J.T.; Dutta, R.; et al. Microglial displacement of inhibitory synapses provides neuroprotection in the adult brain. Nat. Commun. 2014, 5. [Google Scholar] [CrossRef]
- Hu, D.; Huang, L. Negative hemodynamic response in the cortex: Evidence opposing neuronal deactivation revealed via optical imaging and electrophysiological recording. J. Neurophysiol. 2015, 114, 2152–2161. [Google Scholar] [CrossRef]
- Keller, C.J.; Cash, S.S.; Narayanan, S.; Wang, C.; Kuzniecky, R.; Carlson, C.; Devinsky, O.; Thesen, T.; Doyle, W.; Sassaroli, A.; et al. Intracranial microprobe for evaluating neuro-hemodynamic coupling in unanesthetized human neocortex. J. Neurosci. Methods 2009, 179, 208–218. [Google Scholar] [CrossRef][Green Version]
- Tukker, A.M.; Wijnolts, F.M.J.; De Groot, A.; Westerink, R.H.S. Neurotoxicology Human iPSC-derived neuronal models for in vitro neurotoxicity assessment. Neurotoxicology 2018, 67, 215–225. [Google Scholar] [CrossRef] [PubMed]
- Csercsa, R.; Dombovári, B.; Fabó, D.; Wittner, L.; Erss, L.; Entz, L.; Sólyom, A.; Rásonyi, G.; Szcs, A.; Kelemen, A.; et al. Laminar analysis of slow wave activity in humans. Brain 2010, 133, 2814–2829. [Google Scholar] [CrossRef] [PubMed]
- Chan, A.M.; Baker, J.M.; Eskandar, E.; Schomer, D.; Ulbert, I.; Marinkovic, K.; Cash, S.S.; Halgren, E. First-pass selectivity for semantic categories in human anteroventral temporal lobe. J. Neurosci. 2011, 31, 18119–18129. [Google Scholar] [CrossRef] [PubMed]
- Hanrahan, S.J.; Greger, B.; Parker, R.A.; Ogura, T.; Obara, S.; Egan, T.D.; House, P.A. The effects of propofol on local field potential spectra, action potential firing rate, and their temporal relationship in humans and felines. Front. Hum. Neurosci. 2013, 7, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Staba, R.J.; Wilson, C.L.; Bragin, A.; Fried, I.; Engel, J. Sleep states differentiate single neuron activity recorded from human epileptic hippocampus, entorhinal cortex, and subiculum. J. Neurosci. 2002, 22, 5694–5704. [Google Scholar] [CrossRef]
- Staba, R.J.; Wilson, C.L.; Fried, I.; Engel, J. Single neuron burst firing in the human hippocampus during sleep. Hippocampus 2002, 12, 724–734. [Google Scholar] [CrossRef]
- Lewis, L.D.; Weiner, V.S.; Mukamel, E.A.; Donoghue, J.A.; Eskandar, E.N.; Madsen, J.R.; Anderson, W.S.; Hochberg, L.R.; Cash, S.S.; Brown, E.N.; et al. Rapid fragmentation of neuronal networks at the onset of propofol-induced unconsciousness. Proc. Natl. Acad. Sci. USA 2012, 109. [Google Scholar] [CrossRef]
- Le Van Quyen, M.; Muller, L.E.; Telenczuk, B.; Halgren, E.; Cash, S.; Hatsopoulos, N.G.; Dehghani, N.; Destexhe, A. High-frequency oscillations in human and monkey neocortex during the wake-sleep cycle. Proc. Natl. Acad. Sci. USA 2016, 113, 9363–9368. [Google Scholar] [CrossRef]
- Dickey, C.W.; Sargsyan, A.; Madsen, J.R.; Eskandar, E.N.; Cash, S.S.; Halgren, E. Travelling spindles create necessary conditions for spike-timing-dependent plasticity in humans. Nat. Commun. 2021, 12, 1027. [Google Scholar] [CrossRef]
- Schevon, C.A.; Trevelyan, A.J.; Schroeder, C.E.; Goodman, R.R.; McKhann, G.; Emerson, R.G. Spatial characterization of interictal high frequency oscillations in epileptic neocortex. Brain 2009, 132, 3047–3059. [Google Scholar] [CrossRef]
- Stead, M.; Bower, M.; Brinkmann, B.H.; Lee, K.; Marsh, W.R.; Meyer, F.B.; Litt, B.; Van Gompel, J.; Worrell, G.A. Microseizures and the spatiotemporal scales of human partial epilepsy. Brain 2010, 133, 2789–2797. [Google Scholar] [CrossRef] [PubMed]
- Schevon, C.A.; Goodman, R.R.; McKhann, G.; Emerson, R.G. Propagation of epileptiform activity on a submillimeter scale. J. Clin. Neurophysiol. 2010, 27, 406–411. [Google Scholar] [CrossRef] [PubMed]
- Merricks, E.M.; Smith, E.H.; McKhann, G.M.; Goodman, R.R.; Bateman, L.M.; Emerson, R.G.; Schevon, C.A.; Trevelyan, A.J. Single unit action potentials in humans and the effect of seizure activity. Brain 2015, 138, 2891–2906. [Google Scholar] [CrossRef] [PubMed]
- Bower, M.R.; Stead, M.; Meyer, F.B.; Marsh, W.R.; Worrell, G.A. Spatiotemporal neuronal correlates of seizure generation in focal epilepsy. Epilepsia 2012, 53, 807–816. [Google Scholar] [CrossRef] [PubMed]
- Proix, T.; Aghagolzadeh, M.; Madsen, J.R.; Cosgrove, R.; Eskandar, E.; Hochberg, L.R.; Cash, S.S.; Truccolo, W. Intracortical neural activity distal to seizure-onset-areas predicts human focal seizures. PLoS ONE 2019, 14. [Google Scholar] [CrossRef]
- Smith, E.H.; Merricks, E.M.; Liou, J.-Y.; Casadei, C.; Melloni, L.; Thesen, T.; Friedman, D.J.; Doyle, W.K.; Emerson, R.G.; Goodman, R.R.; et al. Dual mechanisms of ictal high frequency oscillations in human rhythmic onset seizures. Sci. Rep. 2020, 10, 19166. [Google Scholar] [CrossRef]
- Dossi, E.; Blauwblomme, T.; Nabbout, R.; Huberfeld, G.; Rouach, N. Multi-electrode array recordings of human epileptic postoperative cortical tissue. J. Vis. Exp. 2014. [Google Scholar] [CrossRef]
- Hsiao, M.C.; Yu, P.N.; Song, D.; Liu, C.Y.; Heck, C.N.; Millett, D.; Berger, T.W. An in vitro seizure model from human hippocampal slices using multi-electrode arrays. J. Neurosci. Methods 2015, 244, 154–163. [Google Scholar] [CrossRef]
- Cohen, I.; Navarro, V.; Clemenceau, S.; Baulac, M.; Miles, R. On the origin of interictal activity in human temporal lobe epilepsy in vitro. Science 2002, 298, 1418–1421. [Google Scholar] [CrossRef]
- Schwarz, N.; Hedrich, U.B.S.; Schwarz, H.; Harshad, P.A.; Dammeier, N.; Auffenberg, E.; Bedogni, F.; Honegger, J.B.; Lerche, H.; Wuttke, T.V.; et al. Human Cerebrospinal fluid promotes long-term neuronal viability and network function in human neocortical organotypic brain slice cultures. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef]
- Wickham, J.; Corna, A.; Schwarz, N.; Uysal, B.; Layer, N.; Honegger, J.B.; Wuttke, T.V.; Koch, H.; Zeck, G. Human Cerebrospinal Fluid Induces Neuronal Excitability Changes in Resected Human Neocortical and Hippocampal Brain Slices. Front. Neurosci. 2020, 14, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Leinonen, V.; Koivisto, A.M.; Savolainen, S.; Rummukainen, J.; Tamminen, J.N.; Tillgren, T.; Vainikka, S.; Pyykkö, O.T.; Mölsä, J.; Fraunberg, M.; et al. Amyloid and tau proteins in cortical brain biopsy and Alzheimer’s disease. Ann. Neurol. 2010, 68, 446–453. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Bartosch, A.M.; Xiao, H.; Maji, S.; Youth, E.H.H.; Flowers, X.; Leskinen, S.; Tomljanovic, Z.; Iodice, G.; Boyett, D.; et al. An immune response characterizes early Alzheimer’s disease pathology and subjective cognitive impairment in hydrocephalus biopsies. Nat. Commun. 2021, 12, 5659. [Google Scholar] [CrossRef] [PubMed]
- Fagerlund, I.; Dougalis, A.; Shakirzyanova, A.; Gómez-Budia, M.; Konttinen, H.; Ohtonen, S.; Fazaludeen, F.; Koskuvi, M.; Kuusisto, J.; Hernández, D.; et al. Microglia orchestrate neuronal activity in brain organoids. Available online: https://www.biorxiv.org/content/10.1101/2020.12.08.416388v1 (accessed on 28 December 2021).
- Millet, L.J.; Gillette, M.U. Over a century of neuron culture: From the hanging drop to microfluidic devices. Yale J. Biol. Med. 2012, 85, 501–521. [Google Scholar] [PubMed]
- Heikkilä, T.J.; Ylä-Outinen, L.; Tanskanen, J.M.A.; Lappalainen, R.S.; Skottman, H.; Suuronen, R.; Mikkonen, J.E.; Hyttinen, J.A.K.; Narkilahti, S. Human embryonic stem cell-derived neuronal cells form spontaneously active neuronal networks in vitro. Exp. Neurol. 2009, 218, 109–116. [Google Scholar] [CrossRef]
- Lappalainen, R.S.; Salomäki, M.; Ylä-Outinen, L.; Heikkilä, T.J.; Hyttinen, J.A.K.; Pihlajamäki, H.; Suuronen, R.; Skottman, H.; Narkilahti, S. Similarly derived and cultured hESC lines show variation in their developmental potential towards neuronal cells in long-term culture. Regen. Med. 2010, 5, 749–762. [Google Scholar] [CrossRef]
- Illes, S.; Fleischer, W.; Siebler, M.; Hartung, H.-P.; Dihné, M. Development and pharmacological modulation of embryonic stem cell-derived neuronal network activity. Exp. Neurol. 2007, 207, 171–176. [Google Scholar] [CrossRef]
- Wagenaar, D.A.; Pine, J.; Potter, S.M. An extremely rich repertoire of bursting patterns during the development of cortical cultures. BMC Neurosci. 2006, 7, 11. [Google Scholar] [CrossRef]
- Toivonen, S.; Ojala, M.; Hyysalo, A.; Ilmarinen, T.; Rajala, K.; Pekkanen-Mattila, M.; Äänismaa, R.; Lundin, K.; Palgi, J.; Weltner, J.; et al. Comparative Analysis of Targeted Differentiation of Human Induced Pluripotent Stem Cells (hiPSCs) and Human Embryonic Stem Cells Reveals Variability Associated With Incomplete Transgene Silencing in Retrovirally Derived hiPSC Lines. Stem Cells Transl. Med. 2013, 2, 83–93. [Google Scholar] [CrossRef]
- Amin, H.; Maccione, A.; Marinaro, F.; Zordan, S.; Nieus, T.; Berdondini, L. Electrical Responses and Spontaneous Activity of Human iPS-Derived Neuronal Networks Characterized for 3-month Culture with 4096-Electrode Arrays. Front. Neurosci. 2016, 10, 121. [Google Scholar] [CrossRef]
- Hyysalo, A.; Ristola, M.; Mäkinen, M.E.L.; Häyrynen, S.; Nykter, M.; Narkilahti, S. Laminin α5 substrates promote survival, network formation and functional development of human pluripotent stem cell-derived neurons in vitro. Stem Cell Res. 2017, 24, 118–127. [Google Scholar] [CrossRef]
- Shi, Y.; Kirwan, P.; Livesey, F.J. Directed differentiation of human pluripotent stem cells to cerebral cortex neurons and neural networks. Nat. Protoc. 2012, 7, 1836–1846. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.S.; Lee, J.S.; Leem, J.W.; Huh, Y.J.; Kim, J.Y.; Kim, H.S.; Park, I.H.; Daley, G.Q.; Hwang, D.Y.; Kim, D.W. Robust enhancement of neural differentiation from human ES and iPS cells regardless of their innate difference in differentiation propensity. Stem Cell Rev. Rep. 2010, 6, 270–281. [Google Scholar] [CrossRef] [PubMed]
- Hyvärinen, T.; Hyysalo, A.; Kapucu, F.E.; Aarnos, L.; Vinogradov, A.; Eglen, S.J.; Ylä-Outinen, L.; Narkilahti, S. Functional characterization of human pluripotent stem cell-derived cortical networks differentiated on laminin-521 substrate: Comparison to rat cortical cultures. Sci. Rep. 2019, 9, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Hyvärinen, T.; Hagman, S.; Ristola, M.; Sukki, L.; Veijula, K.; Kreutzer, J.; Kallio, P.; Narkilahti, S. Co-stimulation with IL-1β and TNF-α induces an inflammatory reactive astrocyte phenotype with neurosupportive characteristics in a human pluripotent stem cell model system. Sci. Rep. 2019, 9, 16944. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Pak, C.H.; Han, Y.; Ahlenius, H.; Zhang, Z.; Chanda, S.; Marro, S.; Patzke, C.; Acuna, C.; Covy, J.; et al. Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron 2013, 78, 785–798. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.P.; Yang, N.; Vierbuchen, T.; Ostermeier, A.; Fuentes, D.R.; Yang, T.Q.; Citri, A.; Sebastiano, V.; Marro, S.; Südhof, T.C.; et al. Induction of human neuronal cells by defined transcription factors. Nature 2011, 476, 220–223. [Google Scholar] [CrossRef]
- Frega, M.; Van Gestel, S.H.C.; Linda, K.; Van Der Raadt, J.; Keller, J.; Van Rhijn, J.R.; Schubert, D.; Albers, C.A.; Kasri, N.N. Rapid neuronal differentiation of induced pluripotent stem cells for measuring network activity on micro-electrode arrays. J. Vis. Exp. 2017, 2017, 1–10. [Google Scholar] [CrossRef]
- Yang, N.; Chanda, S.; Marro, S.; Ng, Y.-H.; Janas, J.A.; Haag, D.; Ang, C.E.; Tang, Y.; Flores, Q.; Mall, M.; et al. Generation of pure GABAergic neurons by transcription factor programming. Nat. Methods 2017, 14, 621–628. [Google Scholar] [CrossRef]
- Ichise, E.; Chiyonobu, T.; Ishikawa, M.; Tanaka, Y.; Shibata, M.; Tozawa, T.; Taura, Y.; Yamashita, S.; Yoshida, M.; Morimoto, M.; et al. Impaired neuronal activity and differential gene expression in STXBP1 encephalopathy patient iPSC-derived GABAergic neurons. Hum. Mol. Genet. 2021, 30, 1337–1348. [Google Scholar] [CrossRef] [PubMed]
- Mossink, B.; van Rhijn, J.-R.; Wang, S.; Linda, K.; Vitale, M.R.; Zöller, J.E.M.; van Hugte, E.J.H.; Bak, J.; Verboven, A.H.A.; Selten, M.; et al. Cadherin-13 is a critical regulator of GABAergic modulation in human stem-cell-derived neuronal networks. Mol. Psychiatry 2021, 2021, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Dionisi, C.; Rai, M.; Chazalon, M.; Schiffmann, S.N.; Pandolfo, M. Primary proprioceptive neurons from human induced pluripotent stem cells: A cell model for afferent ataxias. Sci. Rep. 2020, 10, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Hasselmann, J.; Blurton-Jones, M. Human iPSC-derived microglia: A growing toolset to study the brain’s innate immune cells. Glia 2020, 68, 721–739. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Danés, A.; Richaud-Patin, Y.; Carballo-Carbajal, I.; Jiménez-Delgado, S.; Caig, C.; Mora, S.; Di Guglielmo, C.; Ezquerra, M.; Patel, B.; Giralt, A.; et al. Disease-specific phenotypes in dopamine neurons from human iPS-based models of genetic and sporadic Parkinson’s disease. EMBO Mol. Med. 2012, 4, 380–395. [Google Scholar] [CrossRef] [PubMed]
- Konttinen, H.; Cabral-da-Silva, M.e.C.; Ohtonen, S.; Wojciechowski, S.; Shakirzyanova, A.; Caligola, S.; Giugno, R.; Ishchenko, Y.; Hernández, D.; Fazaludeen, M.F.; et al. PSEN1ΔE9, APPswe, and APOE4 Confer Disparate Phenotypes in Human iPSC-Derived Microglia. Stem Cell Rep. 2019, 13, 669–683. [Google Scholar] [CrossRef]
- Suga, M.; Kondo, T.; Inoue, H. Modeling Neurological Disorders with Human Pluripotent Stem Cell-Derived Astrocytes. Int. J. Mol. Sci. 2019, 20, 3862. [Google Scholar] [CrossRef]
- Cotterill, E.; Charlesworth, P.; Thomas, C.W.; Paulsen, O.; Eglen, S.J. A comparison of computational methods for detecting bursts in neuronal spike trains and their application to human stem cell-derived neuronal networks. J. Neurophysiol. 2016, 116, 306–321. [Google Scholar] [CrossRef]
- Pelkonen, A.; Mzezewa, R.; Sukki, L.; Ryynänen, T.; Kreutzer, J.; Hyvärinen, T.; Vinogradov, A.; Aarnos, L.; Lekkala, J.; Kallio, P.; et al. A modular brain-on-a-chip for modelling epileptic seizures with functionally connected human neuronal networks. Biosens. Bioelectron. 2020, 168, 112553. [Google Scholar] [CrossRef]
- Ronchi, S.; Buccino, A.P.; Prack, G.; Kumar, S.S.; Schröter, M.; Fiscella, M.; Hierlemann, A. Electrophysiological Phenotype Characterization of Human iPSC-Derived Neuronal Cell Lines by Means of High-Density Microelectrode Arrays. Adv. Biol. 2021, 5, e2000223. [Google Scholar] [CrossRef] [PubMed]
- Tang, M.; Li, J.; He, L.; Guo, R.; Yan, X.; Li, D.; Zhang, Y.; Liao, M.; Shao, B.; Hu, Y.; et al. Transcriptomic profiling of neural stem cell differentiation on graphene substrates. Colloids Surf. B Biointerfaces 2019, 182. [Google Scholar] [CrossRef] [PubMed]
- Centeno, E.G.Z.; Cimarosti, H.; Bithell, A. 2D versus 3D human induced pluripotent stem cell-derived cultures for neurodegenerative disease modelling. Mol. Neurodegener. 2018, 13, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Asheghali, D.; Lee, S.J.; Furchner, A.; Gruzd, A.; Larson, S.; Tokarev, A.; Stake, S.; Zhou, X.; Hinrichs, K.; Zhang, L.G.; et al. Enhanced neuronal differentiation of neural stem cells with mechanically enhanced touch-spun nanofibrous scaffolds. Nanomed. Nanotechnol. Biol. Med. 2020, 24, 102152. [Google Scholar] [CrossRef]
- Ben-Ari, Y. Excitatory actions of GABA during development: The nature of the nurture. Nat. Rev. Neurosci. 2002, 3, 728–739. [Google Scholar] [CrossRef]
- Mäkinen, M.E.-L.; Ylä-Outinen, L.; Narkilahti, S. GABA and Gap Junctions in the Development of Synchronized Activity in Human Pluripotent Stem Cell-Derived Neural Networks. Front. Cell. Neurosci. 2018, 12, 56. [Google Scholar] [CrossRef]
- Nimtz, L.; Hartmann, J.; Tigges, J.; Masjosthusmann, S.; Schmuck, M.; Keßel, E.; Theiss, S.; Köhrer, K.; Petzsch, P.; Adjaye, J.; et al. Characterization and application of electrically active neuronal networks established from human induced pluripotent stem cell-derived neural progenitor cells for neurotoxicity evaluation. Stem Cell Res. 2020, 45. [Google Scholar] [CrossRef]
- Hofrichter, M.; Nimtz, L.; Tigges, J.; Kabiri, Y.; Schröter, F.; Royer-Pokora, B.; Hildebrandt, B.; Schmuck, M.; Epanchintsev, A.; Theiss, S.; et al. Comparative performance analysis of human iPSC-derived and primary neural progenitor cells (NPC) grown as neurospheres in vitro. Stem Cell Res. 2017, 25, 72–82. [Google Scholar] [CrossRef]
- Whitesides, G.M. The origins and the future of microfluidics. Nature 2006, 442, 368–373. [Google Scholar] [CrossRef] [PubMed]
- Mofazzal Jahromi, M.A.; Abdoli, A.; Rahmanian, M.; Bardania, H.; Bayandori, M.; Moosavi Basri, S.M.; Kalbasi, A.; Aref, A.R.; Karimi, M.; Hamblin, M.R. Microfluidic Brain-on-a-Chip: Perspectives for Mimicking Neural System Disorders. Mol. Neurobiol. 2019, 56, 8489–8512. [Google Scholar] [CrossRef] [PubMed]
- Berdondini, L.; Chiappalone, M.; Van Der Wal, P.D.; Imfeld, K.; De Rooij, N.F.; Koudelka-Hep, M.; Tedesco, M.; Martinoia, S.; Van Pelt, J.; Le Masson, G.; et al. A microelectrode array (MEA) integrated with clustering structures for investigating in vitro neurodynamics in confined interconnected sub-populations of neurons. Sens. Actuators B Chem. 2006, 114, 530–541. [Google Scholar] [CrossRef]
- Morin, F.; Nishimura, N.; Griscom, L.; LePioufle, B.; Fujita, H.; Takamura, Y.; Tamiya, E. Constraining the connectivity of neuronal networks cultured on microelectrode arrays with microfluidic techniques: A step towards neuron-based functional chips. Biosens. Bioelectron. 2006, 21, 1093–1100. [Google Scholar] [CrossRef]
- Bisio, M.; Bosca, A.; Pasquale, V.; Berdondini, L.; Chiappalone, M. Emergence of bursting activity in connected neuronal sub-populations. PLoS ONE 2014, 9, e107400. [Google Scholar] [CrossRef]
- Le Feber, J.; Postma, W.; de Weerd, E.; Weusthof, M.; Rutten, W.L.C. Barbed channels enhance unidirectional connectivity between neuronal networks cultured on multi electrode arrays. Front. Neurosci. 2015, 9, 1–10. [Google Scholar] [CrossRef]
- Malishev, E.; Pimashkin, A.; Gladkov, A.; Pigareva, Y.; Bukatin, A.; Kazantsev, V.; Mukhina, I.; Dubina, M. Microfluidic device for unidirectional axon growth. J. Phys. Conf. Ser. 2015, 643, 012025. [Google Scholar] [CrossRef]
- Gladkov, A.; Pigareva, Y.; Kutyina, D.; Kolpakov, V.; Bukatin, A.; Mukhina, I.; Kazantsev, V.; Pimashkin, A. Design of Cultured Neuron Networks in vitro with Predefined Connectivity Using Asymmetric Microfluidic Channels. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef]
- DeMarse, T.B.; Pan, L.; Alagapan, S.; Brewer, G.J.; Wheeler, B.C. Feed-Forward Propagation of Temporal and Rate Information between Cortical Populations during Coherent Activation in Engineered In Vitro Networks. Front. Neural Circuits 2016, 10, 32. [Google Scholar] [CrossRef]
- Hong, N.; Joo, S.; Nam, Y. Characterization of Axonal Spikes in Cultured Neuronal Networks Using Microelectrode Arrays and Microchannel Devices. IEEE Trans. Biomed. Eng. 2017, 64, 492–498. [Google Scholar] [CrossRef]
- Toivanen, M.; Pelkonen, A.; Mäkinen, M.; Ylä-Outinen, L.; Sukki, L.; Kallio, P.; Narkilahti, S.; Ristola, M. Optimised PDMS tunnel devices on MEAs increase the probability of detecting electrical activity from human stem cell-derived neuronal networks. Front. Neurosci. 2017, 11, 606. [Google Scholar] [CrossRef]
- Oliva, A.A.; James, C.D.; Kingman, C.E.; Craighead, H.G.; Banker, G.A. Patterning Axonal Guidance Molecules Using a Novel Strategy for Microcontact Printing. Neurochem. Res. 2003, 28, 1639–1648. [Google Scholar] [CrossRef]
- Jang, M.J.; Nam, Y. Aqueous micro-contact printing of cell-adhesive biomolecules for patterning neuronal cell cultures. Biochip J. 2012, 6, 107–113. [Google Scholar] [CrossRef]
- Dauth, S.; Maoz, B.M.; Sheehy, S.P.; Hemphill, M.A.; Murty, T.; Macedonia, M.K.; Greer, A.M.; Budnik, B.; Parker, K.K. Neurons derived from different brain regions are inherently different in vitro: A novel multiregional brain-on-a-chip. J. Neurophysiol. 2017, 117, 1320–1341. [Google Scholar] [CrossRef] [PubMed]
- Honegger, T.; Scott, M.A.; Yanik, M.F.; Voldman, J. Electrokinetic confinement of axonal growth for dynamically configurable neural networks. Lab Chip 2013, 13, 589–598. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Jiang, L.; Li, S.; Deng, J.; Li, Y.; Yao, J.; Li, B.; Zheng, J. Using microfluidic chip to form brain-derived neurotrophic factor concentration gradient for studying neuron axon guidance. Biomicrofluidics 2014, 8, 1–8. [Google Scholar] [CrossRef]
- Dertinger, S.K.W.; Jiang, X.; Li, Z.; Murthy, V.N.; Whitesides, G.M. Gradients of substrate-bound laminin orient axonal specification of neurons. Proc. Natl. Acad. Sci. USA 2002, 99, 12542–12547. [Google Scholar] [CrossRef]
- Dowell-Mesfin, N.M.; Abdul-Karim, M.A.; Turner, A.M.P.; Schanz, S.; Craighead, H.G.; Roysam, B.; Turner, J.N.; Shain, W. Topographically modified surfaces affect orientation and growth of hippocampal neurons. J. Neural Eng. 2004, 1, 78–90. [Google Scholar] [CrossRef]
- Ristola, M.; Fedele, C.; Hagman, S.; Sukki, L.; Kapucu, F.E.; Mzezewa, R.; Hyvärinen, T.; Kallio, P.; Priimagi, A.; Narkilahti, S. Directional Growth of Human Neuronal Axons in a Microfluidic Device with Nanotopography on Azobenzene-Based Material. Adv. Mater. Interfaces 2021, 8, 16–19. [Google Scholar] [CrossRef]
- Roversi, K.; Ebrahimi Orimi, H.; Falchetti, M.; da Rocha, E.; Talbot, S.; Boutopoulos, C. Bioprinting of Adult Dorsal Root Ganglion (DRG) Neurons Using Laser-Induced Side Transfer (LIST). Micromachines 2021, 12. [Google Scholar] [CrossRef]
- Yokoi, R.; Okabe, M.; Matsuda, N.; Odawara, A.; Karashima, A.; Suzuki, I. Impact of sleep–wake-associated neuromodulators and repetitive low-frequency stimulation on human iPSC-derived neurons. Front. Neurosci. 2019, 13, 1–15. [Google Scholar] [CrossRef]
- Bojarskaite, L.; Bjørnstad, D.M.; Pettersen, K.H.; Cunen, C.; Hermansen, G.H.; Åbjørsbråten, K.S.; Chambers, A.R.; Sprengel, R.; Vervaeke, K.; Tang, W.; et al. Astrocytic Ca2+ signaling is reduced during sleep and is involved in the regulation of slow wave sleep. Nat. Commun. 2020, 11, 3240. [Google Scholar] [CrossRef]
- Kayama, T.; Suzuki, I.; Odawara, A.; Sasaki, T.; Ikegaya, Y. Biochemical and Biophysical Research Communications Temporally coordinated spiking activity of human induced pluripotent stem cell-derived neurons co-cultured with astrocytes. Biochem. Biophys. Res. Commun. 2018, 495, 1028–1033. [Google Scholar] [CrossRef]
- Ylä-Outinen, L.; Heikkilä, J.; Skottman, H.; Suuronen, R.; Äänismaa, R.; Narkilahti, S. Human Cell-Based Micro Electrode Array Platform for Studying Neurotoxicity. Front. Neuroeng. 2010, 3, 111. [Google Scholar] [CrossRef]
- Kasteel, E.E.J.; Westerink, R.H.S. Comparison of the acute inhibitory effects of Tetrodotoxin ( TTX ) in rat and human neuronal networks for risk assessment purposes. Toxicol. Lett. 2017, 270, 12–16. [Google Scholar] [CrossRef]
- Pires Monteiro, S.; Voogd, E.; Muzzi, L.; De Vecchis, G.; Mossink, B.; Levers, M.; Hassink, G.; Van Putten, M.; Le Feber, J.; Hofmeijer, J.; et al. Neuroprotective effect of hypoxic preconditioning and neuronal activation in a in vitro human model of the ischemic penumbra. J. Neural Eng. 2021, 18, 36016. [Google Scholar] [CrossRef] [PubMed]
- Frega, M.; Linda, K.; Keller, J.M.; Gümüş-Akay, G.; Mossink, B.; van Rhijn, J.-R.; Negwer, M.; Klein Gunnewiek, T.; Foreman, K.; Kompier, N.; et al. Neuronal network dysfunction in a model for Kleefstra syndrome mediated by enhanced NMDAR signaling. Nat. Commun. 2019, 10, 4928. [Google Scholar] [CrossRef] [PubMed]
- Klein Gunnewiek, T.M.; Van Hugte, E.J.H.; Frega, M.; Guardia, G.S.; Foreman, K.; Panneman, D.; Mossink, B.; Linda, K.; Keller, J.M.; Schubert, D.; et al. m.3243A > G-Induced Mitochondrial Dysfunction Impairs Human Neuronal Development and Reduces Neuronal Network Activity and Synchronicity. Cell Rep. 2020, 31, 107538. [Google Scholar] [CrossRef] [PubMed]
- Linda, K.; Lewerissa, E.I.; Verboven, A.H.A.; Gabriele, M.; Frega, M.; Klein Gunnewiek, T.M.; Devilee, L.; Ulferts, E.; Hommersom, M.; Oudakker, A.; et al. Imbalanced autophagy causes synaptic deficits in a human model for neurodevelopmental disorders. Autophagy 2021, 12, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Simkin, D.; Marshall, K.A.; Vanoye, C.G.; Desai, R.R.; Bustos, B.I.; Piyevsky, B.N.; Ortega, J.A.; Forrest, M.; Robertson, G.L.; Penzes, P.; et al. Dyshomeostatic modulation of ca2+-activated k+ channels in a human neuronal model of kcnq2 encephalopathy. Elife 2021, 10, 1–32. [Google Scholar] [CrossRef]
- Que, Z.; Olivero-Acosta, M.I.; Zhang, J.; Eaton, M.; Tukker, A.M.; Chen, X.; Wu, J.; Xie, J.; Xiao, T.; Wettschurack, K.; et al. Hyperexcitability and pharmacological responsiveness of cortical neurons derived from human iPSCs carrying epilepsy-associated sodium channel Nav1.2-L1342P genetic variant. J. Neurosci. 2021, 49, 10194–10208. [Google Scholar] [CrossRef]
- Wainger, B.J.; Kiskinis, E.; Mellin, C.; Wiskow, O.; Han, S.S.W.; Sandoe, J.; Perez, N.P.; Williams, L.A.; Lee, S.; Boulting, G.; et al. Intrinsic membrane hyperexcitability of amyotrophic lateral sclerosis patient-derived motor neurons. Cell Rep. 2014, 7, 1–11. [Google Scholar] [CrossRef]
- Ghatak, S.; Dolatabadi, N.; Gao, R.; Wu, Y.; Scott, H.; Trudler, D.; Sultan, A.; Ambasudhan, R.; Nakamura, T.; Masliah, E.; et al. NitroSynapsin ameliorates hypersynchronous neural network activity in Alzheimer hiPSC models. Mol. Psychiatry 2020. [Google Scholar] [CrossRef] [PubMed]
- Mason, J.O.; Price, D.J. Building brains in a dish: Prospects for growing cerebral organoids from stem cells. Neuroscience 2016, 334, 105–118. [Google Scholar] [CrossRef]
- Jackson, E.L.; Lu, H. Three-dimensional models for studying development and disease: Moving on from organisms to organs-on-a-chip and organoids. Integr. Biol. 2016, 8, 672–683. [Google Scholar] [CrossRef]
- Ylä-Outinen, L.; Joki, T.; Varjola, M.; Skottman, H.; Narkilahti, S. Three-dimensional growth matrix for human embryonic stem cell-derived neuronal cells. J. Tissue Eng. Regen. Med. 2014, 8, 186–194. [Google Scholar] [CrossRef]
- Innala, M.; Riebe, I.; Kuzmenko, V.; Sundberg, J.; Gatenholm, P.; Hanse, E.; Johannesson, S. 3D culturing and differentiation of SH-SY5Y neuroblastoma cells on bacterial nanocellulose scaffolds. Artif. Cells Nanomed. Biotechnol. 2014, 42, 302–308. [Google Scholar] [CrossRef]
- Smith, I.; Silveirinha, V.; Stein, J.L.; de la Torre-Ubieta, L.; Farrimond, J.A.; Williamson, E.M.; Whalley, B.J. Human neural stem cell-derived cultures in three-dimensional substrates form spontaneously functional neuronal networks. J. Tissue Eng. Regen. Med. 2017, 11, 1022–1033. [Google Scholar] [CrossRef]
- Yin, X.; Mead, B.E.; Safaee, H.; Langer, R.; Karp, J.M.; Levy, O. Engineering Stem Cell Organoids. Cell Stem Cell 2016, 18, 25–38. [Google Scholar] [CrossRef]
- Kaushik, G.; Ponnusamy, M.P.; Batra, S.K. Concise Review: Current Status of Three-Dimensional Organoids as Preclinical Models. Stem Cells 2018, 36, 1329–1340. [Google Scholar] [CrossRef]
- Qian, X.; Song, H.; Ming, G.-L. Brain organoids: Advances, applications and challenges. Development 2019, 146. [Google Scholar] [CrossRef] [PubMed]
- Mich, J.K.; Close, J.L.; Levi, B.P. Putting Two Heads Together to Build a Better Brain. Cell Stem Cell 2017, 21, 289–290. [Google Scholar] [CrossRef]
- Watanabe, K.; Kamiya, D.; Nishiyama, A.; Katayama, T.; Nozaki, S.; Kawasaki, H.; Watanabe, Y.; Mizuseki, K.; Sasai, Y. Directed differentiation of telencephalic precursors from embryonic stem cells. Nat. Neurosci. 2005, 8, 288–296. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.-T.; Bendriem, R.M.; Wu, W.W.; Shen, R.-F. 3D brain Organoids derived from pluripotent stem cells: Promising experimental models for brain development and neurodegenerative disorders. J. Biomed. Sci. 2017, 24, 59. [Google Scholar] [CrossRef] [PubMed]
- Kadoshima, T.; Sakaguchi, H.; Nakano, T.; Soen, M.; Ando, S.; Eiraku, M.; Sasai, Y. Self-organization of axial polarity, inside-out layer pattern, and species-specific progenitor dynamics in human ES cell-derived neocortex. Proc. Natl. Acad. Sci. USA 2013, 110, 20284–20289. [Google Scholar] [CrossRef] [PubMed]
- Papaspyropoulos, A.; Tsolaki, M.; Foroglou, N.; Pantazaki, A.A. Modeling and Targeting Alzheimer’s Disease With Organoids. Front. Pharmacol. 2020, 11, 396. [Google Scholar] [CrossRef]
- Gabriel, E.; Albanna, W.; Pasquini, G.; Ramani, A.; Josipovic, N.; Mariappan, A.; Schinzel, F.; Karch, C.M.; Bao, G.; Gottardo, M.; et al. Human brain organoids assemble functionally integrated bilateral optic vesicles. Cell Stem Cell 2021, 28, 1–18. [Google Scholar] [CrossRef]
- Bagley, J.A.; Reumann, D.; Bian, S.; Lévi-Strauss, J.; Knoblich, J.A. Fused cerebral organoids model interactions between brain regions. Nat. Methods 2017, 14, 743–751. [Google Scholar] [CrossRef]
- Birey, F.; Andersen, J.; Makinson, C.D.; Islam, S.; Wei, W.; Huber, N.; Fan, H.C.; Metzler, K.R.C.; Panagiotakos, G.; Thom, N.; et al. Assembly of functionally integrated human forebrain spheroids. Nature 2017, 545, 54–59. [Google Scholar] [CrossRef]
- Xiang, Y.; Tanaka, Y.; Patterson, B.; Kang, Y.-J.; Govindaiah, G.; Roselaar, N.; Cakir, B.; Kim, K.-Y.; Lombroso, A.P.; Hwang, S.-M.; et al. Fusion of Regionally Specified hPSC-Derived Organoids Models Human Brain Development and Interneuron Migration. Cell Stem Cell 2017, 21, 383–398.e7. [Google Scholar] [CrossRef]
- Marton, R.M.; Miura, Y.; Sloan, S.A.; Li, Q.; Revah, O.; Levy, R.J.; Huguenard, J.R.; Pașca, S.P. Differentiation and maturation of oligodendrocytes in human three-dimensional neural cultures. Nat. Neurosci. 2019, 22, 484–491. [Google Scholar] [CrossRef]
- Madhavan, M.; Nevin, Z.S.; Shick, H.E.; Garrison, E.; Clarkson-Paredes, C.; Karl, M.; Clayton, B.L.L.; Factor, D.C.; Allan, K.C.; Barbar, L.; et al. Induction of myelinating oligodendrocytes in human cortical spheroids. Nat. Methods 2018, 15, 700–706. [Google Scholar] [CrossRef] [PubMed]
- Izsak, J.; Seth, H.; Andersson, M.; Vizlin-Hodzic, D.; Theiss, S.; Hanse, E.; Ågren, H.; Funa, K.; Illes, S. Robust generation of person-specific, synchronously active neuronal networks using purely isogenic human iPSC-3D neural aggregate cultures. Front. Neurosci. 2019, 13, 351. [Google Scholar] [CrossRef]
- Monzel, A.S.; Smits, L.M.; Hemmer, K.; Hachi, S.; Moreno, E.L.; van Wuellen, T.; Jarazo, J.; Walter, J.; Brüggemann, I.; Boussaad, I.; et al. Derivation of Human Midbrain-Specific Organoids from Neuroepithelial Stem Cells. Stem Cell Rep. 2017, 8, 1144–1154. [Google Scholar] [CrossRef] [PubMed]
- Kathuria, A.; Lopez-Lengowski, K.; Jagtap, S.S.; McPhie, D.; Perlis, R.H.; Cohen, B.M.; Karmacharya, R. Transcriptomic Landscape and Functional Characterization of Induced Pluripotent Stem Cell-Derived Cerebral Organoids in Schizophrenia. JAMA Psychiatry 2020, 1, 745–754. [Google Scholar] [CrossRef] [PubMed]
- Muzzi, L.; Falappa, M.; Maccione, A.; Di Lisa, D.; Frega, M.; Martinoia, S. Human derived cortical excitatory neurospheroids showed spontaneous activity on micro electrodes array. In Proceedings of the 2021 10th International IEEE/EMBS Conference on Neural Engineering (NER), 4–6 May 2021; pp. 2–6. [Google Scholar]
- Durens, M.; Nestor, J.; Williams, M.; Herold, K.; Niescier, R.F.; Lunden, J.W.; Phillips, A.W.; Lin, Y.-C.; Dykxhoorn, D.M.; Nestor, M.W. High-throughput screening of human induced pluripotent stem cell-derived brain organoids. J. Neurosci. Methods 2020, 335, 108627. [Google Scholar] [CrossRef] [PubMed]
- Giandomenico, S.L.; Mierau, S.B.; Gibbons, G.M.; Wenger, L.M.D.; Masullo, L.; Sit, T.; Sutcliffe, M.; Boulanger, J.; Tripodi, M.; Derivery, E.; et al. Cerebral organoids at the air–liquid interface generate diverse nerve tracts with functional output. Nat. Neurosci. 2019, 22, 669–679. [Google Scholar] [CrossRef]
- Qian, X.; Su, Y.; Adam, C.D.; Deutschmann, A.U.; Pather, S.R.; Goldberg, E.M.; Su, K.; Li, S.; Lu, L.; Jacob, F.; et al. Sliced Human Cortical Organoids for Modeling Distinct Cortical Layer Formation. Cell Stem Cell 2020, 26, 766–781.e9. [Google Scholar] [CrossRef]
- Szebényi, K.; Wenger, L.M.D.; Sun, Y.; Dunn, A.W.E.; Limegrover, C.A.; Gibbons, G.M.; Conci, E.; Paulsen, O.; Mierau, S.B.; Balmus, G.; et al. Human ALS/FTD brain organoid slice cultures display distinct early astrocyte and targetable neuronal pathology. Nat. Neurosci. 2021, 24, 1542–1554. [Google Scholar] [CrossRef]
- Lam, D.; Fischer, N.O.; Enright, H.A. Probing function in 3D neuronal cultures: A survey of 3D multielectrode array advances. Curr. Opin. Pharmacol. 2021, 60, 255–260. [Google Scholar] [CrossRef]
- Quadrato, G.; Nguyen, T.; Macosko, E.Z.; Sherwood, J.L.; Min Yang, S.; Berger, D.R.; Maria, N.; Scholvin, J.; Goldman, M.; Kinney, J.P.; et al. Cell diversity and network dynamics in photosensitive human brain organoids. Nature 2017, 545, 48–53. [Google Scholar] [CrossRef]
- Yao, Y.; Gulari, M.N.; Wise, K.D. Microassembly Techniques for a Three-Dimensional Neural Stimulating Microelectrode Array. In Proceedings of the 2006 International Conference of the IEEE Engineering in Medicine and Biology Society, New York City, NY, USA, 30 August–3 September 2006; pp. 4643–4646. [Google Scholar]
- Shin, H.; Jeong, S.; Lee, J.-H.; Sun, W.; Choi, N.; Cho, I. 3D high-density microelectrode array with optical stimulation and drug delivery for investigating neural circuit dynamics. Nat. Commun. 2021, 12, 1–18. [Google Scholar] [CrossRef]
- Paşca, A.M.; Sloan, S.A.; Clarke, L.E.; Tian, Y.; Makinson, C.D.; Huber, N.; Kim, C.H.; Park, J.-Y.; O’Rourke, N.A.; Nguyen, K.D.; et al. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat. Methods 2015, 12, 671–678. [Google Scholar] [CrossRef]
- Wilson, E.; Knudson, W.; Newell-Litwa, K. Hyaluronan regulates synapse formation and function in developing neural networks. Sci. Rep. 2020, 10, 16459. [Google Scholar] [CrossRef]
- Qian, X.; Nguyen, H.N.; Song, M.M.; Hadiono, C.; Ogden, S.C.; Hammack, C.; Yao, B.; Hamersky, G.R.; Jacob, F.; Zhong, C.; et al. Brain-Region-Specific Organoids Using Mini-bioreactors for Modeling ZIKV Exposure. Cell 2016, 165, 1238–1254. [Google Scholar] [CrossRef]
- Kathuria, A.; Lopez-Lengowski, K.; Vater, M.; McPhie, D.; Cohen, B.M.; Karmacharya, R. Transcriptome analysis and functional characterization of cerebral organoids in bipolar disorder. Genome Med. 2020, 12, 34. [Google Scholar] [CrossRef] [PubMed]
- Fair, S.R.; Julian, D.; Hartlaub, A.M.; Pusuluri, S.T.; Malik, G.; Summerfied, T.L.; Zhao, G.; Hester, A.B.; Iv, W.E.A.; Hollingsworth, E.W.; et al. Electrophysiological Maturation of Cerebral Organoids Correlates with Dynamic Morphological and Cellular Development. Stem Cell Rep. 2020, 15, 855–868. [Google Scholar] [CrossRef]
- Zhong, S.; Zhang, S.; Fan, X.; Wu, Q.; Yan, L.; Dong, J.; Zhang, H.; Li, L.; Sun, L.; Pan, N.; et al. A single-cell RNA-seq survey of the developmental landscape of the human prefrontal cortex. Nature 2018, 555, 524–528. [Google Scholar] [CrossRef]
- Voytek, B.; Kayser, A.S.; Badre, D.; Fegen, D.; Chang, E.F.; Crone, N.E.; Parvizi, J.; Knight, R.T.; D’Esposito, M. Oscillatory dynamics coordinating human frontal networks in support of goal maintenance. Nat. Neurosci. 2015, 18, 1318–1324. [Google Scholar] [CrossRef]
- Daviaud, N.; Friedel, R.H.; Zou, H. Vascularization and engraftment of transplanted human cerebral organoids in mouse cortex. eNeuro 2018, 5, 6. [Google Scholar] [CrossRef] [PubMed]
- Izsak, J.; Seth, H.; Theiss, S.; Hanse, E.; Illes, S. Human Cerebrospinal Fluid Promotes Neuronal Circuit Maturation of Human Induced Pluripotent Stem Cell-Derived 3D Neural Aggregates. Stem Cell Rep. 2020, 14, 1044–1059. [Google Scholar] [CrossRef] [PubMed]
- Popova, G.; Soliman, S.S.; Kim, C.N.; Keefe, M.G.; Hennick, K.M.; Jain, S.; Li, T.; Tejera, D.; Shin, D.; Chhun, B.B.; et al. Human microglia states are conserved across experimental models and regulate neural stem cell responses in chimeric organoids. Cell Stem Cell 2021, 28, 2153–2166. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, M.; Buth, J.E.; Vishlaghi, N.; de la Torre-Ubieta, L.; Taxidis, J.; Khakh, B.S.; Coppola, G.; Pearson, C.A.; Yamauchi, K.; Gong, D.; et al. Self-Organized Cerebral Organoids with Human-Specific Features Predict Effective Drugs to Combat Zika Virus Infection. Cell Rep. 2017, 21, 517–532. [Google Scholar] [CrossRef]
- Rajamani, U.; Gross, A.R.; Hjelm, B.E.; Sequeira, A.; Vawter, M.P.; Tang, J.; Gangalapudi, V.; Wang, Y.; Andres, A.M.; Gottlieb, R.A.; et al. Super-Obese Patient-Derived iPSC Hypothalamic Neurons Exhibit Obesogenic Signatures and Hormone Responses. Cell Stem Cell 2018, 22, 698–712.e9. [Google Scholar] [CrossRef]
- Yao, H.; Wu, W.; Cerf, I.; Zhao, H.W.; Wang, J.; Negraes, P.D.; Muotri, A.R.; Haddad, G.G. Methadone interrupts neural growth and function in human cortical organoids. Stem Cell Res. 2020, 49, 102065. [Google Scholar] [CrossRef]
- Izsak, J.; Seth, H.; Iljin, M.; Theiss, S.; Ågren, H.; Funa, K.; Aigner, L.; Hanse, E.; Illes, S. Differential acute impact of therapeutically effective and overdose concentrations of lithium on human neuronal single cell and network function. Transl. Psychiatry 2021, 11, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Muffat, J.; Omer, A.; Bosch, I.; Lancaster, M.A.; Sur, M.; Gehrke, L.; Knoblich, J.A.; Jaenisch, R. Induction of Expansion and Folding in Human Cerebral Organoids. Cell Stem Cell 2017, 20, 385–396.e3. [Google Scholar] [CrossRef]
- Karzbrun, E.; Kshirsagar, A.; Cohen, S.R.; Hanna, J.H.; Reiner, O. Human brain organoids on a chip reveal the physics of folding. Nat. Phys. 2018, 14, 515–522. [Google Scholar] [CrossRef] [PubMed]
- Frega, M.; Tedesco, M.; Massobrio, P.; Pesce, M.; Martinoia, S. Network dynamics of 3D engineered neuronal cultures: A new experimental model for in-vitro electrophysiology. Sci. Rep. 2014, 4, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Honkamäki, L.; Joki, T.; Grigoryev, N.A.; Levon, K.; Ylä-Outinen, L.; Narkilahti, S. Novel method to produce a layered 3D scaffold for human pluripotent stem cell-derived neuronal cells. J. Neurosci. Methods 2021, 350, 109043. [Google Scholar] [CrossRef] [PubMed]
- Pham, M.T.; Pollock, K.M.; Rose, M.D.; Cary, W.A.; Stewart, H.R.; Zhou, P.; Nolta, J.A.; Waldau, B. Generation of human vascularized brain organoids. Neuroreport 2018, 29, 588–593. [Google Scholar] [CrossRef] [PubMed]
- Wörsdörfer, P.; Dalda, N.; Kern, A.; Krüger, S.; Wagner, N.; Kwok, C.K.; Henke, E.; Ergün, S. Generation of complex human organoid models including vascular networks by incorporation of mesodermal progenitor cells. Sci. Rep. 2019, 9, 1–13. [Google Scholar] [CrossRef]
- Schwartz, M.P.; Hou, Z.; Propson, N.E.; Zhang, J.; Engstrom, C.J.; Costa, V.S.; Jiang, P.; Nguyen, B.K.; Bolin, J.M.; Daly, W.; et al. Human pluripotent stem cell-derived neural constructs for predicting neural toxicity. Proc. Natl. Acad. Sci. USA 2015, 112, 12516–12521. [Google Scholar] [CrossRef] [PubMed]
- Barateiro, A.; Brites, D.; Fernandes, A. Oligodendrocyte Development and Myelination in Neurodevelopment: Molecular Mechanisms in Health and Disease. Curr. Pharm. Des. 2016, 22, 656–679. [Google Scholar] [CrossRef]
- Cakir, B.; Xiang, Y.; Tanaka, Y.; Kural, M.H.; Parent, M.; Kang, Y.J.; Chapeton, K.; Patterson, B.; Yuan, Y.; He, C.S.; et al. Engineering of human brain organoids with a functional vascular-like system. Nat. Methods 2019, 16, 1169–1175. [Google Scholar] [CrossRef]
- Ormel, P.R.; Vieira de Sá, R.; van Bodegraven, E.J.; Karst, H.; Harschnitz, O.; Sneeboer, M.A.M.; Johansen, L.E.; van Dijk, R.E.; Scheefhals, N.; Berdenis van Berlekom, A.; et al. Microglia innately develop within cerebral organoids. Nat. Commun. 2018, 9, 4167. [Google Scholar] [CrossRef]
- Verkhratsky, A.; Li, Q.; Melino, S.; Melino, G.; Shi, Y. Can COVID-19 pandemic boost the epidemic of neurodegenerative diseases? Biol. Direct 2020, 15, 28. [Google Scholar] [CrossRef] [PubMed]
- Aldana, B.I. Microglia-Specific Metabolic Changes in Neurodegeneration. J. Mol. Biol. 2019, 431, 1830–1842. [Google Scholar] [CrossRef] [PubMed]
- Skardal, A.; Aleman, J.; Forsythe, S.; Rajan, S.; Murphy, S.; Devarasetty, M.; Pourhabibi Zarandi, N.; Nzou, G.; Wicks, R.; Sadri-Ardekani, H.; et al. Drug compound screening in single and integrated multi-organoid body-on-a-chip systems. Biofabrication 2020, 12, 25017. [Google Scholar] [CrossRef]
- Shah, D.; Virtanen, L.; Prajapati, C.; Kiamehr, M.; Gullmets, J.; West, G.; Kreutzer, J.; Pekkanen-Mattila, M.; Heliö, T.; Kallio, P.; et al. Modeling of LMNA-Related Dilated Cardiomyopathy Using Human Induced Pluripotent Stem Cells. Cells 2019, 8, 594. [Google Scholar] [CrossRef]
- Zerti, D.; Hilgen, G.; Dorgau, B.; Collin, J.; Ader, M.; Armstrong, L.; Sernagor, E.; Lako, M. Transplanted pluripotent stem cell-derived photoreceptor precursors elicit conventional and unusual light responses in mice with advanced retinal degeneration. Stem Cells 2021, 39, 882–896. [Google Scholar] [CrossRef]
- Schönecker, S.; Kraushaar, U.; Guenther, E.; Gerst, F.; Ullrich, S.; Häring, H.-U.; Königsrainer, A.; Barthlen, W.; Drews, G.; Krippeit-Drews, P. Human Islets Exhibit Electrical Activity on Microelectrode Arrays (MEA). Exp. Clin. Endocrinol. Diabetes Off. J. Ger. Soc. Endocrinol. Ger. Diabetes Assoc. 2015, 123, 296–298. [Google Scholar] [CrossRef]
- Takayama, Y.; Kida, Y.S. In vitro reconstruction of neuronal networks derived from human iPS cells using microfabricated devices. PLoS ONE 2016, 11, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Duc, P.; Vignes, M.; Hugon, G.; Sebban, A.; Carnac, G.; Malyshev, E.; Charlot, B.; Rage, F. Human neuromuscular junction on micro-structured microfluidic devices implemented with a custom micro electrode array (MEA). Lab Chip 2021, 21, 4223–4236. [Google Scholar] [CrossRef] [PubMed]
- CNET Watch Elon Musk’s ENTIRE Live Neuralink demonstration. Available online: https://youtu.be/iOWFXqT5MZ4 (accessed on 24 September 2021).
- Simeral, J.D.; Hosman, T.; Saab, J.; Flesher, S.N.; Vilela, M.; Franco, B.; Kelemen, J.N.; Brandman, D.M.; Ciancibello, J.G.; Rezaii, P.G.; et al. Home Use of a Percutaneous Wireless Intracortical Brain-Computer Interface by Individuals with Tetraplegia. IEEE Trans. Biomed. Eng. 2021, 68, 2313–2325. [Google Scholar] [CrossRef]
| Reference | Modeled disorder | Affected Gene | Neuron Type | Phenotype on MEA |
|---|---|---|---|---|
| [179] | Koolen-de Vries syndrome | Kansl1 | Glutamatergic | Mean firing rate ↓ Synchronized bursts ↓ |
| [178] | Mitochondrial encephalomyopathy, lactic acidosis, and stroke-like episodes | Mt-tl1 (m.3243A > G) | Glutamatergic | Mean firing rate ↓ Synchronized bursts ↓ |
| [177] | Kleefstra syndrome | Ehmt1 | Glutamatergic | Synchronized bursts ↓ Synchronized burst duration ↑ |
| [180] | Neonatal epileptic encephalopathy | Kcnq2 | Glutamatergic | Bursts ↑ Spikes per burst ↑ |
| [134] | STXBP1 encephalopathy | Stxbp1 | GABAergic | Mean firing rate ↓ Bursts ↓ |
| [181] | Epileptic encephalopathy with intractable seizures | Scn2a (L1342P) | Glutamatergic | Mean firing rate ↑ Bursts ↑ Burst duration ↓ Spike frequency in burst ↑ Synchrony ↑ |
| [182] | Amyotrophic lateral sclerosis (ALS) | Sod1, C9orf72 (hexanucleotide repeat expansion) | Motor neurons | Mean firing rate ↑ |
| [143] | ALS | Tardbp (Q331K) | Motor neurons | Synchronized bursts ↓ Synchronized burst duration ↑ Signal propagation velocity ↑ |
| [143] | Parkinson’s disease (PD) | Snca (A53T) | Dopaminergic | Mean firing rate ↓ Synchronized bursts ↑ Synchronized burst duration ↓ |
| [183] | Alzheimer’s disease (AD) | Psen1 (M146V), App (APPSwe) | Glutamatergic (90–95%) and GABAergic (5–10 %) | Mean firing rate ↑ Synchronized bursts ↑ |
| Reference | Modeled Disorder | Affected Gene | Organoid Type | Phenotype on MEA |
|---|---|---|---|---|
| [218] | Bipolar disorder | Multiple | Cerebral | KCl response ↓ |
| [205] | Schizophrenia | Multiple | Cerebral | KCl response ↓ |
| [183] | Alzheimer’s disease (AD) | Psen1 (M146V), App (APPSwe) | Cerebral | Mean firing rate ↑ Synchronized bursts ↑ |
| [210] | Amyotrophic lateral sclerosis (ALS) and Frontotemporal dementia (FTD) | C9orf72 (hexanucleotide repeat expansion) | Cerebral (grown as slice) | No change |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pelkonen, A.; Pistono, C.; Klecki, P.; Gómez-Budia, M.; Dougalis, A.; Konttinen, H.; Stanová, I.; Fagerlund, I.; Leinonen, V.; Korhonen, P.; et al. Functional Characterization of Human Pluripotent Stem Cell-Derived Models of the Brain with Microelectrode Arrays. Cells 2022, 11, 106. https://doi.org/10.3390/cells11010106
Pelkonen A, Pistono C, Klecki P, Gómez-Budia M, Dougalis A, Konttinen H, Stanová I, Fagerlund I, Leinonen V, Korhonen P, et al. Functional Characterization of Human Pluripotent Stem Cell-Derived Models of the Brain with Microelectrode Arrays. Cells. 2022; 11(1):106. https://doi.org/10.3390/cells11010106
Chicago/Turabian StylePelkonen, Anssi, Cristiana Pistono, Pamela Klecki, Mireia Gómez-Budia, Antonios Dougalis, Henna Konttinen, Iveta Stanová, Ilkka Fagerlund, Ville Leinonen, Paula Korhonen, and et al. 2022. "Functional Characterization of Human Pluripotent Stem Cell-Derived Models of the Brain with Microelectrode Arrays" Cells 11, no. 1: 106. https://doi.org/10.3390/cells11010106
APA StylePelkonen, A., Pistono, C., Klecki, P., Gómez-Budia, M., Dougalis, A., Konttinen, H., Stanová, I., Fagerlund, I., Leinonen, V., Korhonen, P., & Malm, T. (2022). Functional Characterization of Human Pluripotent Stem Cell-Derived Models of the Brain with Microelectrode Arrays. Cells, 11(1), 106. https://doi.org/10.3390/cells11010106






