Statins Decrease Programmed Death-Ligand 1 (PD-L1) by Inhibiting AKT and β-Catenin Signaling
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Reagents
2.2. Protein Extraction and Western Blotting
2.3. RNA Isolation, cDNA Synthesis, and RT-PCR
2.4. Chromatin Immunoprecipitation (ChIP)
2.5. Bioinformatic Analysis
2.6. Statistical Analysis
3. Results and Discussion
3.1. Programmed Death-Ligand 1 (PD-L1) Is Highly Expressed in Various Cancer Cells
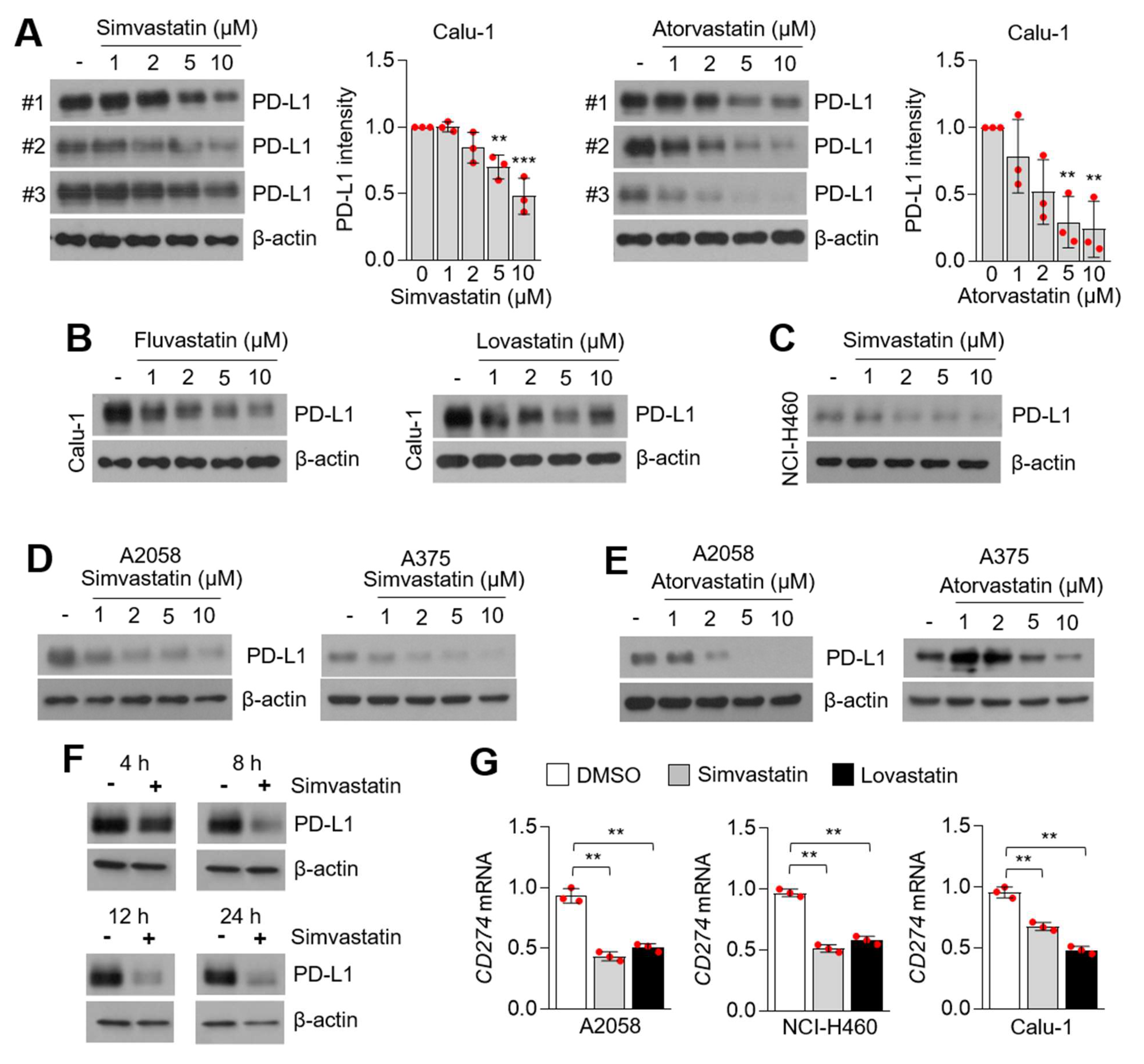
3.2. Statins Suppress PD-L1 Expression in Lung Cancer and Melanoma Cells
3.3. Statins Suppress PD-L1 Expression Independent of Farnesyltransferases (FTase) and Geranylgeranyltransferases (GGTase)
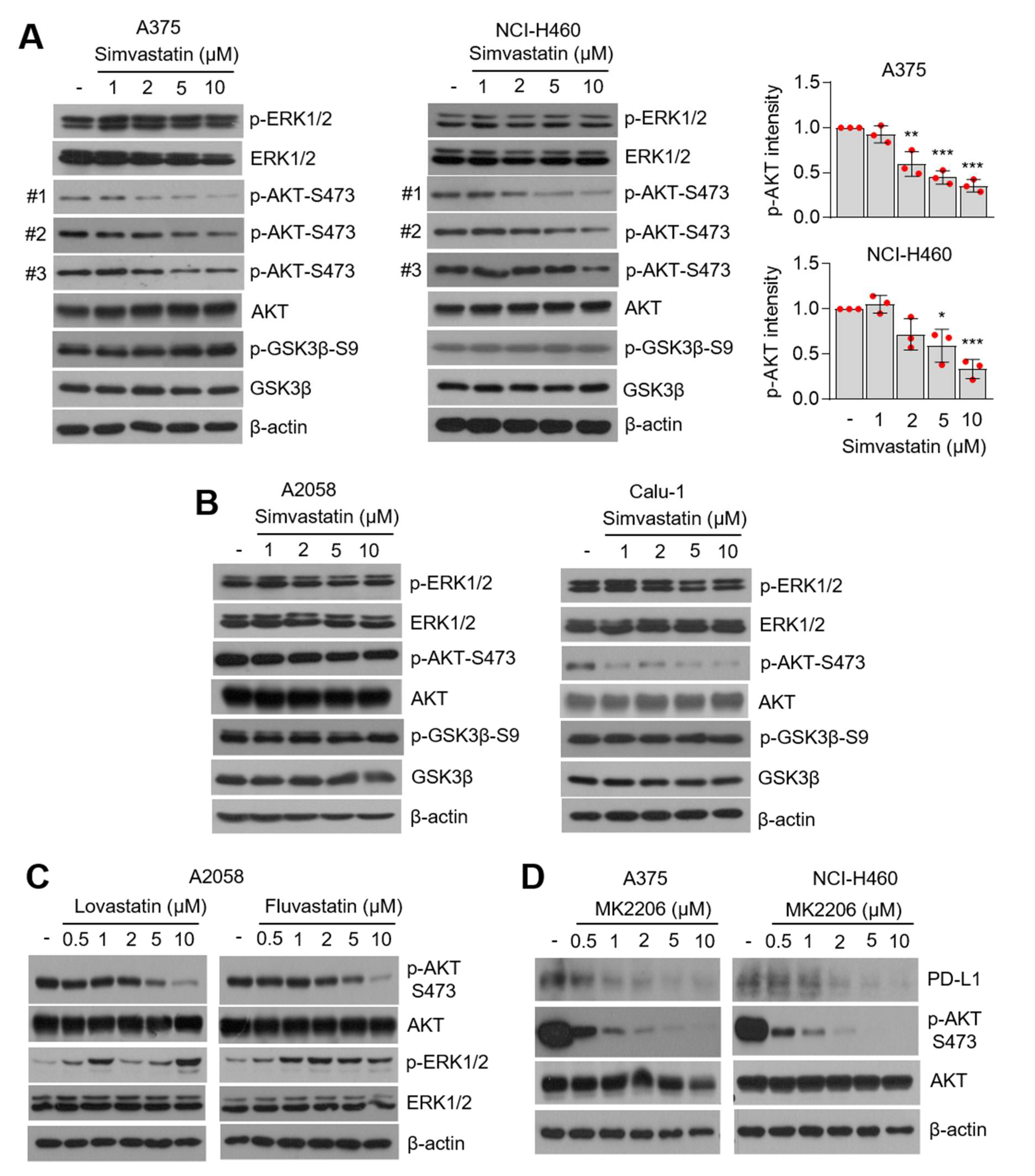
3.4. AKT-Signaling Involves PD-L1 Suppression by Statins
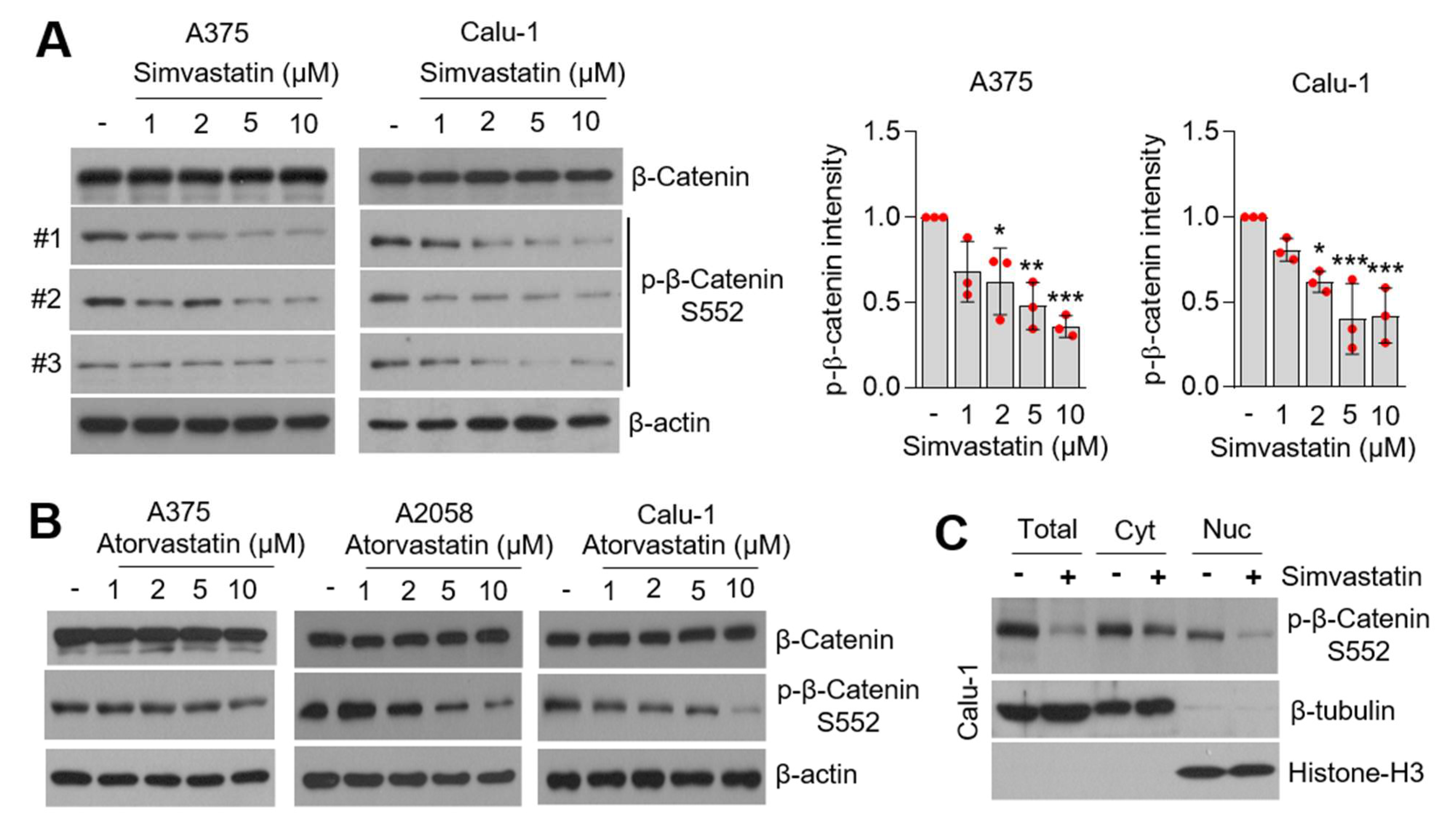
3.5. Statins Suppress β-Catenin Phosphorylation
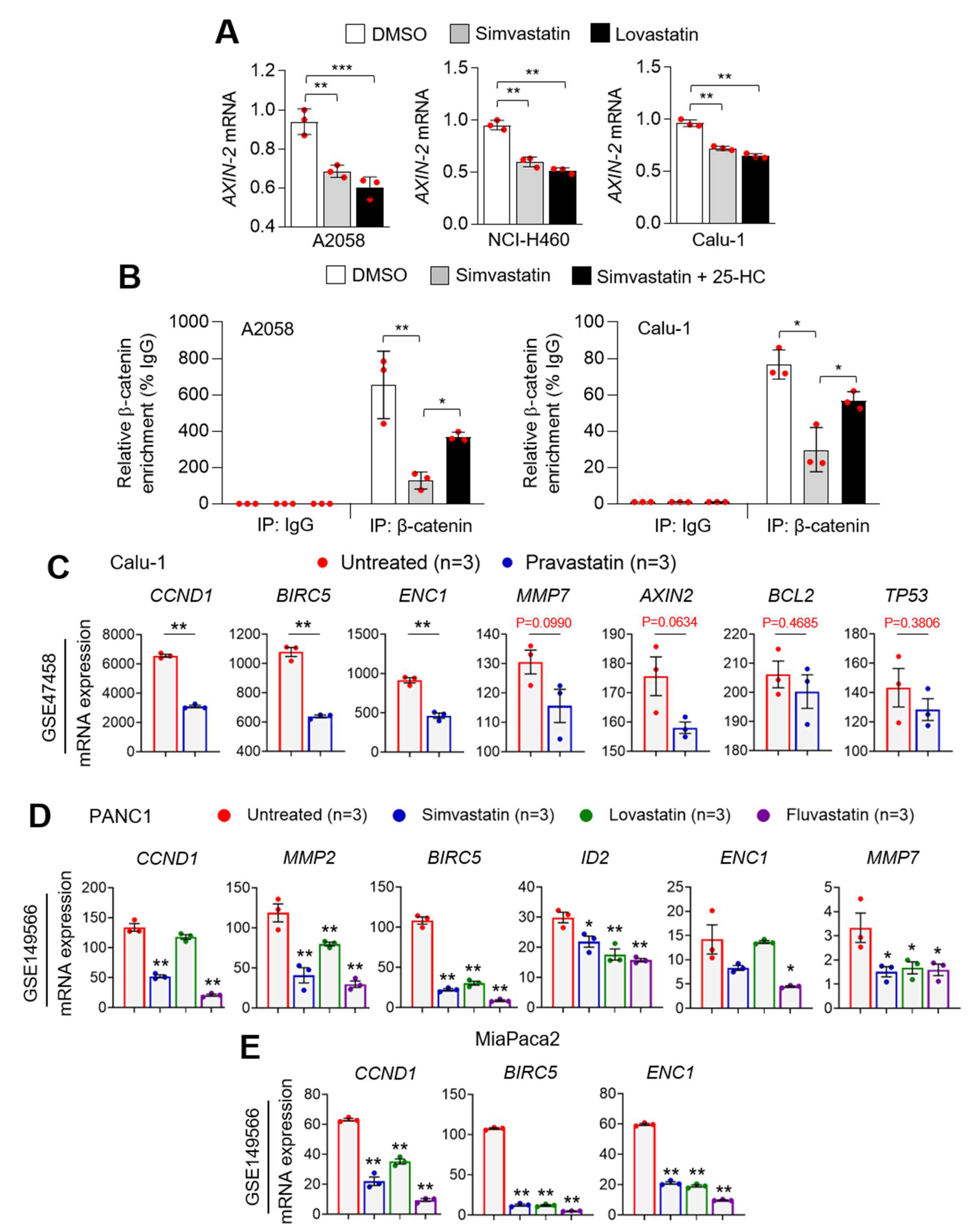
3.6. Statins Suppress PD-L1 at the Transcriptional Level
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Kuzu, O.F.; Noory, M.A.; Robertson, G.P. The Role of Cholesterol in Cancer. Cancer Res. 2016, 76, 2063–2070. [Google Scholar] [CrossRef]
- Ding, X.; Zhang, W.; Li, S.; Yang, H. The role of cholesterol metabolism in cancer. Am. J. Cancer Res. 2019, 9, 219–227. [Google Scholar] [PubMed]
- Huang, P.; Nedelcu, D.; Watanabe, M.; Jao, C.; Kim, Y.; Liu, J.; Salic, A. Cellular Cholesterol Directly Activates Smoothened in Hedgehog Signaling. Cell 2016, 166, 1176–1187. [Google Scholar] [CrossRef]
- Luchetti, G.; Sircar, R.; Kong, J.H.; Nachtergaele, S.; Sagner, A.; Byrne, E.F.; Covey, D.F.; Siebold, C.; Rohatgi, R. Cholesterol activates the G-protein coupled receptor Smoothened to promote Hedgehog signaling. Elife 2016, 5, e20304. [Google Scholar] [CrossRef] [PubMed]
- Codini, M.; Garcia-Gil, M.; Albi, E. Cholesterol and Sphingolipid Enriched Lipid Rafts as Therapeutic Targets in Cancer. Int. J. Mol. Sci. 2021, 22, 726. [Google Scholar] [CrossRef] [PubMed]
- Adhyaru, B.B.; Jacobson, T.A. Safety and efficacy of statin therapy. Nat. Rev. Cardiol. 2018, 15, 757–769. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, S.F.; Nordestgaard, B.G.; Bojesen, S.E. Statin use and reduced cancer-related mortality. N. Engl. J. Med. 2012, 367, 1792–1802. [Google Scholar] [CrossRef] [PubMed]
- Yu, O.; Eberg, M.; Benayoun, S.; Aprikian, A.; Batist, G.; Suissa, S.; Azoulay, L. Use of statins and the risk of death in patients with prostate cancer. J. Clin. Oncol. 2014, 32, 5–11. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.Y.; Hsu, C.H.; Yeh, K.H.; Chen, H.M.; Yeh, Y.C.; Lai, C.L.; Lin, Z.Z.; Cheng, A.L.; Lai, M.S. Statin Use Is Associated With Improved Prognosis of Colorectal Cancer in Taiwan. Clin. Colorectal Cancer 2015, 14, 177–184. [Google Scholar] [CrossRef] [PubMed]
- Viers, B.R.; Houston Thompson, R.; Psutka, S.P.; Lohse, C.M.; Cheville, J.C.; Leibovich, B.C.; Tollefson, M.K.; Boorjian, S.A. The association of statin therapy with clinicopathologic outcomes and survival among patients with localized renal cell carcinoma undergoing nephrectomy. Urol. Oncol. 2015, 33, 311–388. [Google Scholar] [CrossRef]
- Cardwell, C.R.; Hicks, B.M.; Hughes, C.; Murray, L.J. Statin use after diagnosis of breast cancer and survival: A population-based cohort study. Epidemiology 2015, 26, 68–78. [Google Scholar] [CrossRef] [PubMed]
- Habis, M.; Wroblewski, K.; Bradaric, M.; Ismail, N.; Yamada, S.D.; Litchfield, L.; Lengyel, E.; Romero, I.L. Statin therapy is associated with improved survival in patients with non-serous-papillary epithelial ovarian cancer: A retrospective cohort analysis. PLoS ONE 2014, 9, 104521. [Google Scholar] [CrossRef]
- Nowakowski, G.S.; Maurer, M.J.; Habermann, T.M.; Ansell, S.M.; Macon, W.R.; Ristow, K.M.; Allmer, C.; Slager, S.L.; Witzig, T.E.; Cerhan, J.R. Statin use and prognosis in patients with diffuse large B-cell lymphoma and follicular lymphoma in the rituximab era. J. Clin. Oncol. 2010, 28, 412–417. [Google Scholar] [CrossRef]
- Han, J.Y.; Lee, S.H.; Yoo, N.J.; Hyung, L.S.; Moon, Y.J.; Yun, T.; Kim, H.T.; Lee, J.S. A randomized phase II study of gefitinib plus simvastatin versus gefitinib alone in previously treated patients with advanced non-small cell lung cancer. Clin. Cancer Res. 2011, 17, 1553–1560. [Google Scholar] [CrossRef]
- Lee, Y.; Lee, K.H.; Lee, G.K.; Lee, S.H.; Lim, K.Y.; Joo, J.; Go, Y.J.; Lee, J.S.; Han, J.Y. Randomized Phase II Study of Afatinib Plus Simvastatin Versus Afatinib Alone in Previously Treated Patients with Advanced Nonadenocarcinomatous Non-small Cell Lung Cancer. Cancer Res. Treat. 2017, 49, 1001–1011. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.Y.; Nam, E.M.; Lee, J.; Park, J.O.; Lee, S.C.; Song, S.Y.; Choi, S.H.; Heo, J.S.; Park, S.H.; Lim, H.Y.; et al. Randomized double-blinded, placebo-controlled phase II trial of simvastatin and gemcitabine in advanced pancreatic cancer patients. Cancer Chemother. Pharm. 2014, 73, 125–130. [Google Scholar] [CrossRef] [PubMed]
- Jouve, J.L.; Lecomte, T.; Bouche, O.; Barbier, E.; Khemissa Akouz, F.; Riachi, G.; Nguyen Khac, E.; Ollivier-Hourmand, I.; Debette-Gratien, M.; Faroux, R.; et al. Pravastatin combination with sorafenib does not improve survival in advanced hepatocellular carcinoma. J. Hepatol. 2019, 71, 516–522. [Google Scholar] [CrossRef]
- Cantini, L.; Pecci, F.; Hurkmans, D.P.; Belderbos, R.A.; Lanese, A.; Copparoni, C.; Aerts, S.; Cornelissen, R.; Dumoulin, D.W.; Fiordoliva, I.; et al. High-intensity statins are associated with improved clinical activity of PD-1 inhibitors in malignant pleural mesothelioma and advanced non-small cell lung cancer patients. Eur. J. Cancer 2021, 144, 41–48. [Google Scholar] [CrossRef]
- Omori, M.; Okuma, Y.; Hakozaki, T.; Hosomi, Y. Statins improve survival in patients previously treated with nivolumab for advanced non-small cell lung cancer: An observational study. Mol. Clin. Oncol. 2019, 10, 137–143. [Google Scholar] [CrossRef]
- Casey, P.J.; Seabra, M.C. Protein prenyltransferases. J. Biol. Chem. 1996, 271, 5289–5292. [Google Scholar] [CrossRef]
- Whyte, D.B.; Kirschmeier, P.; Hockenberry, T.N.; Nunez-Oliva, I.; James, L.; Catino, J.J.; Bishop, W.R.; Pai, J.K. K- and N-Ras are geranylgeranylated in cells treated with farnesyl protein transferase inhibitors. J. Biol. Chem. 1997, 272, 14459–14464. [Google Scholar] [CrossRef]
- Berndt, N.; Hamilton, A.D.; Sebti, S.M. Targeting protein prenylation for cancer therapy. Nat. Rev. Cancer 2011, 11, 775–791. [Google Scholar] [CrossRef]
- Cha, J.H.; Chan, L.C.; Li, C.W.; Hsu, J.L.; Hung, M.C. Mechanisms Controlling PD-L1 Expression in Cancer. Mol. Cell 2019, 76, 359–370. [Google Scholar] [CrossRef]
- Iwai, Y.; Ishida, M.; Tanaka, Y.; Okazaki, T.; Honjo, T.; Minato, N. Involvement of PD-L1 on tumor cells in the escape from host immune system and tumor immunotherapy by PD-L1 blockade. Proc. Natl. Acad. Sci. USA 2002, 99, 12293–12297. [Google Scholar] [CrossRef]
- Robert, C. A decade of immune-checkpoint inhibitors in cancer therapy. Nat. Commun. 2020, 11, 3801. [Google Scholar] [CrossRef] [PubMed]
- Akinleye, A.; Rasool, Z. Immune checkpoint inhibitors of PD-L1 as cancer therapeutics. J. Hematol. Oncol. 2019, 12, 92. [Google Scholar] [CrossRef]
- Green, M.R.; Monti, S.; Rodig, S.J.; Juszczynski, P.; Currie, T.; O’Donnell, E.; Chapuy, B.; Takeyama, K.; Neuberg, D.; Golub, T.R.; et al. Integrative analysis reveals selective 9p24.1 amplification, increased PD-1 ligand expression, and further induction via JAK2 in nodular sclerosing Hodgkin lymphoma and primary mediastinal large B-cell lymphoma. Blood 2010, 116, 3268–3277. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, S.; Okamoto, T.; Okano, S.; Umemoto, Y.; Tagawa, T.; Morodomi, Y.; Kohno, M.; Shimamatsu, S.; Kitahara, H.; Suzuki, Y.; et al. PD-L1 Is Upregulated by Simultaneous Amplification of the PD-L1 and JAK2 Genes in Non-Small Cell Lung Cancer. J. Thorac. Oncol. 2016, 11, 62–71. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014, 513, 202–209. [Google Scholar] [CrossRef]
- Hogg, S.J.; Vervoort, S.J.; Deswal, S.; Ott, C.J.; Li, J.; Cluse, L.A.; Beavis, P.A.; Darcy, P.K.; Martin, B.P.; Spencer, A.; et al. BET-Bromodomain Inhibitors Engage the Host Immune System and Regulate Expression of the Immune Checkpoint Ligand PD-L1. Cell Rep. 2017, 18, 2162–2174. [Google Scholar] [CrossRef] [PubMed]
- Woods, D.M.; Sodre, A.L.; Villagra, A.; Sarnaik, A.; Sotomayor, E.M.; Weber, J. HDAC Inhibition Upregulates PD-1 Ligands in Melanoma and Augments Immunotherapy with PD-1 Blockade. Cancer Immunol. Res. 2015, 3, 1375–1385. [Google Scholar] [CrossRef] [PubMed]
- Atefi, M.; Avramis, E.; Lassen, A.; Wong, D.J.; Robert, L.; Foulad, D.; Cerniglia, M.; Titz, B.; Chodon, T.; Graeber, T.G.; et al. Effects of MAPK and PI3K pathways on PD-L1 expression in melanoma. Clin. Cancer Res. 2014, 20, 3446–3457. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Liu, F.; Zhang, F.; Zhou, W.; Jiang, X.; Yang, Y.; Qu, K.; Wang, Y.; Ma, Q.; Wang, T.; et al. Genomic ERBB2/ERBB3 mutations promote PD-L1-mediated immune escape in gallbladder cancer: A whole-exome sequencing analysis. Gut 2019, 68, 1024–1033. [Google Scholar] [CrossRef] [PubMed]
- Peng, S.; Wang, R.; Zhang, X.; Ma, Y.; Zhong, L.; Li, K.; Nishiyama, A.; Arai, S.; Yano, S.; Wang, W. EGFR-TKI resistance promotes immune escape in lung cancer via increased PD-L1 expression. Mol. Cancer 2019, 18, 165. [Google Scholar] [CrossRef]
- Sullivan, R.J.; Hamid, O.; Gonzalez, R.; Infante, J.R.; Patel, M.R.; Hodi, F.S.; Lewis, K.D.; Tawbi, H.A.; Hernandez, G.; Wongchenko, M.J.; et al. Atezolizumab plus cobimetinib and vemurafenib in BRAF-mutated melanoma patients. Nat. Med. 2019, 25, 929–935. [Google Scholar] [CrossRef] [PubMed]
- Oh, T.I.; Lee, M.; Lee, Y.M.; Kim, G.H.; Lee, D.; You, J.S.; Kim, S.H.; Choi, M.; Jang, H.; Park, Y.M.; et al. PGC1alpha Loss Promotes Lung Cancer Metastasis through Epithelial-Mesenchymal Transition. Cancers 2021, 13, 1772. [Google Scholar] [CrossRef]
- Luo, C.; Lim, J.H.; Lee, Y.; Granter, S.R.; Thomas, A.; Vazquez, F.; Widlund, H.R.; Puigserver, P. A PGC1alpha-mediated transcriptional axis suppresses melanoma metastasis. Nature 2016, 537, 422–426. [Google Scholar] [CrossRef]
- Du, L.; Lee, J.H.; Jiang, H.; Wang, C.; Wang, S.; Zheng, Z.; Shao, F.; Xu, D.; Xia, Y.; Li, J.; et al. Beta-Catenin induces transcriptional expression of PD-L1 to promote glioblastoma immune evasion. J. Exp. Med. 2020, 217. [Google Scholar] [CrossRef] [PubMed]
- Yan, F.; Pang, J.; Peng, Y.; Molina, J.R.; Yang, P.; Liu, S. Elevated Cellular PD1/PD-L1 Expression Confers Acquired Resistance to Cisplatin in Small Cell Lung Cancer Cells. PLoS ONE 2016, 11, e0162925. [Google Scholar] [CrossRef]
- Wang, X.; Yang, X.; Zhang, C.; Wang, Y.; Cheng, T.; Duan, L.; Tong, Z.; Tan, S.; Zhang, H.; Saw, P.E.; et al. Tumor cell-intrinsic PD-1 receptor is a tumor suppressor and mediates resistance to PD-1 blockade therapy. Proc. Natl. Acad. Sci. USA 2020, 117, 6640–6650. [Google Scholar] [CrossRef] [PubMed]
- Brauner, E.; Gunda, V.; Vanden Borre, P.; Zurakowski, D.; Kim, Y.S.; Dennett, K.V.; Amin, S.; Freeman, G.J.; Parangi, S. Combining BRAF inhibitor and anti PD-L1 antibody dramatically improves tumor regression and anti tumor immunity in an immunocompetent murine model of anaplastic thyroid cancer. Oncotarget 2016, 7, 17194–17211. [Google Scholar] [CrossRef]
- Luo, F.; Luo, M.; Rong, Q.X.; Zhang, H.; Chen, Z.; Wang, F.; Zhao, H.Y.; Fu, L.W. Niclosamide, an antihelmintic drug, enhances efficacy of PD-1/PD-L1 immune checkpoint blockade in non-small cell lung cancer. J. Immunother. Cancer 2019, 7, 245. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.Y.; Kong, P.F.; Xia, L.P.; Huang, Y.; Li, Z.L.; Tang, Y.Y.; Chen, Y.H.; Li, X.; Senthilkumar, R.; Zhang, H.L.; et al. Regorafenib Promotes Antitumor Immunity via Inhibiting PD-L1 and IDO1 Expression in Melanoma. Clin. Cancer Res. 2019, 25, 4530–4541. [Google Scholar] [CrossRef] [PubMed]
- Juarez, D.; Fruman, D.A. Targeting the Mevalonate Pathway in Cancer. Trends Cancer 2021, 7, 525–540. [Google Scholar] [CrossRef]
- Kong, T.; Ahn, R.; Yang, K.; Zhu, X.; Fu, Z.; Morin, G.; Bramley, R.; Cliffe, N.C.; Xue, Y.; Kuasne, H.; et al. CD44 Promotes PD-L1 Expression and Its Tumor-Intrinsic Function in Breast and Lung Cancers. Cancer Res. 2020, 80, 444–457. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Li, X.; Liu, S.; Guo, L.; Zhang, B.; Zhang, J.; Ye, Q. Programmed cell death-1 (PD-1) checkpoint blockade in combination with a mammalian target of rapamycin inhibitor restrains hepatocellular carcinoma growth induced by hepatoma cell-intrinsic PD-1. Hepatology 2017, 66, 1920–1933. [Google Scholar] [CrossRef]
- Simons, K.; Ehehalt, R. Cholesterol, lipid rafts, and disease. J. Clin. Investig. 2002, 110, 597–603. [Google Scholar] [CrossRef]
- Zhuang, L.; Kim, J.; Adam, R.M.; Solomon, K.R.; Freeman, M.R. Cholesterol targeting alters lipid raft composition and cell survival in prostate cancer cells and xenografts. J. Clin. Investig. 2005, 115, 959–968. [Google Scholar] [CrossRef]
- Sanchez-Wandelmer, J.; Davalos, A.; Herrera, E.; Giera, M.; Cano, S.; de la Pena, G.; Lasuncion, M.A.; Busto, R. Inhibition of cholesterol biosynthesis disrupts lipid raft/caveolae and affects insulin receptor activation in 3T3-L1 preadipocytes. Biochim. Biophys. Acta 2009, 1788, 1731–1739. [Google Scholar] [CrossRef]
- Beckwitt, C.H.; Shiraha, K.; Wells, A. Lipophilic statins limit cancer cell growth and survival, via involvement of Akt signaling. PLoS ONE 2018, 13, e0197422. [Google Scholar] [CrossRef]
- Park, Y.H.; Jung, H.H.; Ahn, J.S.; Im, Y.H. Statin induces inhibition of triple negative breast cancer (TNBC) cells via PI3K pathway. Biochem. Biophys. Res. Commun. 2013, 439, 275–279. [Google Scholar] [CrossRef]
- Roudier, E.; Mistafa, O.; Stenius, U. Statins induce mammalian target of rapamycin (mTOR)-mediated inhibition of Akt signaling and sensitize p53-deficient cells to cytostatic drugs. Mol. Cancer Ther. 2006, 5, 2706–2715. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mistafa, O.; Stenius, U. Statins inhibit Akt/PKB signaling via P2X7 receptor in pancreatic cancer cells. Biochem. Pharmacol. 2009, 78, 1115–1126. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Seah, S.; Loh, X.; Chan, C.W.; Hartman, M.; Goh, B.C.; Lee, S.C. Simvastatin-induced breast cancer cell death and deactivation of PI3K/Akt and MAPK/ERK signalling are reversed by metabolic products of the mevalonate pathway. Oncotarget 2016, 7, 2532–2544. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Cao, W.; Yue, P.; Hao, C.; Khuri, F.R.; Sun, S.Y. Celecoxib promotes c-FLIP degradation through Akt-independent inhibition of GSK3. Cancer Res. 2011, 71, 6270–6281. [Google Scholar] [CrossRef]
- Zhang, H.H.; Lipovsky, A.I.; Dibble, C.C.; Sahin, M.; Manning, B.D. S6K1 regulates GSK3 under conditions of mTOR-dependent feedback inhibition of Akt. Mol. Cell 2006, 24, 185–197. [Google Scholar] [CrossRef]
- Mittendorf, E.A.; Philips, A.V.; Meric-Bernstam, F.; Qiao, N.; Wu, Y.; Harrington, S.; Su, X.; Wang, Y.; Gonzalez-Angulo, A.M.; Akcakanat, A.; et al. PD-L1 expression in triple-negative breast cancer. Cancer Immunol. Res. 2014, 2, 361–370. [Google Scholar] [CrossRef]
- Wu, J.; Wong, W.W.; Khosravi, F.; Minden, M.D.; Penn, L.Z. Blocking the Raf/MEK/ERK pathway sensitizes acute myelogenous leukemia cells to lovastatin-induced apoptosis. Cancer Res. 2004, 64, 6461–6468. [Google Scholar] [CrossRef]


| Gene | Forward Primer | Reverse Primer |
|---|---|---|
| Axin-2 | GAGTGGACTTGTGCCGACTTCA | GGTGGCTGGTGCAAAGACATAG |
| CD274 | CTGCACTTTTAGGAGATTAGATC | CTACACCAAGGCATAATAAGATG |
| 36B4 | CATGTTGCTGGCCAATAAGG | TGGTGATACCTAAAGCCTGGAA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lim, W.-J.; Lee, M.; Oh, Y.; Fang, X.-Q.; Lee, S.; Lim, C.-H.; Park, J.; Lim, J.-H. Statins Decrease Programmed Death-Ligand 1 (PD-L1) by Inhibiting AKT and β-Catenin Signaling. Cells 2021, 10, 2488. https://doi.org/10.3390/cells10092488
Lim W-J, Lee M, Oh Y, Fang X-Q, Lee S, Lim C-H, Park J, Lim J-H. Statins Decrease Programmed Death-Ligand 1 (PD-L1) by Inhibiting AKT and β-Catenin Signaling. Cells. 2021; 10(9):2488. https://doi.org/10.3390/cells10092488
Chicago/Turabian StyleLim, Woo-Jin, Mingyu Lee, Yerin Oh, Xue-Quan Fang, Sujin Lee, Chang-Hoon Lim, Jooho Park, and Ji-Hong Lim. 2021. "Statins Decrease Programmed Death-Ligand 1 (PD-L1) by Inhibiting AKT and β-Catenin Signaling" Cells 10, no. 9: 2488. https://doi.org/10.3390/cells10092488
APA StyleLim, W.-J., Lee, M., Oh, Y., Fang, X.-Q., Lee, S., Lim, C.-H., Park, J., & Lim, J.-H. (2021). Statins Decrease Programmed Death-Ligand 1 (PD-L1) by Inhibiting AKT and β-Catenin Signaling. Cells, 10(9), 2488. https://doi.org/10.3390/cells10092488







