Current Applications and Discoveries Related to the Membrane Components of Circulating Tumor Cells and Extracellular Vesicles
Abstract
:1. Introduction
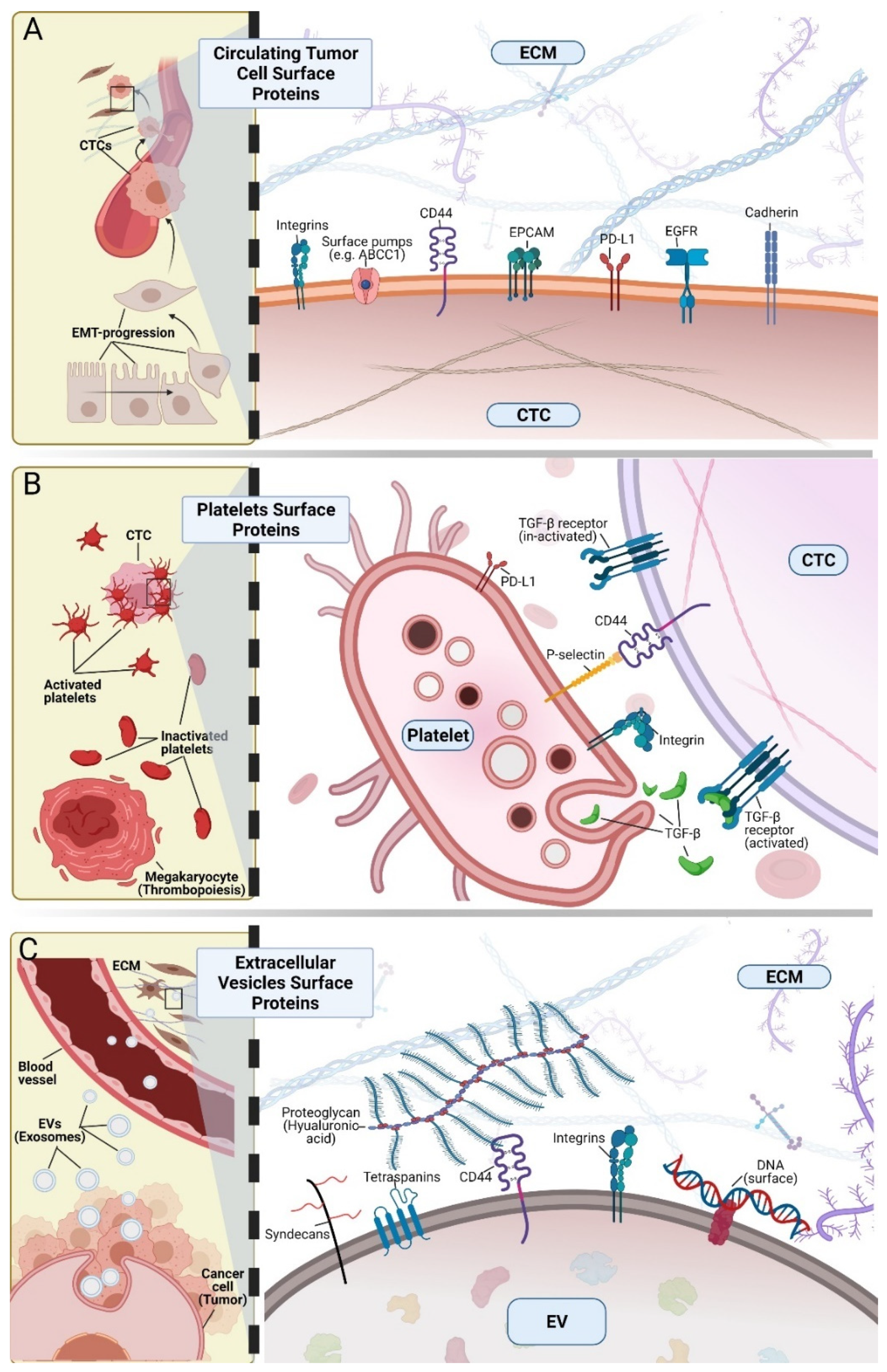
2. Cell Surface Proteins
3. Membrane Lipids
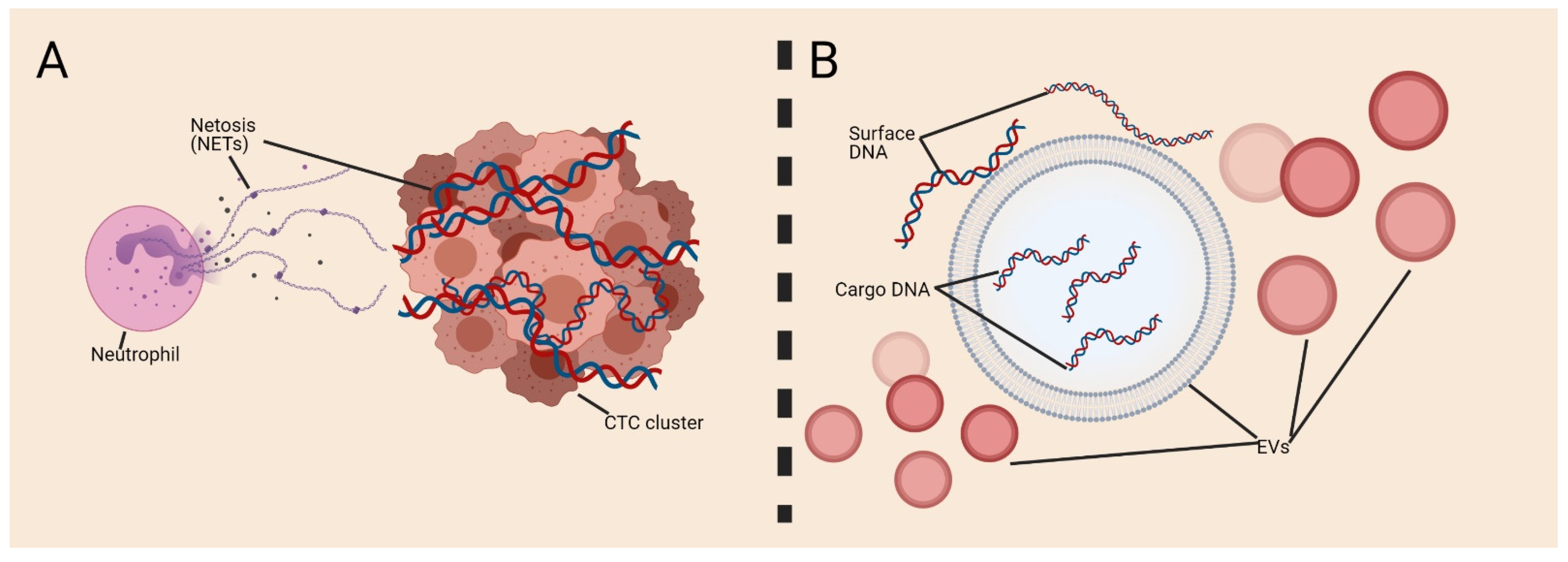
4. Surface Nucleic Acids
5. Complexity and Complementarity of EVs and CTCs
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- McAllister, S.S.; Weinberg, R.A. The tumour-induced systemic environment as a critical regulator of cancer progression and metastasis. Nat. Cell Biol. 2014, 16, 717–727. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eslami-S, Z.; Hernández, L.E.C.; Alix-Panabières, C. The Metastatic Cascade as the Basis for Liquid Biopsy Development. Front. Oncol. 2020, 10, 1055. [Google Scholar] [CrossRef] [PubMed]
- Lambert, A.W.; Pattabiraman, D.R.; Weinberg, R.A. Emerging Biological Principles of Metastasis. Cell 2017, 168, 670–691. [Google Scholar] [CrossRef] [Green Version]
- Alix-Panabières, C.; Pantel, K. Liquid Biopsy: From Discovery to Clinical Application. Cancer Discov. 2021, 11, 858–873. [Google Scholar] [CrossRef]
- Tie, J.; Kinde, I.; Wang, Y.; Wong, H.L.; Roebert, J.; Christie, M.; Tacey, M.; Wong, R.; Singh, M.; Karapetis, C.; et al. Circulating tumor DNA as an early marker of therapeutic response in patients with metastatic colorectal cancer. Ann. Oncol. 2015, 26, 1715–1722. [Google Scholar] [CrossRef]
- Eslami-S, Z.; Cortés-Hernández, L.E.; Cayrefourcq, L.; Alix-Panabières, C. The Different Facets of Liquid Biopsy: A Kaleidoscopic View. Cold Spring Harb. Perspect. Med. 2019, 10, a037333. [Google Scholar] [CrossRef]
- Cristofanilli, M.; Pierga, J.-Y.; Reuben, J.; Rademaker, A.; Davis, A.; Peeters, D.J.; Fehm, T.; Nolé, F.; Gisbert-Criado, R.; Mavroudis, D.; et al. The clinical use of circulating tumor cells (CTCs) enumeration for staging of metastatic breast cancer (MBC): International expert consensus paper. Crit. Rev. Oncol. 2019, 134, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Bidard, F.-C.; Jacot, W.; Kiavue, N.; Dureau, S.; Kadi, A.; Brain, E.; Bachelot, T.; Bourgeois, H.; Gonçalves, A.; Ladoire, S.; et al. Efficacy of Circulating Tumor Cell Count-Driven vs. Clinician-Driven First-line Therapy Choice in Hormone Receptor-Positive, ERBB2-Negative Metastatic Breast Cancer: The STIC CTC Randomized Clinical Trial. JAMA Oncol. 2021, 7, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Locker, G.Y.; Hamilton, S.; Harris, J.; Jessup, J.M.; Kemeny, N.; Macdonald, J.S.; Somerfield, M.R.; Hayes, D.F.; Bast, R.C., Jr. ASCO 2006 Update of Recommendations for the Use of Tumor Markers in Gastrointestinal Cancer. J. Clin. Oncol. 2006, 24, 5313–5327. [Google Scholar] [CrossRef]
- Xue, H.; Lu, B.; Lai, M. The cancer secretome: A reservoir of biomarkers. J. Transl. Med. 2008, 6, 52. [Google Scholar] [CrossRef] [Green Version]
- Sharma, S. Tumor markers in clinical practice: General principles and guidelines. Indian J. Med. Paediatr. Oncol. 2009, 30, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Kim, I.-H.; Lee, J.E.; Yang, J.H.; Jeong, J.W.; Ro, S.; Oh, S.T.; Kim, J.-G.; Choi, M.H.; Lee, M.A. Clinical Significance of Discordance between Carcinoembryonic Antigen Levels and RECIST in Metastatic Colorectal Cancer. Cancer Res. Treat. 2018, 50, 283–292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alix-Panabières, C. The future of liquid biopsy. Nat. Cell Biol. 2020, 579, S9. [Google Scholar] [CrossRef] [Green Version]
- Pantel, K.; Alix-Panabières, C. Circulating tumour cells in cancer patients: Challenges and perspectives. Trends Mol. Med. 2010, 16, 398–406. [Google Scholar] [CrossRef]
- Dell’Olio, F.; Su, J.; Huser, T.; Sottile, V.; Cortés-Hernández, L.E.; Alix-Panabières, C. Liquid Biopsies: Photonic Technologies for Liquid Biopsies: Recent Advances and Open Research Challenges (Laser Photonics Rev. 15(1)/2021). Laser Photon. Rev. 2021, 15, 2170012. [Google Scholar] [CrossRef]
- Thery, C.C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines AU—Théry, Clotilde. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mathieu, M.; Martin-Jaular, L.; Lavieu, G.; Théry, C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell Biol. 2019, 21, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, A.; Costa-Silva, B.; Shen, T.-L.; Rodrigues, G.; Hashimoto, A.; Mark, M.T.; Molina, H.; Kohsaka, S.; Di Giannatale, A.; Ceder, S.; et al. Tumour exosome integrins determine organotropic metastasis. Nature 2015, 527, 329–335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cescon, D.W.; Bratman, S.V.; Chan, S.M.; Siu, L.L. Circulating tumor DNA and liquid biopsy in oncology. Nat. Rev. Cancer 2020, 1, 276–290. [Google Scholar] [CrossRef]
- Jeppesen, D.K.; Franklin, J.L.; Higginbotham, J.N.; Zhang, Q.; Coffey, R.J. Abstract 1985: Reassessment of exosome composition. Tumor Biol. 2019, 177, 428–445. [Google Scholar] [CrossRef]
- Lo, Y.M.D.; Lam, W.K.J. Towards multi-cancer screening using liquid biopsies. Nat. Rev. Clin. Oncol. 2020, 17, 525–526. [Google Scholar] [CrossRef] [PubMed]
- Mattox, A.K.; Bettegowda, C.; Zhou, S.; Papadopoulos, N.; Kinzler, K.W.; Vogelstein, B. Applications of liquid biopsies for cancer. Sci. Transl. Med. 2019, 11, eaay1984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osumi, H.; Shinozaki, E.; Yamaguchi, K.; Zembutsu, H. Early change in circulating tumor DNA as a potential predictor of response to chemotherapy in patients with metastatic colorectal cancer. Sci. Rep. 2019, 9, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Aceto, N. Bring along your friends: Homotypic and heterotypic circulating tumor cell clustering to accelerate metastasis. Biomed. J. 2020, 43, 18–23. [Google Scholar] [CrossRef]
- Mohme, M.; Riethdorf, S.; Pantel, M.M.S.R.K. Circulating and disseminated tumour cells—Mechanisms of immune surveillance and escape. Nat. Rev. Clin. Oncol. 2017, 14, 155–167. [Google Scholar] [CrossRef]
- Costa-Silva, B.; Aiello, N.M.; Ocean, A.J.; Singh, S.; Zhang, H.; Thakur, B.K.; Becker, A.; Hoshino, A.; Mark, M.T.; Molina, H.; et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat. Cell Biol. 2015, 17, 816–826. [Google Scholar] [CrossRef]
- Rodrigues, G.; Hoshino, A.; Kenific, C.M.; Matei, I.R.; Steiner, L.; Freitas, D.; Kim, H.S.; Oxley, P.R.; Scandariato, I.; Casanova-Salas, I.; et al. Tumour exosomal CEMIP protein promotes cancer cell colonization in brain metastasis. Nat. Cell Biol. 2019, 21, 1403–1412. [Google Scholar] [CrossRef]
- Nicolson, G.L. Cell Membrane Fluid–Mosaic Structure and Cancer Metastasis. Cancer Res. 2015, 75, 1169–1176. [Google Scholar] [CrossRef] [Green Version]
- Harayama, T.; Riezman, H. Understanding the diversity of membrane lipid composition. Nat. Rev. Mol. Cell Biol. 2018, 19, 281–296. [Google Scholar] [CrossRef]
- Nicolson, G.L. The Fluid—Mosaic Model of Membrane Structure: Still relevant to understanding the structure, function and dynamics of biological membranes after more than 40years. Biochim. Biophys. Acta Biomembr. 2014, 1838, 1451–1466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hernández, L.E.C.; Eslami-S, Z.; Pantel, K.; Alix-Panabières, C. Molecular and Functional Characterization of Circulating Tumor Cells: From Discovery to Clinical Application. Clin. Chem. 2019, 66, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Keller, L.; Belloum, Y.; Wikman, H.; Pantel, K. Clinical relevance of blood-based ctDNA analysis: Mutation detection and beyond. Br. J. Cancer 2021, 124, 345–358. [Google Scholar] [CrossRef]
- Ignatiadis, M.; Sledge, G.W.; Jeffrey, S.S. Liquid biopsy enters the clinic—Implementation issues and future challenges. Nat. Rev. Clin. Oncol. 2021, 18, 297–312. [Google Scholar] [CrossRef]
- Best, M.G.; Wesseling, P.; Wurdinger, T. Tumor-Educated Platelets as a Noninvasive Biomarker Source for Cancer Detection and Progression Monitoring. Cancer Res. 2018, 78, 3407–3412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, R.; Greening, D.W.; Zhu, H.-J.; Takahashi, N.; Simpson, R.J. Extracellular vesicle isolation and characterization: Toward clinical application. J. Clin. Investig. 2016, 126, 1152–1162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef]
- Zhou, X.; Xie, F.; Wang, L.; Zhang, L.; Zhang, S.; Fang, M.; Zhou, F. The function and clinical application of extracellular vesicles in innate immune regulation. Cell. Mol. Immunol. 2020, 17, 323–334. [Google Scholar] [CrossRef]
- Leckband, D.; De Rooij, J. Cadherin Adhesion and Mechanotransduction. Annu. Rev. Cell Dev. Biol. 2014, 30, 291–315. [Google Scholar] [CrossRef]
- Hynes, R.O. Integrins: Bidirectional, Allosteric Signaling Machines. Cell 2002, 110, 673–687. [Google Scholar] [CrossRef] [Green Version]
- Dongre, A.; Weinberg, R.A. New insights into the mechanisms of epithelial–mesenchymal transition and implications for cancer. Nat. Rev. Mol. Cell Biol. 2019, 20, 69–84. [Google Scholar] [CrossRef]
- Litan, A.; Langhans, S.A. Cancer as a channelopathy: Ion channels and pumps in tumor development and progression. Front. Cell. Neurosci. 2015, 9, 86. [Google Scholar] [CrossRef] [Green Version]
- Selves, J.; Long-Mira, E.; Mathieu, M.-C.; Rochaix, P.; Ilié, M. Immunohistochemistry for Diagnosis of Metastatic Carcinomas of Unknown Primary Site. Cancers 2018, 10, 108. [Google Scholar] [CrossRef] [Green Version]
- Ordóñez, N.G. Broad-spectrum immunohistochemical epithelial markers: A review. Hum. Pathol. 2013, 44, 1195–1215. [Google Scholar] [CrossRef]
- Hernández, L.E.C.; Eslami-S, Z.; Alix-Panabières, C. Circulating tumor cell as the functional aspect of liquid biopsy to understand the metastatic cascade in solid cancer. Mol. Asp. Med. 2020, 72, 100816. [Google Scholar] [CrossRef] [PubMed]
- Allard, W.J.; Matera, J.; Miller, M.C.; Repollet, M.; Connelly, M.C.; Rao, C.; Tibbe, A.G.J.; Uhr, J.W.; Terstappen, L.W.M.M. Tumor Cells Circulate in the Peripheral Blood of All Major Carcinomas but not in Healthy Subjects or Patients with Nonmalignant Diseases. Clin. Cancer Res. 2004, 10, 6897–6904. [Google Scholar] [CrossRef] [Green Version]
- Alix-Panabieres, C.C. EPISPOT assay: Detection of viable DTCs/CTCs in solid tumor patients. Minimal Residual Dis. Circ. Tumor Cells Breast Cancer 2012, 195, 69–76. [Google Scholar] [CrossRef]
- Jacot, W.; Mazel, M.; Mollevi, C.; Pouderoux, S.; D’Hondt, V.; Cayrefourcq, L.; Bourgier, C.; Boissiere-Michot, F.; Berrabah, F.; Lopez-Crapez, E.; et al. Clinical Correlations of Programmed Cell Death Ligand 1 Status in Liquid and Standard Biopsies in Breast Cancer. Clin. Chem. 2020, 66, 1093–1101. [Google Scholar] [CrossRef] [PubMed]
- Eslami-S, Z.; Cortés-Hernández, L.E.; Alix-Panabières, C. Epithelial Cell Adhesion Molecule: An Anchor to Isolate Clinically Relevant Circulating Tumor Cells. Cells 2020, 9, 1836. [Google Scholar] [CrossRef]
- Cohen, S.J.; Punt, C.J.A.; Iannotti, N.; Saidman, B.H.; Sabbath, K.D.; Gabrail, N.Y.; Picus, J.; Morse, M.; Mitchell, E.; Miller, M.C.; et al. Relationship of Circulating Tumor Cells to Tumor Response, Progression-Free Survival, and Overall Survival in Patients with Metastatic Colorectal Cancer. J. Clin. Oncol. 2008, 26, 3213–3221. [Google Scholar] [CrossRef]
- Cristofanilli, M.; Budd, G.T.; Ellis, M.J.; Stopeck, A.; Matera, J.; Miller, M.C.; Reuben, J.M.; Doyle, G.V.; Allard, W.J.; Terstappen, L.W.; et al. Circulating Tumor Cells, Disease Progression, and Survival in Metastatic Breast Cancer. N. Engl. J. Med. 2004, 351, 781–791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bidard, F.C.; Madic, J.; Mariani, P.; Piperno-Neumann, S.; Rampanou, A.; Servois, V.; Cassoux, N.; Desjardins, L.; Milder, M.; Vaucher, I.; et al. Detection rate and prognostic value of circulating tumor cells and circulating tumor DNA in metastatic uveal melanoma. Int. J. Cancer 2014, 134, 1207–1213. [Google Scholar] [CrossRef] [PubMed]
- Nanou, A.; Miller, M.C.; Zeune, L.L.; De Wit, S.; Punt, C.J.A.; Groen, H.J.; Hayes, D.F.; De Bono, J.S.; Terstappen, L.W.M.M. Tumour-derived extracellular vesicles in blood of metastatic cancer patients associate with overall survival. Br. J. Cancer 2020, 122, 801–811. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Antin, P.; Berx, G.; Blanpain, C.; Brabletz, T.; Bronner, M.; Campbell, K.; Cano, A.; Casanova, J.; Christofori, G.; et al. Guidelines and definitions for research on epithelial–mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2020, 21, 341–352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cayrefourcq, L.; Thomas, F.; Mazard, T.; Assenat, E.; Assou, S.; Alix-Panabières, C. Selective treatment pressure in colon cancer drives the molecular profile of resistant circulating tumor cell clones. Mol. Cancer 2021, 20, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Lamouille, S.; Xu, J.; Derynck, R. Molecular mechanisms of epithelial–mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2014, 15, 178–196. [Google Scholar] [CrossRef] [Green Version]
- Loh, C.-Y.; Chai, J.; Tang, T.; Wong, W.; Sethi, G.; Shanmugam, M.; Chong, P.; Looi, C. The E-Cadherin and N-Cadherin Switch in Epithelial-to-Mesenchymal Transition: Signaling, Therapeutic Implications, and Challenges. Cells 2019, 8, 1118. [Google Scholar] [CrossRef] [Green Version]
- Delmonico, L.; Obenauer, J.C.; Qureshi, F.; Alves, G.; Costa, M.A.S.M.; Martin, K.J.; Fournier, M.V. A Novel Panel of 80 RNA Biomarkers with Differential Expression in Multiple Human Solid Tumors against Healthy Blood Samples. Int. J. Mol. Sci. 2019, 20, 4894. [Google Scholar] [CrossRef] [Green Version]
- Gorges, T.M.; Kuske, A.; Röck, K.; Mauermann, O.; Müller, V.; Peine, S.; Verpoort, K.; Novosadova, V.; Kubista, M.; Riethdorf, S.; et al. Accession of Tumor Heterogeneity by Multiplex Transcriptome Profiling of Single Circulating Tumor Cells. Clin. Chem. 2016, 62, 1504–1515. [Google Scholar] [CrossRef]
- Labelle, M.; Begum, S.; Hynes, R.O. Platelets guide the formation of early metastatic niches. Proc. Natl. Acad. Sci. USA 2014, 111, E3053–E3061. [Google Scholar] [CrossRef] [Green Version]
- Labelle, M.; Begum, S.; Hynes, R.O. Direct Signaling between Platelets and Cancer Cells Induces an Epithelial-Mesenchymal-Like Transition and Promotes Metastasis. Cancer Cell 2011, 20, 576–590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, M.S.; Bottsford-Miller, J.; Vasquez, H.G.; Stone, R.; Zand, B.; Kroll, M.H.; Sood, A.K.; Afshar-Kharghan, V. Platelets increase the proliferation of ovarian cancer cells. Blood 2012, 120, 4869–4872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dianat-Moghadam, H.; Azizi, M.; Eslami-S, Z.; Cortés-Hernández, L.E.; Heidarifard, M.; Nouri, M.; Alix-Panabières, C. The Role of Circulating Tumor Cells in the Metastatic Cascade: Biology, Technical Challenges, and Clinical Relevance. Cancers 2020, 12, 867. [Google Scholar] [CrossRef] [Green Version]
- Lindsay, C.; Blackhall, F.; Carmel, A.; Fernandez-Gutierrez, F.; Gazzaniga, P.; Groen, H.; Hiltermann, T.; Krebs, M.; Loges, S.; López-López, R.; et al. EPAC-lung: Pooled analysis of circulating tumour cells in advanced non-small cell lung cancer. Eur. J. Cancer 2019, 117, 60–68. [Google Scholar] [CrossRef] [Green Version]
- Hiltermann, T.J.N.; Pore, M.M.; Berg, A.V.D.; Timens, W.; Boezen, H.M.; Liesker, J.J.W.; Schouwink, J.H.; Wijnands, W.J.A.; Kerner, G.; Kruyt, F.; et al. Circulating tumor cells in small-cell lung cancer: A predictive and prognostic factor. Ann. Oncol. 2012, 23, 2937–2942. [Google Scholar] [CrossRef]
- Campbell, I.D.; Humphries, M. Integrin Structure, Activation, and Interactions. Cold Spring Harb. Perspect. Biol. 2011, 3, a004994. [Google Scholar] [CrossRef] [Green Version]
- Guo, W.; Giancotti, F.G. Integrin signalling during tumour progression. Nat. Rev. Mol. Cell Biol. 2004, 5, 816–826. [Google Scholar] [CrossRef] [PubMed]
- Osmani, N.; Follain, G.; León, M.J.G.; Lefebvre, O.; Busnelli, I.; Larnicol, A.; Harlepp, S.; Goetz, J.G. Metastatic Tumor Cells Exploit Their Adhesion Repertoire to Counteract Shear Forces during Intravascular Arrest. Cell Rep. 2019, 28, 2491–2500.e5. [Google Scholar] [CrossRef]
- Staflin, K.; Krueger, J.S.; Hachmann, J.; Forsyth, J.S.; Lorger, M.; Steiniger, S.C.J.; Mee, J.; Pop, C.; Salvesen, G.S.; Janda, K.D.; et al. Targeting activated integrin alphavbeta3 with patient-derived antibodies impacts late-stage multiorgan metastasis. Clin. Exp. Metastasis 2010, 27, 217–231. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Hoeppner, L.H.; Bach, S.; Guangqi, E.; Guo, Y.; Wang, E.; Wu, J.; Cowley, M.J.; Chang, D.K.; Waddell, N.; et al. Neuropilin-2 Promotes Extravasation and Metastasis by Interacting with Endothelial α5 Integrin. Cancer Res. 2013, 73, 4579–4590. [Google Scholar] [CrossRef] [Green Version]
- Fuentes, P.; Sesé, M.; Guijarro, P.J.; Emperador, M.; Sánchez-Redondo, S.; Peinado, H.; Hümmer, S.; Ramón Y Cajal, S. ITGB3-mediated uptake of small extracellular vesicles facilitates intercellular communication in breast cancer cells. Nat. Commun. 2020, 11, 4261. [Google Scholar] [CrossRef]
- Altei, W.F.; Pachane, B.C.; dos Santos, P.K.; Ribeiro, L.N.M.; Sung, B.H.; Weaver, A.M.; Selistre-De-Araújo, H.S. Inhibition of αvβ3 integrin impairs adhesion and uptake of tumor-derived small extracellular vesicles. Cell Commun. Signal. 2020, 18, 158. [Google Scholar] [CrossRef]
- Guo, Q.; Furuta, K.; Lucien, F.; Sanchez, L.H.G.; Hirsova, P.; Krishnan, A.; Kabashima, A.; Pavelko, K.D.; Madden, B.; Alhuwaish, H.; et al. Integrin β1-enriched extracellular vesicles mediate monocyte adhesion and promote liver inflammation in murine NASH. J. Hepatol. 2019, 71, 1193–1205. [Google Scholar] [CrossRef]
- Kornek, M.; Lynch, M.; Mehta, S.H.; Lai, M.; Exley, M.; Afdhal, N.; Schuppan, D. Circulating Microparticles as Disease-Specific Biomarkers of Severity of Inflammation in Patients With Hepatitis C or Nonalcoholic Steatohepatitis. Gastroenterology 2012, 143, 448–458. [Google Scholar] [CrossRef] [Green Version]
- Hoshino, A.; Kim, H.S.; Bojmar, L.; Gyan, K.E.; Cioffi, M.; Hernandez, J.; Zambirinis, C.P.; Rodrigues, G.; Molina, H.; Heissel, S.; et al. Extracellular vesicle and particle biomarkers define multiple human cancers. Cell 2020, 182, 1044–1061.e18. [Google Scholar] [CrossRef] [PubMed]
- Dardik, R.; Kaufmann, Y.; Savion, N.; Rosenberg, N.; Shenkman, B.; Varon, D. Platelets mediate tumor cell adhesion to the subendothelium under flow conditions: Involvement of platelet GPIIb-IIIa and tumor cell αv integrins. Int. J. Cancer 1997, 70, 201–207. [Google Scholar] [CrossRef]
- Bakewell, S.J.; Nestor, P.; Prasad, S.; Tomasson, M.H.; Dowland, N.; Mehrotra, M.; Scarborough, R.; Kanter, J.; Abe, K.; Phillips, D.; et al. Platelet and osteoclast β3 integrins are critical for bone metastasis. Proc. Natl. Acad. Sci. USA 2003, 100, 14205–14210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, F.; Li, L.; Guan, L.; Yang, H.; Wu, C.; Liu, Y. Roles for GP IIb/IIIa and αvβ3 integrins in MDA-MB-231 cell invasion and shear flow-induced cancer cell mechanotransduction. Cancer Lett. 2014, 344, 62–73. [Google Scholar] [CrossRef] [PubMed]
- Senbanjo, L.T.; Chellaiah, M.A. CD44: A Multifunctional Cell Surface Adhesion Receptor Is a Regulator of Progression and Metastasis of Cancer Cells. Front. Cell Dev. Biol. 2017, 5, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Taftaf, R.; Kawaguchi, M.; Chang, Y.-F.; Chen, W.; Entenberg, D.; Zhang, Y.; Gerratana, L.; Huang, S.; Patel, D.B.; et al. Homophilic CD44 Interactions Mediate Tumor Cell Aggregation and Polyclonal Metastasis in Patient-Derived Breast Cancer Models. Cancer Discov. 2019, 9, 96–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamane, N.; Tsujitani, S.; Makino, M.; Maeta, M.; Kaibara, N. Soluble CD44 Variant 6 as a Prognostic Indicator in Patients with Colorectal Cancer. Oncology 1999, 56, 232–238. [Google Scholar] [CrossRef]
- Gupta, A.; Zhou, C.Q.; Chellaiah, M.A. Osteopontin and MMP9: Associations with VEGF Expression/Secretion and Angiogenesis in PC3 Prostate Cancer Cells. Cancers 2013, 5, 617–638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, C.; Fan, C.; Wang, C.; Huang, Q.; Meng, W.; Yu, Y.; Yang, L.; Peng, Z.; Hu, J.; Li, Y.; et al. CD133+CD54+CD44+ circulating tumor cells as a biomarker of treatment selection and liver metastasis in patients with colorectal cancer. Oncotarget 2016, 7, 77389–77403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nicolazzo, C.; Loreni, F.; Caponnetto, S.; Magri, V.; Vestri, A.R.; Zamarchi, R.; Gradilone, A.; Facchinetti, A.; Rossi, E.; Cortesi, E.; et al. Baseline CD44v6-positive circulating tumor cells to predict first-line treatment failure in patients with metastatic colorectal cancer. Oncotarget 2020, 11, 4115–4122. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Wang, C.; Zhu, H.; Wang, Y.; Wang, X.; Cheng, X.; Ge, W.; Lu, W. Exosome-mediated transfer of CD44 from high-metastatic ovarian cancer cells promotes migration and invasion of low-metastatic ovarian cancer cells. J. Ovarian Res. 2021, 14, 1–11. [Google Scholar] [CrossRef]
- Wang, Z.; Von Au, A.; Schnölzer, M.; Hackert, T.; Zöller, M. CD44v6-competent tumor exosomes promote motility, invasion and cancer-initiating cell marker expression in pancreatic and colorectal cancer cells. Oncotarget 2016, 7, 55409–55436. [Google Scholar] [CrossRef] [Green Version]
- Hur, Y.H.; Feng, S.; Wilson, K.F.; Cerione, R.A.; Antonyak, M.A. Embryonic Stem Cell-Derived Extracellular Vesicles Maintain ESC Stemness by Activating FAK. Dev. Cell 2020, 56, 277–291.e6. [Google Scholar] [CrossRef]
- Wang, M.; Ji, S.; Shao, G.; Zhang, J.; Zhao, K.; Wang, Z.; Wu, A. Effect of exosome biomarkers for diagnosis and prognosis of breast cancer patients. Clin. Transl. Oncol. 2018, 20, 906–911. [Google Scholar] [CrossRef]
- Alves, C.S.; Burdick, M.M.; Thomas, S.N.; Pawar, P.; Konstantopoulos, K. The dual role of CD44 as a functional P-selectin ligand and fibrin receptor in colon carcinoma cell adhesion. Am. J. Physiol.-Cell Physiol. 2008, 294, C907–C916. [Google Scholar] [CrossRef] [PubMed]
- Burdick, M.; Chu, J.T.; Godar, S.; Sackstein, R. HCELL Is the Major E- and L-selectin Ligand Expressed on LS174T Colon Carcinoma Cells. J. Biol. Chem. 2006, 281, 13899–13905. [Google Scholar] [CrossRef] [Green Version]
- Orellana, R.; Kato, S.; Erices, R.; Bravo, M.L.; Gonzalez, P.; Oliva, B.; Cubillos, S.; Valdivia, A.; Ibañez, C.; Brañes, J.; et al. Platelets enhance tissue factor protein and metastasis initiating cell markers, and act as chemoattractants increasing the migration of ovarian cancer cells. BMC Cancer 2015, 15, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malehmir, M.; Pfister, D.; Gallage, S.; Szydlowska, M.; Inverso, D.; Kotsiliti, E.; Leone, V.; Peiseler, M.; Surewaard, B.G.J.; Rath, D.; et al. Platelet GPIbα is a mediator and potential interventional target for NASH and subsequent liver cancer. Nat. Med. 2019, 25, 641–655. [Google Scholar] [CrossRef] [Green Version]
- Wong, K.; Ma, J.; Rothnie, A.; Biggin, P.; Kerr, I.D. Towards understanding promiscuity in multidrug efflux pumps. Trends Biochem. Sci. 2014, 39, 8–16. [Google Scholar] [CrossRef]
- Steinbach, D.; Legrand, O. ABC transporters and drug resistance in leukemia: Was P-gp nothing but the first head of the Hydra? Leukemia 2007, 21, 1172–1176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kerr, I.D.; Jones, P.M.; George, A.M. Multidrug efflux pumps: The structures of prokaryotic ATP-binding cassette transporter efflux pumps and implications for our understanding of eukaryotic P-glycoproteins and homologues. FEBS J. 2009, 277, 550–563. [Google Scholar] [CrossRef] [Green Version]
- Abdallah, E.A.; Fanelli, M.F.; Souza E Silva, V.; Machado Netto, M.C.; Gasparini Junior, J.L.; Araújo, D.V.; Ocea, L.M.M.; Buim, M.E.C.; Tariki, M.S.; Alves, V.D.S.; et al. MRP1 expression in CTCs confers resistance to irinotecan-based chemotherapy in metastatic colorectal cancer. Int. J. Cancer 2016, 139, 890–898. [Google Scholar] [CrossRef] [PubMed]
- Lv, M.-M.; Zhu, X.-Y.; Chen, W.-X.; Zhong, S.; Hu, Q.; Ma, T.-F.; Zhang, J.; Chen, L.; Tang, J.-H.; Zhao, J.-H. Exosomes mediate drug resistance transfer in MCF-7 breast cancer cells and a probable mechanism is delivery of P-glycoprotein. Tumor Biol. 2014, 35, 10773–10779. [Google Scholar] [CrossRef]
- Corcoran, C.; Rani, S.; O’Driscoll, L.; O’Brien, K.; O’Neill, A.; Prencipe, M.; Sheikh, R.; Webb, G.; McDermott, R.; Watson, W.; et al. Docetaxel-resistance in prostate cancer: Evaluating associated phenotypic changes and potential for resistance transfer via exosomes. PLoS ONE 2012, 7, e50999. [Google Scholar] [CrossRef]
- Soekmadji, C.; Nelson, C.C. The Emerging Role of Extracellular Vesicle-Mediated Drug Resistance in Cancers: Implications in Advanced Prostate Cancer. BioMed Res. Int. 2015, 2015, 454837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mostafazadeh, M.; Samadi, N.; Kahroba, H.; Baradaran, B.; Haiaty, S.; Nouri, M. Potential roles and prognostic significance of exosomes in cancer drug resistance. Cell Biosci. 2021, 11, 1–15. [Google Scholar] [CrossRef]
- Ciravolo, V.; Huber, V.; Ghedini, G.C.; Venturelli, E.; Bianchi, F.; Campiglio, M.; Morelli, D.; Villa, A.; Della Mina, P.; Menard, S.; et al. Potential role of HER2-overexpressing exosomes in countering trastuzumab-based therapy. J. Cell. Physiol. 2012, 227, 658–667. [Google Scholar] [CrossRef] [PubMed]
- Mazel, M.; Jacot, W.; Pantel, K.; Bartkowiak, K.; Topart, D.; Cayrefourcq, L.; Rossille, D.; Maudelonde, T.; Fest, T.; Alix-Panabières, C. Frequent expression of PD-L1 on circulating breast cancer cells. Mol. Oncol. 2015, 9, 1773–1782. [Google Scholar] [CrossRef] [Green Version]
- Sinoquet, L.; Jacot, W.; Gauthier, L.; Pouderoux, S.; Viala, M.; Cayrefourcq, L.; Quantin, X.; Alix-Panabières, C. Programmed Cell Death Ligand 1-Expressing Circulating Tumor Cells: A New Prognostic Biomarker in Non-Small Cell Lung Cancer. Clin. Chem. 2021. [Google Scholar] [CrossRef] [PubMed]
- Zaslavsky, A.B.; Adams, M.P.; Cao, X.; Maj, T.; Choi, J.E.; Stangl-Kremser, J.; Patel, S.; Putelo, A.; Lee, S.K.; Nallandhighal, S.; et al. Platelet PD-L1 suppresses anti-cancer immune cell activity in PD-L1 negative tumors. Sci. Rep. 2020, 10, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Del Re, M.; van Schaik, R.H.N.; Fogli, S.; Mathijssen, R.H.J.; Cucchiara, F.; Capuano, A.; Scavone, C.; Jenster, G.W.; Danesi, R. Blood-based PD-L1 analysis in tumor-derived extracellular vesicles: Applications for optimal use of anti-PD-1/PD-L1 axis inhibitors. Biochim. Biophys. Acta Rev. Cancer 2021, 1875, 188463. [Google Scholar] [CrossRef]
- Chen, G.; Huang, A.C.; Zhang, W.; Zhang, G.; Wu, M.; Xu, W.; Yu, Z.; Yang, J.; Wang, B.; Sun, H.; et al. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature 2018, 560, 382–386. [Google Scholar] [CrossRef]
- Maheswaran, S.S.; Sequist, L.V.; Nagrath, S.; Ulkus, L.; Brannigan, B.; Collura, C.V.; Inserra, E.; Diederichs, S.; Iafrate, A.J.; Bell, D.W.; et al. Detection of mutations in EGFR in circulating lung-cancer cells. N. Engl. J. Med. 2008, 359, 366–377. [Google Scholar] [CrossRef] [Green Version]
- Sundaresan, T.K.; Sequist, L.V.; Heymach, J.V.; Riely, G.J.; Jänne, P.A.; Koch, W.H.; Sullivan, J.P.; Fox, D.B.; Maher, R.C.; Muzikansky, A.; et al. Detection of T790M, the Acquired Resistance EGFR Mutation, by Tumor Biopsy versus Noninvasive Blood-Based Analyses. Clin. Cancer Res. 2016, 22, 1103–1110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marchetti, A.; Del Grammastro, M.; Felicioni, L.; Malatesta, S.; Filice, G.; Centi, I.; De Pas, T.; Santoro, A.; Chella, A.; Brandes, A.A.; et al. Assessment of EGFR Mutations in Circulating Tumor Cell Preparations from NSCLC Patients by Next Generation Sequencing: Toward a Real-Time Liquid Biopsy for Treatment. PLoS ONE 2014, 9, e103883. [Google Scholar] [CrossRef]
- Castellanos-Rizaldos, E.; Grimm, D.; Tadigotla, V.; Hurley, J.; Healy, J.; Neal, P.L.; Sher, M.; Venkatesan, R.; Karlovich, C.; Raponi, M.; et al. Exosome-Based Detection of EGFR T790M in Plasma from Non–Small Cell Lung Cancer Patients. Clin. Cancer Res. 2018, 24, 2944–2950. [Google Scholar] [CrossRef] [Green Version]
- Krug, A.K.; Enderle, D.; Karlovich, C.; Priewasser, T.; Bentink, S.; Spiel, A.; Brinkmann, K.; Emenegger, J.; Grimm, D.G.; Castellanos-Rizaldos, E.; et al. Improved EGFR mutation detection using combined exosomal RNA and circulating tumor DNA in NSCLC patient plasma. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2018, 29, 700–706. [Google Scholar] [CrossRef] [Green Version]
- Skotland, T.; Sagini, K.; Sandvig, K.; Llorente, A. An emerging focus on lipids in extracellular vesicles. Adv. Drug Deliv. Rev. 2020, 159, 308–321. [Google Scholar] [CrossRef]
- Takimoto, C.; Chao, M.; Gibbs, C.; McCamish, M.; Liu, J.; Chen, J.; Majeti, R.; Weissman, I. The Macrophage ‘Do not eat me’ signal, CD47, is a clinically validated cancer immunotherapy target. Ann. Oncol. 2019, 30, 486–489. [Google Scholar] [CrossRef]
- Llorente, A.; Skotland, T.; Sylvänne, T.; Kauhanen, D.; Róg, T.; Orlowski, A.; Vattulainen, I.; Ekroos, K.; Sandvig, K. Molecular lipidomics of exosomes released by PC-3 prostate cancer cells. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2013, 1831, 1302–1309. [Google Scholar] [CrossRef] [PubMed]
- Skotland, T.; Sandvig, K.; Llorente, A. Lipids in exosomes: Current knowledge and the way forward. Prog. Lipid Res. 2017, 66, 30–41. [Google Scholar] [CrossRef]
- Min, H.K.; Lim, S.; Chung, B.C.; Moon, M.H. Shotgun lipidomics for candidate biomarkers of urinary phospholipids in prostate cancer. Anal. Bioanal. Chem. 2010, 399, 823–830. [Google Scholar] [CrossRef]
- Kim, H.; Min, H.K.; Kong, G.; Moon, M.H. Quantitative analysis of phosphatidylcholines and phosphatidylethanolamines in urine of patients with breast cancer by nanoflow liquid chromatography/tandem mass spectrometry. Anal. Bioanal. Chem. 2009, 393, 1649–1656. [Google Scholar] [CrossRef]
- Del Boccio, P.; Raimondo, F.; Pieragostino, D.; Morosi, L.; Cozzi, G.; Sacchetta, P.; Magni, F.; Pitto, M.; Urbani, A. A hyphenated microLC-Q-TOF-MS platform for exosomal lipidomics investigations: Application to RCC urinary exosomes. Electrophoresis 2012, 33, 689–696. [Google Scholar] [CrossRef]
- Kowal, J.; Arras, G.; Colombo, M.; Jouve, M.; Morath, J.P.; Primdal-Bengtson, B.; Dingli, F.; Loew, D.; Tkach, M.; Théry, C. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. USA 2016, 113, E968–E977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sódar, B.W.; Kittel, Á.; Pálóczi, K.; Vukman, K.V.; Osteikoetxea, X.; Szabó-Taylor, K.; Németh, A.; Sperlágh, B.; Baranyai, T.; Giricz, Z.; et al. Low-density lipoprotein mimics blood plasma-derived exosomes and microvesicles during isolation and detection. Sci. Rep. 2016, 6, 24316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hessvik, N.P.; Øverbye, A.; Brech, A.; Torgersen, M.L.; Jakobsen, I.S.; Sandvig, K.; Llorente, A. PIKfyve inhibition increases exosome release and induces secretory autophagy. Cell. Mol. Life Sci. 2016, 73, 4717–4737. [Google Scholar] [CrossRef] [PubMed]
- Maheswaran, S.; Haber, D.A. Ex Vivo Culture of CTCs: An Emerging Resource to Guide Cancer Therapy. Cancer Res. 2015, 75, 2411–2415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pantel, K.; Alix-Panabières, C. Liquid biopsy and minimal residual disease—Latest advances and implications for cure. Nat. Rev. Clin. Oncol. 2019, 16, 409–424. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.-Y.; Lee, K.-M.; Kim, S.-H.; Kwon, Y.-J.; Chun, Y.-J.; Choi, H.-K. Comparative metabolic and lipidomic profiling of human breast cancer cells with different metastatic potentials. Oncotarget 2016, 7, 67111–67128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naik, J.; Hau, C.M.; Bloemendaal, L.T.; Mok, K.S.; Hajji, N.; Wehman, A.M.; Meisner, S.; Muncan, V.; Paauw, N.J.; De Vries, H.E.; et al. The P4-ATPase ATP9A is a novel determinant of exosome release. PLoS ONE 2019, 14, e0213069. [Google Scholar] [CrossRef] [PubMed]
- Nagata, S.; Suzuki, J.; Segawa, K.; Fujii, T. Exposure of phosphatidylserine on the cell surface. Cell Death Differ. 2016, 23, 952–961. [Google Scholar] [CrossRef] [Green Version]
- Utsugi, T.; Schroit, A.J.; Connor, J.; Bucana, C.D.; Fidler, I.J. Elevated expression of phosphatidylserine in the outer membrane leaflet of human tumor cells and recognition by activated human blood monocytes. Cancer Res. 1991, 51, 3062–3066. [Google Scholar] [PubMed]
- Riedl, S.; Rinner, B.; Asslaber, M.; Schaider, H.; Walzer, S.M.; Novak, A.; Lohner, K.; Zweytick, D. In search of a novel target—Phosphatidylserine exposed by non-apoptotic tumor cells and metastases of malignancies with poor treatment efficacy. Biochim. Biophys. Acta Biomembr. 2011, 1808, 2638–2645. [Google Scholar] [CrossRef] [Green Version]
- Lentz, B.R. Exposure of platelet membrane phosphatidylserine regulates blood coagulation. Prog. Lipid Res. 2003, 42, 423–438. [Google Scholar] [CrossRef]
- Koseoglu, S.; Meyer, A.F.; Kim, D.; Meyer, B.M.; Wang, Y.; Dalluge, J.J.; Haynes, C.L. Analytical Characterization of the Role of Phospholipids in Platelet Adhesion and Secretion. Anal. Chem. 2015, 87, 413–421. [Google Scholar] [CrossRef] [Green Version]
- Szczerba, B.M.; Castro-Giner, F.; Vetter, M.; Krol, I.; Gkountela, S.; Landin, J.; Scheidmann, M.C.; Donato, C.; Scherrer, R.; Singer, J.; et al. Neutrophils escort circulating tumour cells to enable cell cycle progression. Nat. Cell Biol. 2019, 566, 553–557. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Wysocki, R.; Amoozgar, Z.; Maiorino, L.; Fein, M.R.; Jorns, J.; Schott, A.F.; Kinugasa-Katayama, Y.; Lee, Y.; Won, N.H.; et al. Cancer cells induce metastasis-supporting neutrophil extracellular DNA traps. Sci. Transl. Med. 2016, 8, 361ra138. [Google Scholar] [CrossRef] [Green Version]
- Tohme, S.; Yazdani, H.O.; Al-Khafaji, A.B.; Chidi, A.P.; Loughran, P.; Mowen, K.A.; Wang, Y.; Simmons, R.L.; Huang, H.; Tsung, A. Neutrophil extracellular traps promote the development and progression of liver metastases after surgical stress. Cancer Res. 2016, 76, 1367–1380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cools-Lartigue, J.; Spicer, J.; McDonald, B.; Gowing, S.; Chow, S.; Giannias, B.; Bourdeau, F.; Kubes, P.; Ferri, L. Neutrophil extracellular traps sequester circulating tumor cells and promote metastasis. J. Clin. Investig. 2013, 123, 3446–3458. [Google Scholar] [CrossRef] [PubMed]
- Zucoloto, A.Z.; Jenne, C.N. Platelet-Neutrophil Interplay: Insights into Neutrophil Extracellular Trap (NET)-Driven Coagulation in Infection. Front. Cardiovasc. Med. 2019, 6, 85. [Google Scholar] [CrossRef] [Green Version]
- Dyer, M.R.; Chen, Q.; Haldeman, S.; Yazdani, H.; Hoffman, R.; Loughran, P.; Tsung, A.; Zuckerbraun, B.S.; Simmons, R.L.; Neal, M.D. Deep vein thrombosis in mice is regulated by platelet HMGB1 through release of neutrophil-extracellular traps and DNA. Sci. Rep. 2018, 8, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Németh, A.; Orgovan, N.; Sódar, B.W.; Osteikoetxea, X.; Pálóczi, K.; Szabó-Taylor, K.É.; Vukman, K.V.; Kittel, Á.; Turiák, L.; Wiener, Z.; et al. Antibiotic-induced release of small extracellular vesicles (exosomes) with surface-associated DNA. Sci. Rep. 2017, 7, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Malkin, E.Z.; Bratman, S. V Bioactive DNA from extracellular vesicles and particles. Cell Death Dis. 2020, 11, 584. [Google Scholar] [CrossRef]
- Huang, N.; Fan, X.; Zaleta-Rivera, K.; Nguyen, T.C.; Zhou, J.; Luo, Y.; Gao, J.; Fang, R.H.; Yan, Z.; Chen, Z.B.; et al. Natural display of nuclear-encoded RNA on the cell surface and its impact on cell interaction. Genome Biol. 2020, 21, 1–23. [Google Scholar] [CrossRef]
- Flynn, R.A.; Pedram, K.; Malaker, S.A.; Batista, P.J.; Smith, B.A.; Johnson, A.G.; George, B.M.; Majzoub, K.; Villalta, P.W.; Carette, J.E.; et al. Small RNAs are modified with N-glycans and displayed on the surface of living cells. Cell 2021, 184, 3109–3124.e22. [Google Scholar] [CrossRef] [PubMed]
- Pantel, K.; Hille, C.; Scher, H.I. Circulating Tumor Cells in Prostate Cancer: From Discovery to Clinical Utility. Clin. Chem. 2019, 65, 87–99. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Freitas, D.; Kim, H.S.; Fabijanic, K.; Li, Z.; Chen, H.; Mark, M.T.; Molina, H.; Benito-Martin, A.; Bojmar, L.; et al. Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field-flow fractionation. Nat. Cell Biol. 2018, 20, 332–343. [Google Scholar] [CrossRef] [PubMed]
- Cheung, K.J.; Padmanaban, V.; Silvestri, V.; Schipper, K.; Cohen, J.D.; Fairchild, A.N.; Gorin, M.A.; Verdone, J.E.; Pienta, K.J.; Bader, J.S.; et al. Polyclonal breast cancer metastases arise from collective dissemination of keratin 14-expressing tumor cell clusters. Proc. Natl. Acad. Sci. USA 2016, 113, E854–E863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aceto, N.; Bardia, A.; Miyamoto, D.T.; Donaldson, M.C.; Wittner, B.S.; Spencer, J.A.; Yu, M.; Pely, A.; Engstrom, A.; Zhu, H.; et al. Circulating Tumor Cell Clusters Are Oligoclonal Precursors of Breast Cancer Metastasis. Cell 2014, 158, 1110–1122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gkountela, S.; Castro-Giner, F.; Szczerba, B.M.; Vetter, M.; Landin, J.; Scherrer, R.; Krol, I.; Scheidmann, M.C.; Beisel, C.; Stirnimann, C.; et al. Circulating Tumor Cell Clustering Shapes DNA Methylation to Enable Metastasis Seeding. Cell 2019, 176, 98–112.e14. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cortés-Hernández, L.E.; Eslami-S, Z.; Costa-Silva, B.; Alix-Panabières, C. Current Applications and Discoveries Related to the Membrane Components of Circulating Tumor Cells and Extracellular Vesicles. Cells 2021, 10, 2221. https://doi.org/10.3390/cells10092221
Cortés-Hernández LE, Eslami-S Z, Costa-Silva B, Alix-Panabières C. Current Applications and Discoveries Related to the Membrane Components of Circulating Tumor Cells and Extracellular Vesicles. Cells. 2021; 10(9):2221. https://doi.org/10.3390/cells10092221
Chicago/Turabian StyleCortés-Hernández, Luis Enrique, Zahra Eslami-S, Bruno Costa-Silva, and Catherine Alix-Panabières. 2021. "Current Applications and Discoveries Related to the Membrane Components of Circulating Tumor Cells and Extracellular Vesicles" Cells 10, no. 9: 2221. https://doi.org/10.3390/cells10092221
APA StyleCortés-Hernández, L. E., Eslami-S, Z., Costa-Silva, B., & Alix-Panabières, C. (2021). Current Applications and Discoveries Related to the Membrane Components of Circulating Tumor Cells and Extracellular Vesicles. Cells, 10(9), 2221. https://doi.org/10.3390/cells10092221






