Functional Roles of FGF Signaling in Early Development of Vertebrate Embryos
Abstract
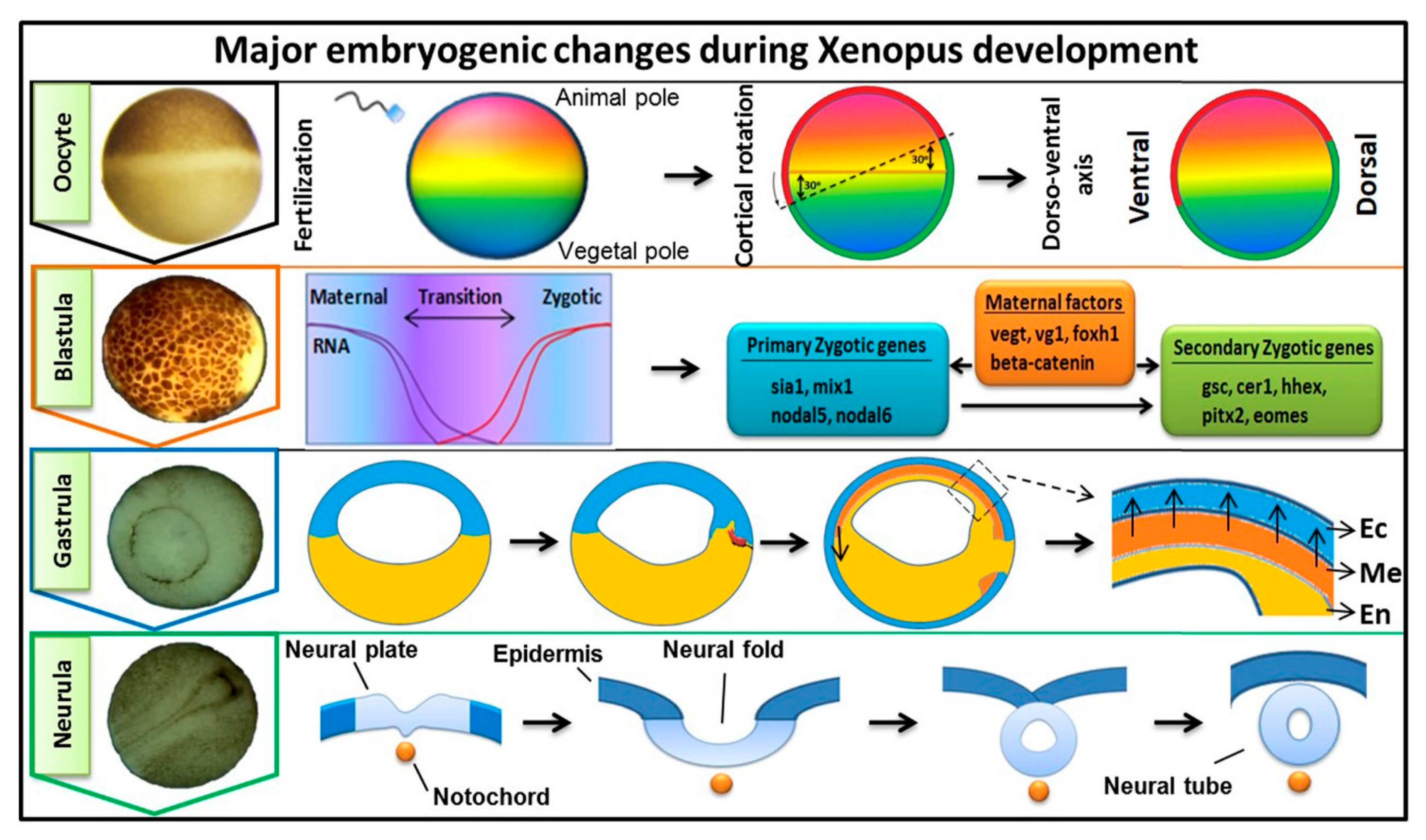
1. Introduction
2. FGF and FGFR Families and Signal Transduction
2.1. FGF Ligands
2.2. FGF Receptors
2.3. An Overview of FGFR Signaling Pathways
| Human | Mouse | Xenopus | ||||
|---|---|---|---|---|---|---|
| Subfamily | Members (Other Name) | Ref. | Members (Other Name) | Ref. | Members (Other Name) | Ref. |
| FGF1 | FGF1 (aFGF) | [31] | FGF1 (FGFa) | [32] | xFGF1 | [33] |
| FGF2 (bFGF/FGF-β) | [4] | FGF2 | [34] | xFGF2 (bFGF) | [35] | |
| FGF4 | FGF4 (eFGF) | [36] | FGF4 (KFGF) | [37] | xFGF4 (eFGF, fgf4-a, fgf4-b) | [38] |
| FGF5 | [39] | FGF5 | [40] | xFGF5 | [33,41] | |
| FGF6 (HST2, HSTF2) | [42] | FGF6 (HSTF2) | [43] | xFGF6 | [33,41] | |
| FGF7 | FGF3 | [44] | FGF3 (Int-2) | [45] | xFGF3 (INT-2, FGF3A) | [46] |
| FGF7 (KGF) | [47] | FGF7 (KGF) | [48] | * | ||
| FGF10 | [49] | FGF10 | [50] | xFGF10 | [51] | |
| FGF22 (UNQ2500/PRO5800) | [52] | FGF22 | [52] | xFGF22 | [41] | |
| FGF8 | FGF8 (AIGF) | [53] | FGF8 (AIGF) | [54] | xFGF8 (FGF8a, FGF8b) | [55] |
| FGF17 (UNQ161/PRO187) | [56] | FGF17 | [56] | * | ||
| FGF18 (UNQ420/PRO856) | [57] | FGF18 | [57] | * | ||
| FGF9 | FGF9 | [58] | FGF9 | [59] | xFGF9 (GAF, HBGF9) | [60] |
| FGF16 | [61] | FGF16 | [62] | xFGF16 | [41] | |
| FGF20 | [63] | FGF20 | [64] | xFGF20 | [65] | |
| FGF11 | FGF11 (FHF3) | [66] | FGF11 (FHF3) | [66] | xFGF11 | [41] |
| FGF12 (FGF12B, FHF1) | [66] | FGF12 (FHF1) | [67] | xFGF12 | [68] | |
| FGF13 (FHF2) | [69] | FGF13 (FHF2) | [67] | xFGF13 | [70] | |
| FGF14 (FHF4) | [71] | FGF14 (FHF4) | [72] | xFGF14 (FHF4) | [33] | |
| FGF19 | * | FGF15 | [73] | * | ||
| FGF19 (UNQ334/PRO533) | [74] | * | xFGF19 | [41] | ||
| FGF21 (UNQ3115/PRO10196) | [75] | FGF21 | [75] | * | ||
| FGF23 (HYPF, UNQ3027/PRO9828) | [76] | FGF23 | [76] | xFGF23 (fgf23.1, FGF23.2) | [41] | |
| FGF receptor family | ||||||
| FGFR1 | [31] | FGFR1 | [77] | xFGFR1 | [78] | |
| FGFR2 | [79] | FGFR2 | [80] | xFGFR2 | [81] | |
| FGFR3 | [82] | FGFR3 | [83] | xFGFR3 | [84] | |
| FGFR4 | [85] | FGFR4 | [86] | xFGFR4 | [87] | |
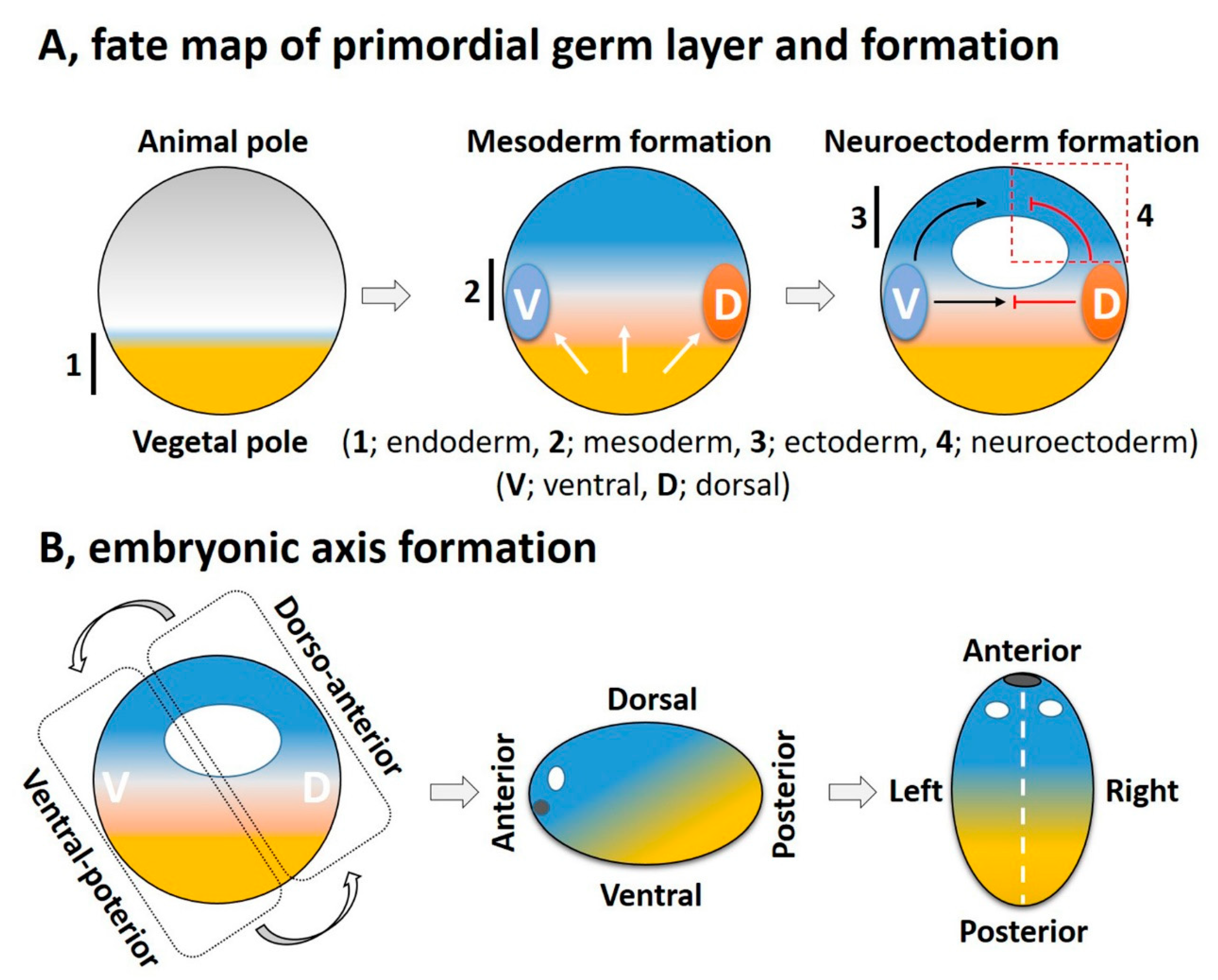
3. FGF Signaling in Embryonic Germ Layer Formation
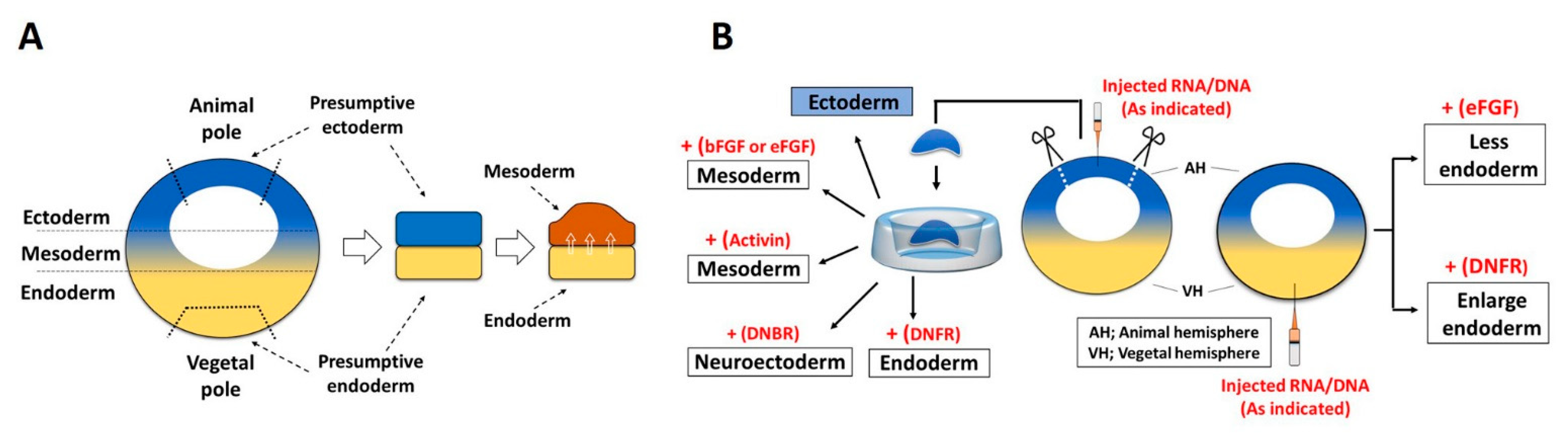
3.1. Role of FGF Signaling in Endoderm Formation
3.2. FGF in Mesoderm Induction
3.3. FGF in Ectoderm Specification
3.4. Role of FGF in Neural Induction and Neuroectoderm Formation
4. FGF Signaling Integrates with Other Signaling Pathways in a Crosstalk for Embryonic Axis Formation
4.1. The DV Axis
4.2. The AP Axis
4.3. The LR Axis
5. Regulation of FGF Signaling
5.1. Endogenous Protein Activators
5.2. Endogenous Protein Inhibitors
5.3. FGF Modulators in Developmental Biology
6. Conclusions and Prospects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bottcher, R.T.; Niehrs, C. Fibroblast growth factor signaling during early vertebrate development. Endocr. Rev. 2005, 26, 63–77. [Google Scholar] [CrossRef]
- Dorey, K.; Amaya, E. FGF signalling: Diverse roles during early vertebrate embryogenesis. Development 2010, 137, 3731–3742. [Google Scholar] [CrossRef]
- Pownall, M.E.; Isaacs, H.V. FGF signalling in vertebrate development. Colloq. Ser. Dev. Biol. 2010, 1, 1–75. [Google Scholar] [CrossRef]
- Florkiewicz, R.Z.; Shibata, F.; Barankiewicz, T.; Baird, A.; Gonzalez, A.M.; Florkiewicz, E.; Shah, N. Basic fibroblast growth factor gene expression. Ann. N. Y. Acad. Sci. 1991, 638, 109–126. [Google Scholar] [CrossRef]
- Xie, Y.; Su, N.; Yang, J.; Tan, Q.; Huang, S.; Jin, M.; Ni, Z.; Zhang, B.; Zhang, D.; Luo, F.; et al. FGF/FGFR signaling in health and disease. Signal. Transduct. Target. Ther. 2020, 5, 181. [Google Scholar] [CrossRef]
- Kato, S.; Sekine, K. FGF-FGFR signaling in vertebrate organogenesis. Cell Mol. Biol. 1999, 45, 631–638. [Google Scholar] [PubMed]
- Katoh, M.; Nakagama, H. FGF receptors: Cancer biology and therapeutics. Med. Res. Rev. 2014, 34, 280–300. [Google Scholar] [CrossRef] [PubMed]
- Coumoul, X.; Deng, C.X. Roles of FGF receptors in mammalian development and congenital diseases. Birth Defects Res. C Embryo Today 2003, 69, 286–304. [Google Scholar] [CrossRef] [PubMed]
- Moosa, S.; Wollnik, B. Altered FGF signalling in congenital craniofacial and skeletal disorders. Semin. Cell Dev. Biol. 2016, 53, 115–125. [Google Scholar] [CrossRef]
- Itoh, N.; Ornitz, D.M. Evolution of the Fgf and Fgfr gene families. Trends Genet. 2004, 20, 563–569. [Google Scholar] [CrossRef]
- Ornitz, D.M.; Itoh, N. Fibroblast growth factors. Genome Biol. 2001, 2, REVIEWS3005. [Google Scholar] [CrossRef] [PubMed]
- Hui, Q.; Jin, Z.; Li, X.; Liu, C.; Wang, X. FGF Family: From Drug Development to Clinical Application. Int. J. Mol. Sci. 2018, 19, 1875. [Google Scholar] [CrossRef]
- Olsen, S.K.; Garbi, M.; Zampieri, N.; Eliseenkova, A.V.; Ornitz, D.M.; Goldfarb, M.; Mohammadi, M. Fibroblast growth factor (FGF) homologous factors share structural but not functional homology with FGFs. J. Biol. Chem. 2003, 278, 34226–34236. [Google Scholar] [CrossRef] [PubMed]
- Schoorlemmer, J.; Goldfarb, M. Fibroblast growth factor homologous factors are intracellular signaling proteins. Curr. Biol. 2001, 11, 793–797. [Google Scholar] [CrossRef]
- Yan, H.; Pablo, J.L.; Pitt, G.S. FGF14 regulates presynaptic Ca2+ channels and synaptic transmission. Cell Rep. 2013, 4, 66–75. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.F.; Yang, L.; Li, S.; Wang, Q.; Yuan, X.B.; Gao, X.; Bao, L.; Zhang, X. Fibroblast growth factor 13 is a microtubule-stabilizing protein regulating neuronal polarization and migration. Cell 2012, 149, 1549–1564. [Google Scholar] [CrossRef]
- Ornitz, D.M.; Xu, J.; Colvin, J.S.; McEwen, D.G.; MacArthur, C.A.; Coulier, F.; Gao, G.; Goldfarb, M. Receptor specificity of the fibroblast growth factor family. J. Biol. Chem. 1996, 271, 15292–15297. [Google Scholar] [CrossRef]
- Givol, D.; Yayon, A. Complexity of FGF receptors: Genetic basis for structural diversity and functional specificity. FASEB J. 1992, 6, 3362–3369. [Google Scholar] [CrossRef]
- Farrell, B.; Breeze, A.L. Structure, activation and dysregulation of fibroblast growth factor receptor kinases: Perspectives for clinical targeting. Biochem. Soc. Trans. 2018, 46, 1753–1770. [Google Scholar] [CrossRef]
- Brown, A.; Robinson, C.J.; Gallagher, J.T.; Blundell, T.L. Cooperative heparin-mediated oligomerization of fibroblast growth factor-1 (FGF1) precedes recruitment of FGFR2 to ternary complexes. Biophys. J. 2013, 104, 1720–1730. [Google Scholar] [CrossRef] [PubMed]
- Sarabipour, S.; Hristova, K. Mechanism of FGF receptor dimerization and activation. Nat. Commun. 2016, 7, 10262. [Google Scholar] [CrossRef] [PubMed]
- Werner, S.; Duan, D.S.; de Vries, C.; Peters, K.G.; Johnson, D.E.; Williams, L.T. Differential splicing in the extracellular region of fibroblast growth factor receptor 1 generates receptor variants with different ligand-binding specificities. Mol. Cell Biol. 1992, 12, 82–88. [Google Scholar] [CrossRef] [PubMed]
- Latko, M.; Czyrek, A.; Porebska, N.; Kucinska, M.; Otlewski, J.; Zakrzewska, M.; Opalinski, L. Cross-Talk between Fibroblast Growth Factor Receptors and Other Cell Surface Proteins. Cells 2019, 8, 455. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Tang, P.M.K.; Zhou, Y.; Cheng, A.S.L.; Yu, J.; Kang, W.; To, K.F. Targeting the Oncogenic FGF-FGFR Axis in Gastric Carcinogenesis. Cells 2019, 8, 637. [Google Scholar] [CrossRef]
- Zhang, Y.; McKeehan, K.; Lin, Y.; Zhang, J.; Wang, F. Fibroblast growth factor receptor 1 (FGFR1) tyrosine phosphorylation regulates binding of FGFR substrate 2alpha (FRS2alpha) but not FRS2 to the receptor. Mol. Endocrinol. 2008, 22, 167–175. [Google Scholar] [CrossRef]
- Goetz, R.; Mohammadi, M. Exploring mechanisms of FGF signalling through the lens of structural biology. Nat. Rev. Mol. Cell Biol. 2013, 14, 166–180. [Google Scholar] [CrossRef]
- Dey, J.H.; Bianchi, F.; Voshol, J.; Bonenfant, D.; Oakeley, E.J.; Hynes, N.E. Targeting fibroblast growth factor receptors blocks PI3K/AKT signaling, induces apoptosis, and impairs mammary tumor outgrowth and metastasis. Cancer Res. 2010, 70, 4151–4162. [Google Scholar] [CrossRef]
- Turner, N.; Grose, R. Fibroblast growth factor signalling: From development to cancer. Nat. Rev. Cancer 2010, 10, 116–129. [Google Scholar] [CrossRef]
- Eswarakumar, V.P.; Lax, I.; Schlessinger, J. Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev. 2005, 16, 139–149. [Google Scholar] [CrossRef]
- Tsang, M.; Dawid, I.B. Promotion and attenuation of FGF signaling through the Ras-MAPK pathway. Sci. STKE 2004, 2004, pe17. [Google Scholar] [CrossRef]
- Dionne, C.A.; Crumley, G.; Bellot, F.; Kaplow, J.M.; Searfoss, G.; Ruta, M.; Burgess, W.H.; Jaye, M.; Schlessinger, J. Cloning and expression of two distinct high-affinity receptors cross-reacting with acidic and basic fibroblast growth factors. EMBO J. 1990, 9, 2685–2692. [Google Scholar] [CrossRef] [PubMed]
- Madiai, F.; Hackshaw, K.V.; Chiu, I.M. Cloning and characterization of the mouse Fgf-1 gene. Gene 1996, 179, 231–236. [Google Scholar] [CrossRef]
- Lea, R.; Papalopulu, N.; Amaya, E.; Dorey, K. Temporal and spatial expression of FGF ligands and receptors during Xenopus development. Dev. Dyn. 2009, 238, 1467–1479. [Google Scholar] [CrossRef]
- Hebert, J.M.; Basilico, C.; Goldfarb, M.; Haub, O.; Martin, G.R. Isolation of cDNAs encoding four mouse FGF family members and characterization of their expression patterns during embryogenesis. Dev. Biol. 1990, 138, 454–463. [Google Scholar] [CrossRef]
- Kimelman, D.; Kirschner, M. Synergistic induction of mesoderm by FGF and TGF-beta and the identification of an mRNA coding for FGF in the early Xenopus embryo. Cell 1987, 51, 869–877. [Google Scholar] [CrossRef]
- Feldman, B.; Poueymirou, W.; Papaioannou, V.E.; DeChiara, T.M.; Goldfarb, M. Requirement of FGF-4 for postimplantation mouse development. Science 1995, 267, 246–249. [Google Scholar] [CrossRef]
- Brookes, S.; Smith, R.; Thurlow, J.; Dickson, C.; Peters, G. The mouse homologue of hst/k-FGF: Sequence, genome organization and location relative to int-2. Nucleic Acids Res. 1989, 17, 4037–4045. [Google Scholar] [CrossRef]
- Isaacs, H.V.; Tannahill, D.; Slack, J.M. Expression of a novel FGF in the Xenopus embryo. A new candidate inducing factor for mesoderm formation and anteroposterior specification. Development 1992, 114, 711–720. [Google Scholar] [CrossRef] [PubMed]
- Zhan, X.; Bates, B.; Hu, X.G.; Goldfarb, M. The human FGF-5 oncogene encodes a novel protein related to fibroblast growth factors. Mol. Cell Biol. 1988, 8, 3487–3495. [Google Scholar] [CrossRef]
- Ozawa, K.; Suzuki, S.; Asada, M.; Tomooka, Y.; Li, A.J.; Yoneda, A.; Komi, A.; Imamura, T. An alternatively spliced fibroblast growth factor (FGF)-5 mRNA is abundant in brain and translates into a partial agonist/antagonist for FGF-5 neurotrophic activity. J. Biol. Chem. 1998, 273, 29262–29271. [Google Scholar] [CrossRef]
- Session, A.M.; Uno, Y.; Kwon, T.; Chapman, J.A.; Toyoda, A.; Takahashi, S.; Fukui, A.; Hikosaka, A.; Suzuki, A.; Kondo, M.; et al. Genome evolution in the allotetraploid frog Xenopus laevis. Nature 2016, 538, 336–343. [Google Scholar] [CrossRef]
- Iida, S.; Yoshida, T.; Naito, K.; Sakamoto, H.; Katoh, O.; Hirohashi, S.; Sato, T.; Onda, M.; Sugimura, T.; Terada, M. Human hst-2 (FGF-6) oncogene: cDNA cloning and characterization. Oncogene 1992, 7, 303–309. [Google Scholar]
- De Lapeyriere, O.; Rosnet, O.; Benharroch, D.; Raybaud, F.; Marchetto, S.; Planche, J.; Galland, F.; Mattei, M.G.; Copeland, N.G.; Jenkins, N.A.; et al. Structure, chromosome mapping and expression of the murine Fgf-6 gene. Oncogene 1990, 5, 823–831. [Google Scholar] [PubMed]
- Brookes, S.; Smith, R.; Casey, G.; Dickson, C.; Peters, G. Sequence organization of the human int-2 gene and its expression in teratocarcinoma cells. Oncogene 1989, 4, 429–436. [Google Scholar] [PubMed]
- Dickson, C.; Acland, P.; Smith, R.; Dixon, M.; Deed, R.; MacAllan, D.; Walther, W.; Fuller-Pace, F.; Kiefer, P.; Peters, G. Characterization of int-2: A member of the fibroblast growth factor family. J. Cell Sci. Suppl. 1990, 13, 87–96. [Google Scholar] [CrossRef]
- Kiefer, P.; Mathieu, M.; Close, M.J.; Peters, G.; Dickson, C. FGF3 from Xenopus laevis. EMBO J. 1993, 12, 4159–4168. [Google Scholar] [CrossRef]
- Finch, P.W.; Rubin, J.S.; Miki, T.; Ron, D.; Aaronson, S.A. Human KGF is FGF-related with properties of a paracrine effector of epithelial cell growth. Science 1989, 245, 752–755. [Google Scholar] [CrossRef] [PubMed]
- Mason, I.J.; Fuller-Pace, F.; Smith, R.; Dickson, C. FGF-7 (keratinocyte growth factor) expression during mouse development suggests roles in myogenesis, forebrain regionalisation and epithelial-mesenchymal interactions. Mech. Dev. 1994, 45, 15–30. [Google Scholar] [CrossRef]
- Emoto, H.; Tagashira, S.; Mattei, M.G.; Yamasaki, M.; Hashimoto, G.; Katsumata, T.; Negoro, T.; Nakatsuka, M.; Birnbaum, D.; Coulier, F.; et al. Structure and expression of human fibroblast growth factor-10. J. Biol. Chem. 1997, 272, 23191–23194. [Google Scholar] [CrossRef] [PubMed]
- Tagashira, S.; Harada, H.; Katsumata, T.; Itoh, N.; Nakatsuka, M. Cloning of mouse FGF10 and up-regulation of its gene expression during wound healing. Gene 1997, 197, 399–404. [Google Scholar] [CrossRef]
- Yokoyama, H.; Ide, H.; Tamura, K. FGF-10 stimulates limb regeneration ability in Xenopus laevis. Dev. Biol. 2001, 233, 72–79. [Google Scholar] [CrossRef] [PubMed]
- Nakatake, Y.; Hoshikawa, M.; Asaki, T.; Kassai, Y.; Itoh, N. Identification of a novel fibroblast growth factor, FGF-22, preferentially expressed in the inner root sheath of the hair follicle. Biochim. Biophys. Acta 2001, 1517, 460–463. [Google Scholar] [CrossRef]
- Tanaka, A.; Miyamoto, K.; Matsuo, H.; Matsumoto, K.; Yoshida, H. Human androgen-induced growth factor in prostate and breast cancer cells: Its molecular cloning and growth properties. FEBS Lett. 1995, 363, 226–230. [Google Scholar] [CrossRef]
- Tanaka, A.; Miyamoto, K.; Minamino, N.; Takeda, M.; Sato, B.; Matsuo, H.; Matsumoto, K. Cloning and characterization of an androgen-induced growth factor essential for the androgen-dependent growth of mouse mammary carcinoma cells. Proc. Natl. Acad. Sci. USA 1992, 89, 8928–8932. [Google Scholar] [CrossRef]
- Fletcher, R.B.; Baker, J.C.; Harland, R.M. FGF8 spliceforms mediate early mesoderm and posterior neural tissue formation in Xenopus. Development 2006, 133, 1703–1714. [Google Scholar] [CrossRef]
- Hoshikawa, M.; Ohbayashi, N.; Yonamine, A.; Konishi, M.; Ozaki, K.; Fukui, S.; Itoh, N. Structure and expression of a novel fibroblast growth factor, FGF-17, preferentially expressed in the embryonic brain. Biochem. Biophys. Res. Commun. 1998, 244, 187–191. [Google Scholar] [CrossRef]
- Hu, M.C.; Qiu, W.R.; Wang, Y.P.; Hill, D.; Ring, B.D.; Scully, S.; Bolon, B.; DeRose, M.; Luethy, R.; Simonet, W.S.; et al. FGF-18, a novel member of the fibroblast growth factor family, stimulates hepatic and intestinal proliferation. Mol. Cell Biol. 1998, 18, 6063–6074. [Google Scholar] [CrossRef]
- Miyamoto, M.; Naruo, K.; Seko, C.; Matsumoto, S.; Kondo, T.; Kurokawa, T. Molecular cloning of a novel cytokine cDNA encoding the ninth member of the fibroblast growth factor family, which has a unique secretion property. Mol. Cell Biol. 1993, 13, 4251–4259. [Google Scholar] [CrossRef]
- Santos-Ocampo, S.; Colvin, J.S.; Chellaiah, A.; Ornitz, D.M. Expression and biological activity of mouse fibroblast growth factor-9. J. Biol. Chem. 1996, 271, 1726–1731. [Google Scholar] [CrossRef]
- Song, J.; Slack, J.M. XFGF-9: A new fibroblast growth factor from Xenopus embryos. Dev. Dyn. 1996, 206, 427–436. [Google Scholar] [CrossRef]
- Miyake, A.; Konishi, M.; Martin, F.H.; Hernday, N.A.; Ozaki, K.; Yamamoto, S.; Mikami, T.; Arakawa, T.; Itoh, N. Structure and expression of a novel member, FGF-16, on the fibroblast growth factor family. Biochem. Biophys. Res. Commun. 1998, 243, 148–152. [Google Scholar] [CrossRef]
- Sontag, D.P.; Cattini, P.A. Cloning and bacterial expression of postnatal mouse heart FGF-16. Mol. Cell Biochem. 2003, 242, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Kirikoshi, H.; Sagara, N.; Saitoh, T.; Tanaka, K.; Sekihara, H.; Shiokawa, K.; Katoh, M. Molecular cloning and characterization of human FGF-20 on chromosome 8p21.3-p22. Biochem. Biophys. Res. Commun. 2000, 274, 337–343. [Google Scholar] [CrossRef]
- Hajihosseini, M.K.; Heath, J.K. Expression patterns of fibroblast growth factors-18 and -20 in mouse embryos is suggestive of novel roles in calvarial and limb development. Mech. Dev. 2002, 113, 79–83. [Google Scholar] [CrossRef]
- Koga, C.; Adati, N.; Nakata, K.; Mikoshiba, K.; Furuhata, Y.; Sato, S.; Tei, H.; Sakaki, Y.; Kurokawa, T.; Shiokawa, K.; et al. Characterization of a novel member of the FGF family, XFGF-20, in Xenopus laevis. Biochem. Biophys. Res. Commun. 1999, 261, 756–765. [Google Scholar] [CrossRef]
- Smallwood, P.M.; Munoz-Sanjuan, I.; Tong, P.; Macke, J.P.; Hendry, S.H.; Gilbert, D.J.; Copeland, N.G.; Jenkins, N.A.; Nathans, J. Fibroblast growth factor (FGF) homologous factors: New members of the FGF family implicated in nervous system development. Proc. Natl. Acad. Sci. USA 1996, 93, 9850–9857. [Google Scholar] [CrossRef]
- Hartung, H.; Feldman, B.; Lovec, H.; Coulier, F.; Birnbaum, D.; Goldfarb, M. Murine FGF-12 and FGF-13: Expression in embryonic nervous system, connective tissue and heart. Mech. Dev. 1997, 64, 31–39. [Google Scholar] [CrossRef]
- Fukui, L.; Henry, J.J. FGF signaling is required for lens regeneration in Xenopus laevis. Biol. Bull. 2011, 221, 137–145. [Google Scholar] [CrossRef]
- Gecz, J.; Baker, E.; Donnelly, A.; Ming, J.E.; McDonald-McGinn, D.M.; Spinner, N.B.; Zackai, E.H.; Sutherland, G.R.; Mulley, J.C. Fibroblast growth factor homologous factor 2 (FHF2): Gene structure, expression and mapping to the Borjeson-Forssman-Lehmann syndrome region in Xq26 delineated by a duplication breakpoint in a BFLS-like patient. Hum. Genet. 1999, 104, 56–63. [Google Scholar] [CrossRef]
- Nishimoto, S.; Nishida, E. Fibroblast growth factor 13 is essential for neural differentiation in Xenopus early embryonic development. J. Biol. Chem. 2007, 282, 24255–24261. [Google Scholar] [CrossRef]
- Goetz, R.; Dover, K.; Laezza, F.; Shtraizent, N.; Huang, X.; Tchetchik, D.; Eliseenkova, A.V.; Xu, C.F.; Neubert, T.A.; Ornitz, D.M.; et al. Crystal structure of a fibroblast growth factor homologous factor (FHF) defines a conserved surface on FHFs for binding and modulation of voltage-gated sodium channels. J. Biol. Chem. 2009, 284, 17883–17896. [Google Scholar] [CrossRef]
- Yamamoto, S.; Mikami, T.; Konishi, M.; Itoh, N. Stage-specific expression of a novel isoform of mouse FGF-14 (FHF-4) in spermatocytes. Biochim. Biophys. Acta 2000, 1490, 121–124. [Google Scholar] [CrossRef]
- Vergnes, L.; Lee, J.M.; Chin, R.G.; Auwerx, J.; Reue, K. Diet1 functions in the FGF15/19 enterohepatic signaling axis to modulate bile acid and lipid levels. Cell Metab. 2013, 17, 916–928. [Google Scholar] [CrossRef]
- Nishimura, T.; Utsunomiya, Y.; Hoshikawa, M.; Ohuchi, H.; Itoh, N. Structure and expression of a novel human FGF, FGF-19, expressed in the fetal brain. Biochim. Biophys. Acta 1999, 1444, 148–151. [Google Scholar] [CrossRef]
- Nishimura, T.; Nakatake, Y.; Konishi, M.; Itoh, N. Identification of a novel FGF, FGF-21, preferentially expressed in the liver. Biochim. Biophys. Acta 2000, 1492, 203–206. [Google Scholar] [CrossRef]
- Yamashita, T.; Yoshioka, M.; Itoh, N. Identification of a novel fibroblast growth factor, FGF-23, preferentially expressed in the ventrolateral thalamic nucleus of the brain. Biochem. Biophys. Res. Commun. 2000, 277, 494–498. [Google Scholar] [CrossRef]
- Reid, H.H.; Wilks, A.F.; Bernard, O. Two forms of the basic fibroblast growth factor receptor-like mRNA are expressed in the developing mouse brain. Proc. Natl. Acad. Sci. USA 1990, 87, 1596–1600. [Google Scholar] [CrossRef]
- Friesel, R.; Dawid, I.B. cDNA cloning and developmental expression of fibroblast growth factor receptors from Xenopus laevis. Mol. Cell Biol. 1991, 11, 2481–2488. [Google Scholar] [CrossRef][Green Version]
- Zhang, Y.; Gorry, M.C.; Post, J.C.; Ehrlich, G.D. Genomic organization of the human fibroblast growth factor receptor 2 (FGFR2) gene and comparative analysis of the human FGFR gene family. Gene 1999, 230, 69–79. [Google Scholar] [CrossRef]
- Orr-Urtreger, A.; Bedford, M.T.; Burakova, T.; Arman, E.; Zimmer, Y.; Yayon, A.; Givol, D.; Lonai, P. Developmental localization of the splicing alternatives of fibroblast growth factor receptor-2 (FGFR2). Dev. Biol. 1993, 158, 475–486. [Google Scholar] [CrossRef] [PubMed]
- Friesel, R.; Brown, S.A. Spatially restricted expression of fibroblast growth factor receptor-2 during Xenopus development. Development 1992, 116, 1051–1058. [Google Scholar] [CrossRef] [PubMed]
- Keegan, K.; Johnson, D.E.; Williams, L.T.; Hayman, M.J. Isolation of an additional member of the fibroblast growth factor receptor family, FGFR-3. Proc. Natl. Acad. Sci. USA 1991, 88, 1095–1099. [Google Scholar] [CrossRef] [PubMed]
- Chellaiah, A.T.; McEwen, D.G.; Werner, S.; Xu, J.; Ornitz, D.M. Fibroblast growth factor receptor (FGFR) 3. Alternative splicing in immunoglobulin-like domain III creates a receptor highly specific for acidic FGF/FGF-1. J. Biol. Chem. 1994, 269, 11620–11627. [Google Scholar] [CrossRef]
- Hongo, I.; Kengaku, M.; Okamoto, H. FGF signaling and the anterior neural induction in Xenopus. Dev. Biol. 1999, 216, 561–581. [Google Scholar] [CrossRef][Green Version]
- Partanen, J.; Makela, T.P.; Eerola, E.; Korhonen, J.; Hirvonen, H.; Claesson-Welsh, L.; Alitalo, K. FGFR-4, a novel acidic fibroblast growth factor receptor with a distinct expression pattern. EMBO J. 1991, 10, 1347–1354. [Google Scholar] [CrossRef]
- Stark, K.L.; McMahon, J.A.; McMahon, A.P. FGFR-4, a new member of the fibroblast growth factor receptor family, expressed in the definitive endoderm and skeletal muscle lineages of the mouse. Development 1991, 113, 641–651. [Google Scholar] [CrossRef]
- Riou, J.F.; Clavilier, L.; Boucaut, J.C. Early regionalized expression of a novel Xenopus fibroblast growth factor receptor in neuroepithelium. Biochem. Biophys. Res. Commun. 1996, 218, 198–204. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Houston, D.W.; King, M.L.; Payne, C.; Wylie, C.; Heasman, J. The role of maternal VegT in establishing the primary germ layers in Xenopus embryos. Cell 1998, 94, 515–524. [Google Scholar] [CrossRef]
- Joseph, E.M.; Melton, D.A. Mutant Vg1 ligands disrupt endoderm and mesoderm formation in Xenopus embryos. Development 1998, 125, 2677–2685. [Google Scholar] [CrossRef]
- Kofron, M.; Demel, T.; Xanthos, J.; Lohr, J.; Sun, B.; Sive, H.; Osada, S.; Wright, C.; Wylie, C.; Heasman, J. Mesoderm induction in Xenopus is a zygotic event regulated by maternal VegT via TGFbeta growth factors. Development 1999, 126, 5759–5770. [Google Scholar] [CrossRef]
- Cha, S.W.; Hwang, Y.S.; Chae, J.P.; Lee, S.Y.; Lee, H.S.; Daar, I.; Park, M.J.; Kim, J. Inhibition of FGF signaling causes expansion of the endoderm in Xenopus. Biochem. Biophys. Res. Commun. 2004, 315, 100–106. [Google Scholar] [CrossRef]
- Umair, Z.; Kumar, S.; Rafiq, K.; Kumar, V.; Reman, Z.U.; Lee, S.H.; Kim, S.; Lee, J.Y.; Lee, U.; Kim, J. Dusp1 modulates activin/smad2 mediated germ layer specification via FGF signal inhibition in Xenopus embryos. Anim. Cells Syst. 2020, 24, 359–370. [Google Scholar] [CrossRef]
- Brown, J.L.; Snir, M.; Noushmehr, H.; Kirby, M.; Hong, S.K.; Elkahloun, A.G.; Feldman, B. Transcriptional profiling of endogenous germ layer precursor cells identifies dusp4 as an essential gene in zebrafish endoderm specification. Proc. Natl. Acad. Sci. USA 2008, 105, 12337–12342. [Google Scholar] [CrossRef]
- Poulain, M.; Furthauer, M.; Thisse, B.; Thisse, C.; Lepage, T. Zebrafish endoderm formation is regulated by combinatorial Nodal, FGF and BMP signalling. Development 2006, 133, 2189–2200. [Google Scholar] [CrossRef]
- Sui, L.; Mfopou, J.K.; Geens, M.; Sermon, K.; Bouwens, L. FGF signaling via MAPK is required early and improves Activin A-induced definitive endoderm formation from human embryonic stem cells. Biochem. Biophys. Res. Commun. 2012, 426, 380–385. [Google Scholar] [CrossRef]
- Paraiso, K.D.; Cho, J.S.; Yong, J.; Cho, K.W.Y. Early Xenopus gene regulatory programs, chromatin states, and the role of maternal transcription factors. Curr. Top. Dev. Biol. 2020, 139, 35–60. [Google Scholar] [CrossRef]
- Nieuwkoop, P.D. The formation of the mesoderm in urodelean amphibians: I. Induction by the endoderm. Wilhelm Roux Arch. Entwickl Mech. Org. 1969, 162, 341–373. [Google Scholar] [CrossRef]
- Slack, J.M.; Darlington, B.G.; Heath, J.K.; Godsave, S.F. Mesoderm induction in early Xenopus embryos by heparin-binding growth factors. Nature 1987, 326, 197–200. [Google Scholar] [CrossRef] [PubMed]
- Dvorak, P.; Flechon, J.E.; Thompson, E.M.; Horak, V.; Adenot, P.; Renard, J.P. Embryoglycans regulate FGF-2-mediated mesoderm induction in the rabbit embryo. J. Cell Sci. 1997, 110 Pt 1, 1–10. [Google Scholar] [CrossRef]
- Burdsal, C.A.; Flannery, M.L.; Pedersen, R.A. FGF-2 alters the fate of mouse epiblast from ectoderm to mesoderm in vitro. Dev. Biol. 1998, 198, 231–244. [Google Scholar] [CrossRef][Green Version]
- Amaya, E.; Musci, T.J.; Kirschner, M.W. Expression of a dominant negative mutant of the FGF receptor disrupts mesoderm formation in Xenopus embryos. Cell 1991, 66, 257–270. [Google Scholar] [CrossRef]
- Yamaguchi, T.P.; Harpal, K.; Henkemeyer, M.; Rossant, J. fgfr-1 is required for embryonic growth and mesodermal patterning during mouse gastrulation. Genes Dev. 1994, 8, 3032–3044. [Google Scholar] [CrossRef] [PubMed]
- LaBonne, C.; Whitman, M. Mesoderm induction by activin requires FGF-mediated intracellular signals. Development 1994, 120, 463–472. [Google Scholar] [CrossRef] [PubMed]
- Amaya, E.; Stein, P.A.; Musci, T.J.; Kirschner, M.W. FGF signalling in the early specification of mesoderm in Xenopus. Development 1993, 118, 477–487. [Google Scholar] [CrossRef] [PubMed]
- Dawid, I.B.; Taira, M.; Good, P.J.; Rebagliati, M.R. The role of growth factors in embryonic induction in Xenopus laevis. Mol. Reprod. Dev. 1992, 32, 136–144. [Google Scholar] [CrossRef] [PubMed]
- Takenaga, M.; Fukumoto, M.; Hori, Y. Regulated Nodal signaling promotes differentiation of the definitive endoderm and mesoderm from ES cells. J. Cell Sci. 2007, 120, 2078–2090. [Google Scholar] [CrossRef]
- Agius, E.; Oelgeschlager, M.; Wessely, O.; Kemp, C.; de Robertis, E.M. Endodermal Nodal-related signals and mesoderm induction in Xenopus. Development 2000, 127, 1173–1183. [Google Scholar] [CrossRef] [PubMed]
- Feldman, B.; Gates, M.A.; Egan, E.S.; Dougan, S.T.; Rennebeck, G.; Sirotkin, H.I.; Schier, A.F.; Talbot, W.S. Zebrafish organizer development and germ-layer formation require nodal-related signals. Nature 1998, 395, 181–185. [Google Scholar] [CrossRef]
- Jones, C.M.; Kuehn, M.R.; Hogan, B.L.; Smith, J.C.; Wright, C.V. Nodal-related signals induce axial mesoderm and dorsalize mesoderm during gastrulation. Development 1995, 121, 3651–3662. [Google Scholar] [CrossRef]
- Kumar, V.; Umair, Z.; Kumar, S.; Lee, U.; Kim, J. Smad2 and Smad3 differentially modulate chordin transcription via direct binding on the distal elements in gastrula Xenopus embryos. Biochem. Biophys. Res. Commun. 2021, 559, 168–175. [Google Scholar] [CrossRef]
- Cornell, R.A.; Musci, T.J.; Kimelman, D. FGF is a prospective competence factor for early activin-type signals in Xenopus mesoderm induction. Development 1995, 121, 2429–2437. [Google Scholar] [CrossRef]
- Tannahill, D.; Isaacs, H.V.; Close, M.J.; Peters, G.; Slack, J.M. Developmental expression of the Xenopus int-2 (FGF-3) gene: Activation by mesodermal and neural induction. Development 1992, 115, 695–702. [Google Scholar] [CrossRef]
- Branney, P.A.; Faas, L.; Steane, S.E.; Pownall, M.E.; Isaacs, H.V. Characterisation of the fibroblast growth factor dependent transcriptome in early development. PLoS ONE 2009, 4, e4951. [Google Scholar] [CrossRef]
- Fletcher, R.B.; Harland, R.M. The role of FGF signaling in the establishment and maintenance of mesodermal gene expression in Xenopus. Dev. Dyn. 2008, 237, 1243–1254. [Google Scholar] [CrossRef]
- Christen, B.; Slack, J.M. FGF-8 is associated with anteroposterior patterning and limb regeneration in Xenopus. Dev. Biol. 1997, 192, 455–466. [Google Scholar] [CrossRef]
- Isaacs, H.V. New perspectives on the role of the fibroblast growth factor family in amphibian development. Cell Mol. Life Sci. 1997, 53, 350–361. [Google Scholar] [CrossRef]
- Conlon, F.L.; Sedgwick, S.G.; Weston, K.M.; Smith, J.C. Inhibition of Xbra transcription activation causes defects in mesodermal patterning and reveals autoregulation of Xbra in dorsal mesoderm. Development 1996, 122, 2427–2435. [Google Scholar] [CrossRef] [PubMed]
- Schulte-Merker, S.; Smith, J.C. Mesoderm formation in response to Brachyury requires FGF signalling. Curr. Biol. 1995, 5, 62–67. [Google Scholar] [CrossRef]
- Ryan, K.; Garrett, N.; Mitchell, A.; Gurdon, J.B. Eomesodermin, a key early gene in Xenopus mesoderm differentiation. Cell 1996, 87, 989–1000. [Google Scholar] [CrossRef]
- Kumar, S.; Umair, Z.; Yoon, J.; Lee, U.; Kim, S.C.; Park, J.B.; Lee, J.Y.; Kim, J. Xbra and Smad-1 cooperate to activate the transcription of neural repressor ventx1.1 in Xenopus embryos. Sci. Rep. 2018, 8, 11391. [Google Scholar] [CrossRef]
- Brewster, R.; Mullor, J.L.; Ruiz i Altaba, A. Gli2 functions in FGF signaling during antero-posterior patterning. Development 2000, 127, 4395–4405. [Google Scholar] [CrossRef]
- Kim, J.; Lin, J.J.; Xu, R.H.; Kung, H.F. Mesoderm induction by heterodimeric AP-1 (c-Jun and c-Fos) and its involvement in mesoderm formation through the embryonic fibroblast growth factor/Xbra autocatalytic loop during the early development of Xenopus embryos. J. Biol. Chem. 1998, 273, 1542–1550. [Google Scholar] [CrossRef]
- Yoon, J.; Kim, J.H.; Lee, S.Y.; Kim, S.; Park, J.B.; Lee, J.Y.; Kim, J. PV. 1 induced by FGF-Xbra functions as a repressor of neurogenesis in Xenopus embryos. BMB Rep. 2014, 47, 673–678. [Google Scholar] [CrossRef]
- Rao, Y. Conversion of a mesodermalizing molecule, the Xenopus Brachyury gene, into a neuralizing factor. Genes Dev. 1994, 8, 939–947. [Google Scholar] [CrossRef] [PubMed]
- Isaacs, H.V.; Pownall, M.E.; Slack, J.M. eFGF regulates Xbra expression during Xenopus gastrulation. EMBO J. 1994, 13, 4469–4481. [Google Scholar] [CrossRef]
- Kuroda, H.; Wessely, O.; de Robertis, E.M. Neural induction in Xenopus: Requirement for ectodermal and endomesodermal signals via Chordin, Noggin, beta-Catenin, and Cerberus. PLoS Biol. 2004, 2, E92. [Google Scholar] [CrossRef]
- Delaune, E.; Lemaire, P.; Kodjabachian, L. Neural induction in Xenopus requires early FGF signalling in addition to BMP inhibition. Development 2005, 132, 299–310. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, T.S.; Sheets, M.D. The FGFR pathway is required for the trunk-inducing functions of Spemann’s organizer. Dev. Biol. 2001, 237, 295–305. [Google Scholar] [CrossRef] [PubMed]
- Shiotsugu, J.; Katsuyama, Y.; Arima, K.; Baxter, A.; Koide, T.; Song, J.; Chandraratna, R.A.; Blumberg, B. Multiple points of interaction between retinoic acid and FGF signaling during embryonic axis formation. Development 2004, 131, 2653–2667. [Google Scholar] [CrossRef]
- Kumar, S.; Umair, Z.; Kumar, V.; Lee, U.; Choi, S.C.; Kim, J. Ventx1.1 competes with a transcriptional activator Xcad2 to regulate negatively its own expression. BMB Rep. 2019, 52, 403–408. [Google Scholar] [CrossRef]
- Keenan, I.D.; Sharrard, R.M.; Isaacs, H.V. FGF signal transduction and the regulation of Cdx gene expression. Dev. Biol. 2006, 299, 478–488. [Google Scholar] [CrossRef] [PubMed]
- Levy, V.; Marom, K.; Zins, S.; Koutsia, N.; Yelin, R.; Fainsod, A. The competence of marginal zone cells to become Spemann’s organizer is controlled by Xcad2. Dev. Biol. 2002, 248, 40–51. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kolpakova, A.; Katz, S.; Keren, A.; Rojtblat, A.; Bengal, E. Transcriptional regulation of mesoderm genes by MEF2D during early Xenopus development. PLoS ONE 2013, 8, e69693. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nentwich, O.; Dingwell, K.S.; Nordheim, A.; Smith, J.C. Downstream of FGF during mesoderm formation in Xenopus: The roles of Elk-1 and Egr-1. Dev. Biol. 2009, 336, 313–326. [Google Scholar] [CrossRef]
- Reichert, S.; Randall, R.A.; Hill, C.S. A BMP regulatory network controls ectodermal cell fate decisions at the neural plate border. Development 2013, 140, 4435–4444. [Google Scholar] [CrossRef]
- Lee, H.S.; Lee, S.Y.; Lee, H.; Hwang, Y.S.; Cha, S.W.; Park, S.; Lee, J.Y.; Park, J.B.; Kim, S.; Park, M.J.; et al. Direct response elements of BMP within the PV.1A promoter are essential for its transcriptional regulation during early Xenopus development. PLoS ONE 2011, 6, e22621. [Google Scholar] [CrossRef]
- Umair, Z.; Kumar, S.; Kim, D.H.; Rafiq, K.; Kumar, V.; Kim, S.; Park, J.B.; Lee, J.Y.; Lee, U.; Kim, J. Ventx1.1 as a Direct Repressor of Early Neural Gene zic3 in Xenopus laevis. Mol. Cells 2018, 41, 1061–1071. [Google Scholar] [CrossRef]
- Ault, K.T.; Dirksen, M.L.; Jamrich, M. A novel homeobox gene PV.1 mediates induction of ventral mesoderm in Xenopus embryos. Proc. Natl. Acad. Sci. USA 1996, 93, 6415–6420. [Google Scholar] [CrossRef]
- Marchal, L.; Luxardi, G.; Thome, V.; Kodjabachian, L. BMP inhibition initiates neural induction via FGF signaling and Zic genes. Proc. Natl. Acad. Sci. USA 2009, 106, 17437–17442. [Google Scholar] [CrossRef]
- Furthauer, M.; van Celst, J.; Thisse, C.; Thisse, B. Fgf signalling controls the dorsoventral patterning of the zebrafish embryo. Development 2004, 131, 2853–2864. [Google Scholar] [CrossRef]
- Smith, J.C.; Slack, J.M. Dorsalization and neural induction: Properties of the organizer in Xenopus laevis. J. Embryol. Exp. Morphol. 1983, 78, 299–317. [Google Scholar]
- Leikola, A. Hensen’s node—the ‘organizer’ of the amniote embryo. Experientia 1976, 32, 269–277. [Google Scholar] [CrossRef]
- Shih, J.; Fraser, S.E. Characterizing the zebrafish organizer: Microsurgical analysis at the early-shield stage. Development 1996, 122, 1313–1322. [Google Scholar] [CrossRef]
- Smith, W.C.; Harland, R.M. Expression cloning of noggin, a new dorsalizing factor localized to the Spemann organizer in Xenopus embryos. Cell 1992, 70, 829–840. [Google Scholar] [CrossRef]
- Sasal, Y.; Lu, B.; Steinbelsser, H.; de Robertis, E.M. Regulation of neural induction by the Chd and Bmp-4 antagonistic patterning signals in Xenopus. Nature 1995, 378, 419. [Google Scholar] [CrossRef] [PubMed]
- Lemaire, P.; Kodjabachian, L. The vertebrate organizer: Structure and molecules. Trends Genet. 1996, 12, 525–531. [Google Scholar] [CrossRef]
- Stern, C.D. Neural induction: 10 years on since the ‘default model’. Curr. Opin. Cell Biol. 2006, 18, 692–697. [Google Scholar] [CrossRef]
- Khokha, M.K.; Yeh, J.; Grammer, T.C.; Harland, R.M. Depletion of three BMP antagonists from Spemann’s organizer leads to a catastrophic loss of dorsal structures. Dev. Cell 2005, 8, 401–411. [Google Scholar] [CrossRef]
- Linker, C.; Stern, C.D. Neural induction requires BMP inhibition only as a late step, and involves signals other than FGF and Wnt antagonists. Development 2004, 131, 5671–5681. [Google Scholar] [CrossRef]
- Stern, C.D. Neural induction: Old problem, new findings, yet more questions. Development 2005, 132, 2007–2021. [Google Scholar] [CrossRef] [PubMed]
- Sasai, Y.; Lu, B.; Piccolo, S.; de Robertis, E.M. Endoderm induction by the organizer-secreted factors chordin and noggin in Xenopus animal caps. EMBO J. 1996, 15, 4547–4555. [Google Scholar] [CrossRef] [PubMed]
- Launay, C.; Fromentoux, V.; Shi, D.L.; Boucaut, J.C. A truncated FGF receptor blocks neural induction by endogenous Xenopus inducers. Development 1996, 122, 869–880. [Google Scholar] [CrossRef]
- Kumar, S.; Umair, Z.; Kumar, V.; Kumar, S.; Lee, U.; Kim, J. Foxd4l1.1 negatively regulates transcription of neural repressor ventx1.1 during neuroectoderm formation in Xenopus embryos. Sci. Rep. 2020, 10, 16780. [Google Scholar] [CrossRef]
- Hardcastle, Z.; Chalmers, A.D.; Papalopulu, N. FGF-8 stimulates neuronal differentiation through FGFR-4a and interferes with mesoderm induction in Xenopus embryos. Curr. Biol. 2000, 10, 1511–1514. [Google Scholar] [CrossRef]
- Kang, W.; Hebert, J.M. FGF Signaling is Necessary for Neurogenesis in Young Mice and Sufficient to Reverse Its Decline in Old Mice. J. Neurosci. 2015, 35, 10217–10223. [Google Scholar] [CrossRef]
- Streit, A.; Berliner, A.J.; Papanayotou, C.; Sirulnik, A.; Stern, C.D. Initiation of neural induction by FGF signalling before gastrulation. Nature 2000, 406, 74–78. [Google Scholar] [CrossRef]
- Crossley, P.H.; Martinez, S.; Martin, G.R. Midbrain development induced by FGF8 in the chick embryo. Nature 1996, 380, 66–68. [Google Scholar] [CrossRef]
- Da Silva, S.; Cepko, C.L. Fgf8 Expression and Degradation of Retinoic Acid are Required for Patterning a High-Acuity Area in the Retina. Dev. Cell 2017, 42, 68–81.e66. [Google Scholar] [CrossRef]
- Rogers, C.D.; Moody, S.A.; Casey, E.S. Neural induction and factors that stabilize a neural fate. Birth Defects Res. C Embryo Today 2009, 87, 249–262. [Google Scholar] [CrossRef]
- Pera, E.M.; Ikeda, A.; Eivers, E.; de Robertis, E.M. Integration of IGF, FGF, and anti-BMP signals via Smad1 phosphorylation in neural induction. Genes Dev. 2003, 17, 3023–3028. [Google Scholar] [CrossRef]
- Suzuki, A.; Thies, R.S.; Yamaji, N.; Song, J.J.; Wozney, J.M.; Murakami, K.; Ueno, N. A truncated bone morphogenetic protein receptor affects dorsal-ventral patterning in the early Xenopus embryo. Proc. Natl. Acad. Sci. USA 1994, 91, 10255–10259. [Google Scholar] [CrossRef]
- Hwang, Y.S.; Seo, J.J.; Cha, S.W.; Lee, H.S.; Lee, S.Y.; Roh, D.H.; Kung Hf, H.F.; Kim, J.; Ja Park, M. Antimorphic PV.1 causes secondary axis by inducing ectopic organizer. Biochem. Biophys. Res. Commun. 2002, 292, 1081–1086. [Google Scholar] [CrossRef]
- Kumano, G.; Ezal, C.; Smith, W.C. Boundaries and functional domains in the animal/vegetal axis of Xenopus gastrula mesoderm. Dev. Biol. 2001, 236, 465–477. [Google Scholar] [CrossRef][Green Version]
- Curran, K.L.; Grainger, R.M. Expression of activated MAP kinase in Xenopus laevis embryos: Evaluating the roles of FGF and other signaling pathways in early induction and patterning. Dev. Biol. 2000, 228, 41–56. [Google Scholar] [CrossRef]
- Christen, B.; Slack, J.M. Spatial response to fibroblast growth factor signalling in Xenopus embryos. Development 1999, 126, 119–125. [Google Scholar] [CrossRef]
- De Robertis, E.M.; Kuroda, H. Dorsal-ventral patterning and neural induction in Xenopus embryos. Annu Rev. Cell Dev. Biol. 2004, 20, 285–308. [Google Scholar] [CrossRef]
- Munoz-Sanjuan, I.; Brivanlou, A.H. Neural induction, the default model and embryonic stem cells. Nat. Rev. NeuroSci. 2002, 3, 271–280. [Google Scholar] [CrossRef]
- Cox, W.G.; Hemmati-Brivanlou, A. Caudalization of neural fate by tissue recombination and bFGF. Development 1995, 121, 4349–4358. [Google Scholar] [CrossRef] [PubMed]
- Lamb, T.M.; Harland, R.M. Fibroblast growth factor is a direct neural inducer, which combined with noggin generates anterior-posterior neural pattern. Development 1995, 121, 3627–3636. [Google Scholar] [CrossRef] [PubMed]
- Ribisi, S., Jr.; Mariani, F.V.; Aamar, E.; Lamb, T.M.; Frank, D.; Harland, R.M. Ras-mediated FGF signaling is required for the formation of posterior but not anterior neural tissue in Xenopus laevis. Dev. Biol. 2000, 227, 183–196. [Google Scholar] [CrossRef] [PubMed]
- Pownall, M.E.; Tucker, A.S.; Slack, J.M.; Isaacs, H.V. eFGF, Xcad3 and Hox genes form a molecular pathway that establishes the anteroposterior axis in Xenopus. Development 1996, 122, 3881–3892. [Google Scholar] [CrossRef] [PubMed]
- Sunmonu, N.A.; Li, K.; Li, J.Y. Numerous isoforms of Fgf8 reflect its multiple roles in the developing brain. J. Cell Physiol. 2011, 226, 1722–1726. [Google Scholar] [CrossRef]
- Shim, S.; Bae, N.; Park, S.Y.; Kim, W.S.; Han, J.K. Isolation of Xenopus FGF-8b and comparison with FGF-8a. Mol. Cells 2005, 19, 310–317. [Google Scholar] [PubMed]
- Liu, A.; Losos, K.; Joyner, A.L. FGF8 can activate Gbx2 and transform regions of the rostral mouse brain into a hindbrain fate. Development 1999, 126, 4827–4838. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.M.; Danielian, P.S.; Fritzsch, B.; McMahon, A.P. Evidence that FGF8 signalling from the midbrain-hindbrain junction regulates growth and polarity in the developing midbrain. Development 1997, 124, 959–969. [Google Scholar] [CrossRef]
- Martin, B.L.; Kimelman, D. Wnt signaling and the evolution of embryonic posterior development. Curr. Biol. 2009, 19, R215–R219. [Google Scholar] [CrossRef]
- Epstein, M.; Pillemer, G.; Yelin, R.; Yisraeli, J.K.; Fainsod, A. Patterning of the embryo along the anterior-posterior axis: The role of the caudal genes. Development 1997, 124, 3805–3814. [Google Scholar] [CrossRef]
- Marom, K.; Shapira, E.; Fainsod, A. The chicken caudal genes establish an anterior-posterior gradient by partially overlapping temporal and spatial patterns of expression. Mech. Dev. 1997, 64, 41–52. [Google Scholar] [CrossRef]
- Bayha, E.; Jorgensen, M.C.; Serup, P.; Grapin-Botton, A. Retinoic acid signaling organizes endodermal organ specification along the entire antero-posterior axis. PLoS ONE 2009, 4, e5845. [Google Scholar] [CrossRef]
- Kiecker, C.; Niehrs, C. A morphogen gradient of Wnt/beta-catenin signalling regulates anteroposterior neural patterning in Xenopus. Development 2001, 128, 4189–4201. [Google Scholar] [CrossRef]
- Ikeya, M.; Takada, S. Wnt-3a is required for somite specification along the anteroposterior axis of the mouse embryo and for regulation of cdx-1 expression. Mech. Dev. 2001, 103, 27–33. [Google Scholar] [CrossRef]
- Doniach, T. Basic FGF as an inducer of anteroposterior neural pattern. Cell 1995, 83, 1067–1070. [Google Scholar] [CrossRef]
- Fisher, M.E.; Isaacs, H.V.; Pownall, M.E. eFGF is required for activation of XmyoD expression in the myogenic cell lineage of Xenopus laevis. Development 2002, 129, 1307–1315. [Google Scholar] [CrossRef]
- Hoppler, S.; Brown, J.D.; Moon, R.T. Expression of a dominant-negative Wnt blocks induction of MyoD in Xenopus embryos. Genes Dev. 1996, 10, 2805–2817. [Google Scholar] [CrossRef]
- Bel-Vialar, S.; Itasaki, N.; Krumlauf, R. Initiating Hox gene expression: In the early chick neural tube differential sensitivity to FGF and RA signaling subdivides the HoxB genes in two distinct groups. Development 2002, 129, 5103–5115. [Google Scholar] [CrossRef]
- Wilson, V.; Olivera-Martinez, I.; Storey, K.G. Stem cells, signals and vertebrate body axis extension. Development 2009, 136, 1591–1604. [Google Scholar] [CrossRef]
- Diez del Corral, R.; Storey, K.G. Opposing FGF and retinoid pathways: A signalling switch that controls differentiation and patterning onset in the extending vertebrate body axis. Bioessays 2004, 26, 857–869. [Google Scholar] [CrossRef]
- Shiratori, H.; Hamada, H. The left-right axis in the mouse: From origin to morphology. Development 2006, 133, 2095–2104. [Google Scholar] [CrossRef] [PubMed]
- Fischer, A.; Viebahn, C.; Blum, M. FGF8 acts as a right determinant during establishment of the left-right axis in the rabbit. Curr. Biol. 2002, 12, 1807–1816. [Google Scholar] [CrossRef]
- Meyers, E.N.; Martin, G.R. Differences in left-right axis pathways in mouse and chick: Functions of FGF8 and SHH. Science 1999, 285, 403–406. [Google Scholar] [CrossRef] [PubMed]
- Levin, M.; Johnson, R.L.; Stern, C.D.; Kuehn, M.; Tabin, C. A molecular pathway determining left-right asymmetry in chick embryogenesis. Cell 1995, 82, 803–814. [Google Scholar] [CrossRef]
- Okada, Y.; Takeda, S.; Tanaka, Y.; Belmonte, J.I.; Hirokawa, N. Mechanism of nodal flow: A conserved symmetry breaking event in left-right axis determination. Cell 2005, 121, 633–644. [Google Scholar] [CrossRef] [PubMed]
- Albertson, R.C.; Yelick, P.C. Roles for fgf8 signaling in left-right patterning of the visceral organs and craniofacial skeleton. Dev. Biol. 2005, 283, 310–321. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Okada, Y.; Hirokawa, N. FGF-induced vesicular release of Sonic hedgehog and retinoic acid in leftward nodal flow is critical for left-right determination. Nature 2005, 435, 172–177. [Google Scholar] [CrossRef] [PubMed]
- Neugebauer, J.M.; Amack, J.D.; Peterson, A.G.; Bisgrove, B.W.; Yost, H.J. FGF signalling during embryo development regulates cilia length in diverse epithelia. Nature 2009, 458, 651–654. [Google Scholar] [CrossRef]
- Hong, S.K.; Dawid, I.B. FGF-dependent left-right asymmetry patterning in zebrafish is mediated by Ier2 and Fibp1. Proc. Natl. Acad. Sci. USA 2009, 106, 2230–2235. [Google Scholar] [CrossRef]
- Eblaghie, M.C.; Lunn, J.S.; Dickinson, R.J.; Munsterberg, A.E.; Sanz-Ezquerro, J.J.; Farrell, E.R.; Mathers, J.; Keyse, S.M.; Storey, K.; Tickle, C. Negative feedback regulation of FGF signaling levels by Pyst1/MKP3 in chick embryos. Curr. Biol. 2003, 13, 1009–1018. [Google Scholar] [CrossRef]
- Casci, T.; Vinos, J.; Freeman, M. Sprouty, an intracellular inhibitor of Ras signaling. Cell 1999, 96, 655–665. [Google Scholar] [CrossRef]
- Kovalenko, D.; Yang, X.; Nadeau, R.J.; Harkins, L.K.; Friesel, R. Sef inhibits fibroblast growth factor signaling by inhibiting FGFR1 tyrosine phosphorylation and subsequent ERK activation. J. Biol. Chem. 2003, 278, 14087–14091. [Google Scholar] [CrossRef]
- Sivak, J.M.; Petersen, L.F.; Amaya, E. FGF signal interpretation is directed by Sprouty and Spred proteins during mesoderm formation. Dev. Cell 2005, 8, 689–701. [Google Scholar] [CrossRef]
- Lax, I.; Wong, A.; Lamothe, B.; Lee, A.; Frost, A.; Hawes, J.; Schlessinger, J. The docking protein FRS2alpha controls a MAP kinase-mediated negative feedback mechanism for signaling by FGF receptors. Mol. Cell 2002, 10, 709–719. [Google Scholar] [CrossRef]
- Fico, A.; Maina, F.; Dono, R. Fine-tuning of cell signaling by glypicans. Cell Mol. Life Sci. 2011, 68, 923–929. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Garcia, M.J.; Anderson, K.V. Essential role of glycosaminoglycans in Fgf signaling during mouse gastrulation. Cell 2003, 114, 727–737. [Google Scholar] [CrossRef]
- Galli, A.; Roure, A.; Zeller, R.; Dono, R. Glypican 4 modulates FGF signalling and regulates dorsoventral forebrain patterning in Xenopus embryos. Development 2003, 130, 4919–4929. [Google Scholar] [CrossRef]
- Korsensky, L.; Ron, D. Regulation of FGF signaling: Recent insights from studying positive and negative modulators. Semin. Cell Dev. Biol. 2016, 53, 101–114. [Google Scholar] [CrossRef] [PubMed]
- Saydmohammed, M.; Vollmer, L.L.; Onuoha, E.O.; Vogt, A.; Tsang, M. A high-content screening assay in transgenic zebrafish identifies two novel activators of fgf signaling. Birth Defects Res. C Embryo Today 2011, 93, 281–287. [Google Scholar] [CrossRef] [PubMed]
- Jaeger, I.; Arber, C.; Risner-Janiczek, J.R.; Kuechler, J.; Pritzsche, D.; Chen, I.C.; Naveenan, T.; Ungless, M.A.; Li, M. Temporally controlled modulation of FGF/ERK signaling directs midbrain dopaminergic neural progenitor fate in mouse and human pluripotent stem cells. Development 2011, 138, 4363–4374. [Google Scholar] [CrossRef]
| Modulators | Target | Effect on FGF Signaling |
|---|---|---|
| Anosmin-1 | Extracellular domain of FGFR | Positive |
| L1CAM | Extracellular domain of FGFR | Positive |
| HSPGs | FGFs or extracellular domain of FGFR | Positive |
| FLRT3 | Intracellular domain of FGFR | Positive |
| Sprouty | Grb/Raf | Negative |
| Sef’s/Spred/Pyst1/Dusp’s | Modification of several kinases activity like (MAPK, MEK, ERK, and PKB/Akt) | Negative |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumar, V.; Goutam, R.S.; Park, S.; Lee, U.; Kim, J. Functional Roles of FGF Signaling in Early Development of Vertebrate Embryos. Cells 2021, 10, 2148. https://doi.org/10.3390/cells10082148
Kumar V, Goutam RS, Park S, Lee U, Kim J. Functional Roles of FGF Signaling in Early Development of Vertebrate Embryos. Cells. 2021; 10(8):2148. https://doi.org/10.3390/cells10082148
Chicago/Turabian StyleKumar, Vijay, Ravi Shankar Goutam, Soochul Park, Unjoo Lee, and Jaebong Kim. 2021. "Functional Roles of FGF Signaling in Early Development of Vertebrate Embryos" Cells 10, no. 8: 2148. https://doi.org/10.3390/cells10082148
APA StyleKumar, V., Goutam, R. S., Park, S., Lee, U., & Kim, J. (2021). Functional Roles of FGF Signaling in Early Development of Vertebrate Embryos. Cells, 10(8), 2148. https://doi.org/10.3390/cells10082148








