The Role of microRNAs in Pulp Inflammation
Abstract
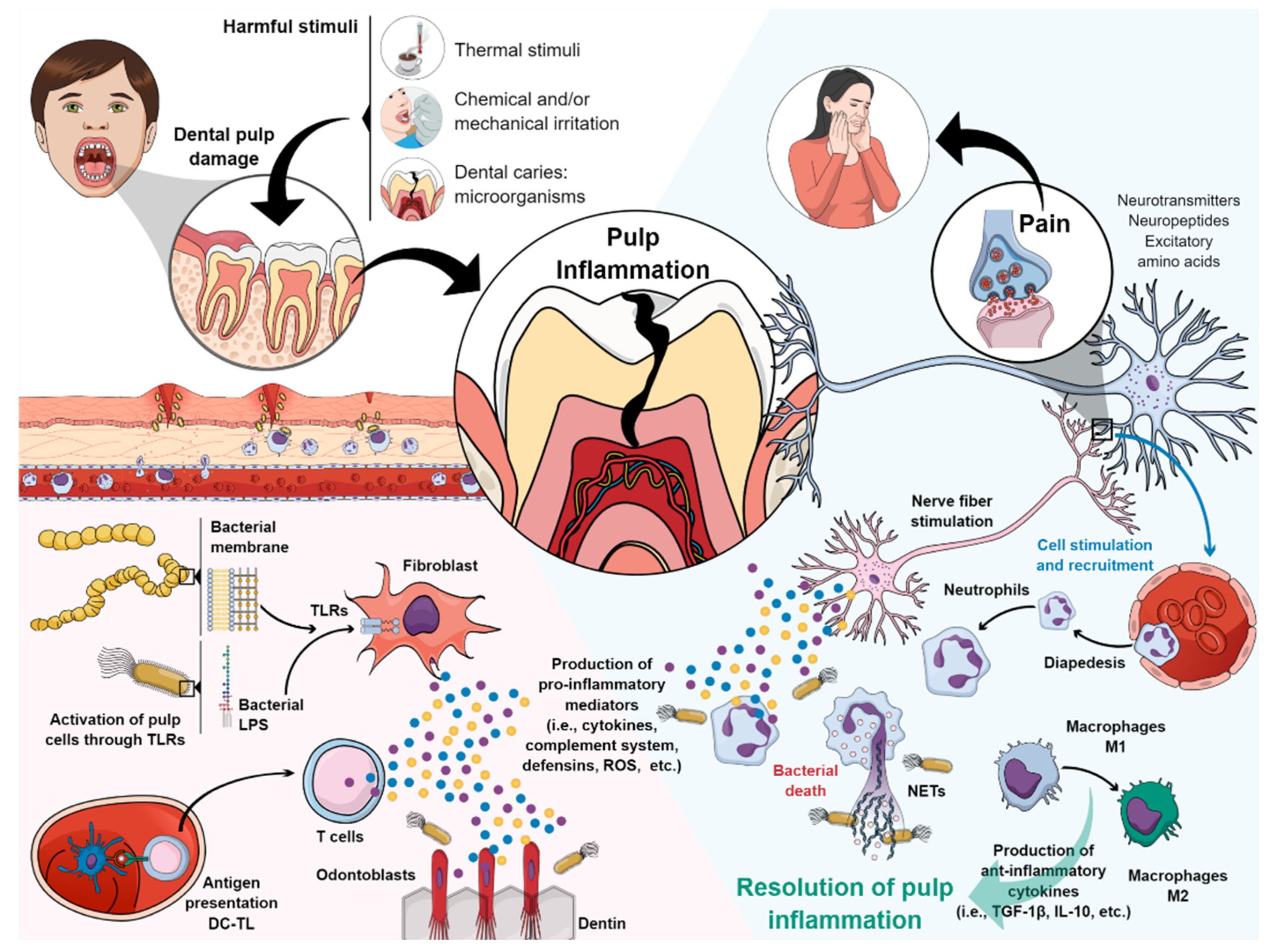
:1. Introduction
2. Pulp Inflammation
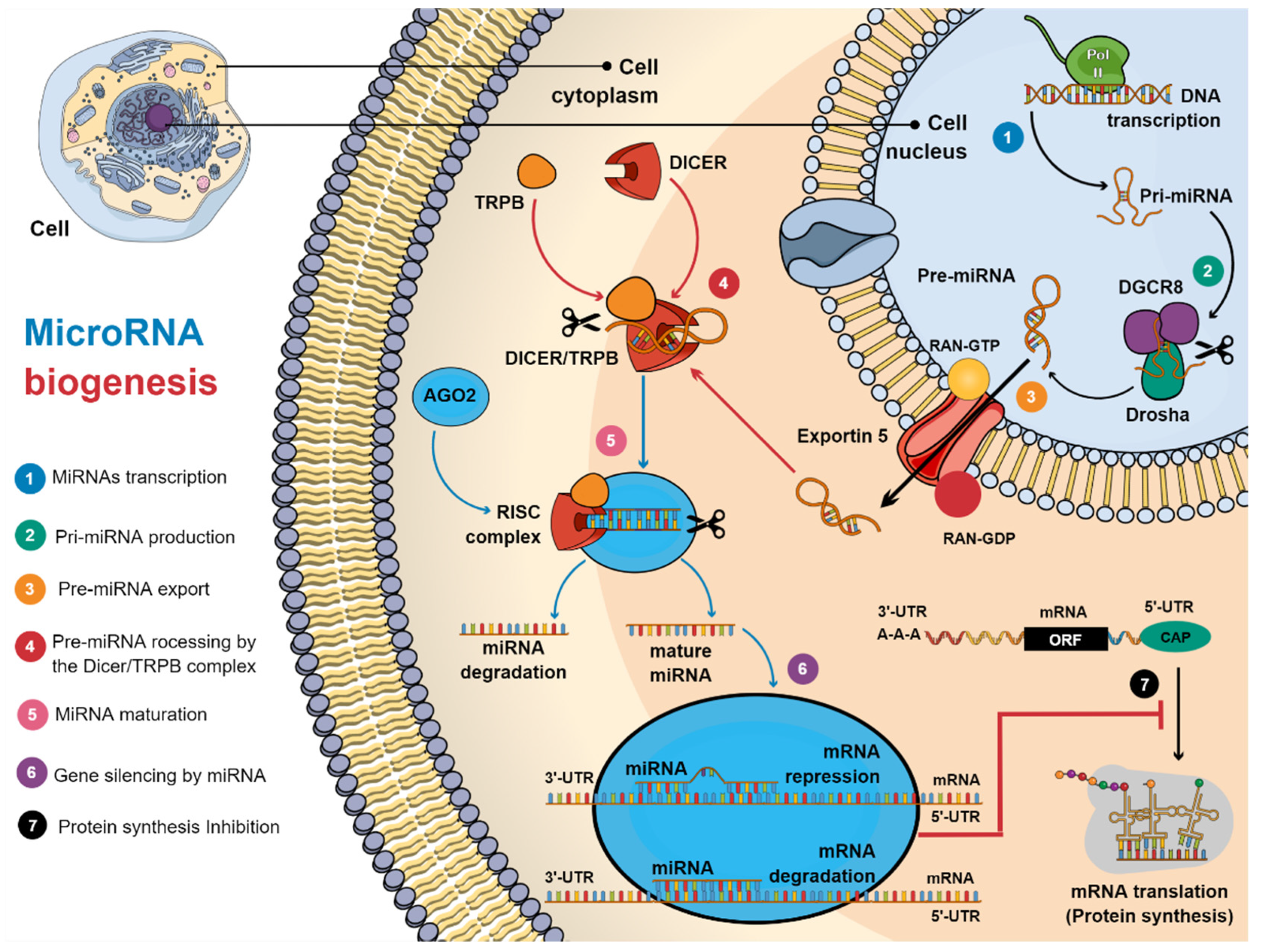
3. Biogenesis of MicroRNAs
4. Upregulated miRNAs in Pulp Inflammation
4.1. MiR-21
4.2. MiR-146a (miR-146a-5b)
4.3. MiR-150
4.4. MiR-223
4.5. MiR-506
4.6. MiR-584
4.7. MiR-766
5. Downregulated miRNAs in Pulp Inflammation
5.1. MiR-Let-7c
5.2. MiR-125b
5.3. MiR-30b
5.4. MiR-152
5.5. MiR-181
5.6. MiR-204
5.7. MiR-221
5.8. MiR-410
| miRNA | Target Gene | MiRNA Function | Ref |
|---|---|---|---|
| miR-Let-7c | IL-13, IκB-α 1, IKK-β1, IGF-1R, DMP-1 1, HMGA2/PI3K/Akt 1 | Osteo/odontogenic differentiation in DPMSCs Response to mechanical stimuli Pro-inflammatory Anti-inflammatory 1 Promoting osteogenic differentiation 1 | [51,94,128,129,130] |
| miR-25b | TNF-α, Fyn | Development and maintenance of orofacial inflammatory pain | [94,132,135,136] |
| Anti-inflammatory and improve odontogenic differentiation | |||
| miR-30b | IL-6R | Attenuate the phagocytosis 2 Anti-inflammatory activity 2 | [142] |
| miR-152 | IL-6, TLR-4, MAPK8, SIRT7 | Negative regulator of the innate immune response Protect against neuroinflammation 3 Anti-inflammatory 3 Response to cold, heat, and mechanical stimuli | [51,52,146,147,148] |
| miR-181 family | IL-6, IL-2, TGFB1, STAT1 CCL8, MMP9, IL-8 | Pro-inflammatory Response to thermal and mechanical stimuli | [51,155,156,157,158,159,160,161] |
| miR-204 | IL-6R, MM9 | Anti-inflammatory activity | [163] |
| miR-221 | Stem cell factor receptor c-Kit, p27 | Inhibits endothelial cell migration, proliferation, and angiogenesis Antiapoptotic Anti-inflammatory 4 | [166,167,168,169] |
| miR-410 | HMGB1 | Anti-inflammatory 5 | [176,177] |
6. Clinical Implications and Future Perspectives
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yu, C.; Abbott, P. An overview of the dental pulp: Its functions and responses to injury. Aust. Dent. J. 2007, 52, S4–S6. [Google Scholar] [CrossRef]
- Goldberg, M.; Hirata, A. The Dental Pulp: Composition, Properties and Functions. JSM Dent. 2017, 5, 1079. [Google Scholar]
- Lv, G.; Zhu, G.; Xu, M.; Gao, X.; Xiao, Q. Inhibition of carrageenan-induced dental inflammatory responses owing to decreased TRPV1 activity by Dexmedetomidine. J. Inflamm. 2020, 17, 18. [Google Scholar] [CrossRef]
- Zheng, J.; Wu, Z.; Niu, K.; Xie, Y.; Hu, X.; Fu, J.; Tian, D.; Fu, K.; Zhao, B.; Kong, W.; et al. Microbiome of Deep Dentinal Caries from Reversible Pulpitis to Irreversible Pulpitis. J. Endod. 2019, 45, 302–309.e1. [Google Scholar] [CrossRef] [PubMed]
- Park, S.H.; Ye, L.; Love, R.M.; Farges, J.C.; Yumoto, H. Inflammation of the Dental Pulp. Mediat. Inflamm. 2015, 2015, 980196. [Google Scholar] [CrossRef] [PubMed]
- Cooper, P.R.; Holder, M.J.; Smith, A.J. Inflammation and regeneration in the dentin-pulp complex: A double-edged sword. J. Endod. 2014, 40, S46–S51. [Google Scholar] [CrossRef] [PubMed]
- Shah, D.; Lynd, T.; Ho, D.; Chen, J.; Vines, J.; Jung, H.-D.; Kim, J.-H.; Zhang, P.; Wu, H.; Jun, H.-W.; et al. Pulp–Dentin Tissue Healing Response: A Discussion of Current Biomedical Approaches. J. Clin. Med. 2020, 9, 434. [Google Scholar] [CrossRef] [Green Version]
- Kokkas, A.; Goulas, A.; Stavrianos, C.; Anogianakis, G. The role of cytokines in pulp inflammation. J. Biol. Regul. Homeost. Agents 2011, 25, 303–311. [Google Scholar]
- Boyle, M.; Chun, C.; Strojny, C.; Narayanan, R.; Bartholomew, A.; Sundivakkam, P.; Alapati, S. Chronic Inflammation and Angiogenic Signaling Axis Impairs Differentiation of Dental-Pulp Stem Cells. PLoS ONE 2014, 9, e113419. [Google Scholar] [CrossRef] [Green Version]
- Zanini, M.; Meyer, E.; Simon, S. Pulp Inflammation Diagnosis from Clinical to Inflammatory Mediators: A Systematic Review. J. Endod. 2017, 43, 1033–1051. [Google Scholar] [CrossRef]
- Konicke, K.; López-Luna, A.; Muñoz-Carrillo, J.L.; Servín-González, L.S.; de la Torre, A.F.; Olasz, E.; Lazarova, Z. The microRNA landscape of cutaneous squamous cell carcinoma. Drug Discov. Today 2018, 23, 864–870. [Google Scholar] [CrossRef] [PubMed]
- miRBase. Available online: https://www.mirbase.org/index.shtml (accessed on 30 July 2021).
- Gaudet, A.D.; Fonken, L.K.; Watkins, L.R.; Nelson, R.J.; Popovich, P.G. MicroRNAs: Roles in Regulating Neuroinflammation. Neurosci 2018, 24, 221–245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sonkoly, E.; Pivarcsi, A. microRNAs in Inflammation. Int. Rev. Immunol. 2009, 28, 535–561. [Google Scholar] [CrossRef]
- Contreras, J.; Rao, D.S. MicroRNAs in inflammation and immune responses. Leukemia 2012, 26, 404–413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pogribny, I.P. MicroRNAs as biomarkers for clinical studies. Exp. Biol. Med. 2018, 243, 283–290. [Google Scholar] [CrossRef]
- Meyle, J.; Dommisch, H.; Groeger, S.; Giacaman, R.A.; Costalonga, M.; Herzberg, M. The innate host response in caries and periodontitis. J. Clin. Periodontol. 2017, 44, 1215–1225. [Google Scholar] [CrossRef]
- Farges, J.-C.; Alliot-Licht, B.; Baudouin, C.; Msika, P.; Bleicher, F.; Carrouel, F. Odontoblast control of dental pulp inflammation triggered by cariogenic bacteria. Front. Physiol. 2013, 4, 326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yumoto, H.; Hirao, K.; Hosokawa, Y.; Kuramoto, H.; Takegawa, D.; Nakanishi, T.; Matsuo, T. The roles of odontoblasts in dental pulp innate immunity. Jpn. Dent. Sci. Rev. 2018, 54, 105–117. [Google Scholar] [CrossRef]
- Bergmann, M.; Jeanneau, C.; Giraud, T.; Richard, G.; About, I. Complement activation links inflammation to dental tissue regeneration. Clin. Oral Investig. 2020, 24, 4185–4196. [Google Scholar] [CrossRef]
- Cooper, P.R.; Chicca, I.J.; Holder, M.J.; Milward, M.R. Inflammation and Regeneration in the Dentin-pulp Complex: Net Gain or Net Loss? J. Endod. 2017, 43, S87–S94. [Google Scholar] [CrossRef]
- Khorasani, M.M.Y.; Hassanshahi, G.; Brodzikowska, A.; Khorramdelazad, H. Role(s) of cytokines in pulpitis: Latest evidence and therapeutic approaches. Cytokine 2020, 126, 154896. [Google Scholar] [CrossRef]
- Arora, S.; Cooper, P.R.; Friedlander, L.T.; Rizwan, S.; Seo, B.; Rich, A.M.; Hussaini, H.M. Potential application of immunotherapy for modulation of pulp inflammation: Opportunities for vital pulp treatment. Int. Endod. J. 2021, 54, 1263–1274. [Google Scholar] [CrossRef]
- Zhan, C.; Huang, M.; Yang, X.; Hou, J. Dental nerves: A neglected mediator of pulpitis. Int. Endod. J. 2021, 54, 85–99. [Google Scholar] [CrossRef] [PubMed]
- El-Sayed, K.M.F.; Elsalawy, R.; Ibrahim, N.; Gadalla, M.; Albargasy, H.; Zahra, N.; Mokhtar, S.; El Nahhas, N.; El Kaliouby, Y.; Dörfer, C.E. The Dental Pulp Stem/Progenitor Cells-Mediated Inflammatory-Regenerative Axis. Tissue Eng. Part. B Rev. 2019, 25, 445–460. [Google Scholar] [CrossRef]
- Kaplan, M.J.; Radic, M. Neutrophil Extracellular Traps: Double-Edged Swords of Innate Immunity. J. Immunol. 2012, 189, 2689–2695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duncan, H.F.; Cooper, P.R. Pulp Innate Immune Defense: Translational Opportunities. J. Endod. 2020, 46, S10–S18. [Google Scholar] [CrossRef] [PubMed]
- Swirski, F.K.; Nahrendorf, M.; Etzrodt, M.; Wildgruber, M.; Cortez-Retamozo, V.; Panizzi, P.; Figueiredo, J.L.; Kohler, R.H.; Chudnovskiy, A.; Waterman, P.; et al. Identification of splenic reservoir monocytes and their deployment to inflammatory sites. Science 2009, 325, 612–616. [Google Scholar] [CrossRef] [Green Version]
- Zaky, S.H.; Shehabeldin, M.; Ray, H.; Sfeir, C. The role of inflammation modulation in dental pulp regeneration. Eur. Cell. Mater. 2021, 41, 184–193. [Google Scholar] [CrossRef]
- Elliott, M.R.; Koster, K.M.; Murphy, P.S. Efferocytosis Signaling in the Regulation of Macrophage Inflammatory Responses. J. Immunol. 2017, 198, 1387–1394. [Google Scholar] [CrossRef]
- Gordon, S.; Martinez, F.O. Alternative activation of macrophages: Mechanism and functions. Immunity 2010, 32, 593–604. [Google Scholar] [CrossRef] [Green Version]
- Farges, J.-C.; Alliot-Licht, B.; Renard, E.; Ducret, M.; Gaudin, A.; Smith, A.J.; Cooper, P.R. Dental Pulp Defence and Repair Mechanisms in Dental Caries. Mediators Inflamm. 2015, 2015, 230251. [Google Scholar] [CrossRef] [Green Version]
- Korkmaz, Y.; Lang, H.; Beikler, T.; Cho, B.; Behrends, S.; Bloch, W.; Addicks, K.; Raab, W.H. Irreversible inflammation is associated with decreased levels of the alpha1-, beta1-, and alpha2-subunits of sGC in human odontoblasts. J. Dent. Res. 2011, 90, 517–522. [Google Scholar] [CrossRef] [PubMed]
- Adams, B.D.; Parsons, C.; Walker, L.; Zhang, W.C.; Slack, F.J. Targeting noncoding RNAs in disease. J. Clin. Investig. 2017, 127, 761–771. [Google Scholar] [CrossRef]
- Winkle, M.; El-Daly, S.M.; Fabbri, M.; Calin, G.A. Noncoding RNA therapeutics—Challenges and potential solutions. Nat. Rev. Drug Discov. 2021, 20, 629–651. [Google Scholar] [CrossRef]
- Winter, J.; Jung, S.; Keller, S.; Gregory, R.I.; Diederichs, S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat. Cell Biol. 2009, 11, 228–234. [Google Scholar] [CrossRef]
- Saliminejad, K.; Khorshid, H.R.K.; Fard, S.S.; Ghaffari, S.H. An overview of microRNAs: Biology, functions, therapeutics, and analysis methods. J. Cell. Physiol. 2019, 234, 5451–5465. [Google Scholar] [CrossRef]
- Yao, Q.; Chen, Y.; Zhou, X. The roles of microRNAs in epigenetic regulation. Curr. Opin. Chem. Biol. 2019, 51, 11–17. [Google Scholar] [CrossRef]
- Vinchure, O.S.; Kulshreshtha, R. miR-490: A potential biomarker and therapeutic target in cancer and other diseases. J. Cell. Physiol. 2021, 236, 3178–3193. [Google Scholar] [CrossRef]
- de Rie, D.; Abugessaisa, I.; Alam, T.; Arner, E.; Arner, P.; Ashoor, H.; Åström, G.; Babina, M.; Bertin, N.; Burroughs, A.M.; et al. An integrated expression atlas of miRNAs and their promoters in human and mouse. Nat. Biotechnol. 2017, 35, 872–878. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-K.; Kim, V.N. Processing of intronic microRNAs. EMBO J. 2007, 26, 775–783. [Google Scholar] [CrossRef] [Green Version]
- Tanzer, A.; Stadler, P.F. Molecular Evolution of a MicroRNA Cluster. J. Mol. Biol. 2004, 339, 327–335. [Google Scholar] [CrossRef]
- Dexheimer, P.J.; Cochella, L. MicroRNAs: From Mechanism to Organism. Front. Cell Dev. Biol. 2020, 8, 409. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzuki, H.I.; Miyazono, K. Emerging complexity of microRNA generation cascades. J. Biochem. 2011, 149, 15–25. [Google Scholar] [CrossRef] [Green Version]
- Davis-Dusenbery, B.N.; Hata, A. MicroRNA in Cancer: The Involvement of Aberrant MicroRNA Biogenesis Regulatory Pathways. Genes Cancer 2010, 1, 1100–1114. [Google Scholar] [CrossRef]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Blelloch, R. Cell Cycle Regulation by microRNAs in Stem Cells. In Results and Problems in Cell Differentiation; Springer: Berlin/Heidelberg, Germany, 2011; Volume 53, pp. 459–472. [Google Scholar]
- Bartel, D.P. Metazoan MicroRNAs. Cell 2018, 173, 20–51. [Google Scholar] [CrossRef] [Green Version]
- Ohtsuka, M.; Ling, H.; Doki, Y.; Mori, M.; Calin, G. MicroRNA Processing and Human Cancer. J. Clin. Med. 2015, 4, 1651–1667. [Google Scholar] [CrossRef] [Green Version]
- Zhong, S.; Zhang, S.; Bair, E.; Nares, S.; Khan, A.A. Differential Expression of MicroRNAs in Normal and Inflamed Human Pulps. J. Endod. 2012, 38, 746–752. [Google Scholar] [CrossRef]
- Hui, T.; Wang, C.; Chen, D.; Zheng, L.; Huang, D.; Ye, L. Epigenetic regulation in dental pulp inflammation. Oral Dis. 2017, 23, 22–28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sheedy, F.J.; Palsson-McDermott, E.; Hennessy, E.J.; Martin, C.; O’Leary, J.J.; Ruan, Q.; Johnson, D.S.; Chen, Y.; O’Neill, L.A.J. Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nat. Immunol. 2010, 11, 141–147. [Google Scholar] [CrossRef]
- Nara, K.; Kawashima, N.; Noda, S.; Fujii, M.; Hashimoto, K.; Tazawa, K.; Okiji, T. Anti-inflammatory roles of microRNA 21 in lipopolysaccharide-stimulated human dental pulp cells. J. Cell. Physiol. 2019, 234, 21331–21341. [Google Scholar] [CrossRef]
- Liu, T.; Zhang, L.; Joo, D.; Sun, S.-C. NF-κB signaling in inflammation. Signal. Transduct. Target. Ther. 2017, 2, 17023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, K.; Chen, Q.; Li, M.; He, L.; Riaz, F.; Zhang, T.; Li, D. Programmed cell death factor 4 (PDCD4), a novel therapy target for metabolic diseases besides cancer. Free. Radic. Biol. Med. 2020, 159, 150–163. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Wu, Q.; Jiang, J.; Sun, D.; Wang, F.; Xin, B.; Cui, Q. Berberine reduces inflammation of human dental pulp fibroblast via miR-21/KBTBD7 axis. Arch. Oral Biol. 2020, 110, 104630. [Google Scholar] [CrossRef]
- Quinn, S.R.; O’Neill, L.A. A trio of microRNAs that control Toll-like receptor signalling. Int. Immunol. 2011, 23, 421–425. [Google Scholar] [CrossRef] [Green Version]
- Taganov, K.D.; Boldin, M.P.; Chang, K.-J.; Baltimore, D. NF-B-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. USA 2006, 103, 12481–12486. [Google Scholar] [CrossRef] [Green Version]
- Yan, F.; Wufuer, D.; Ding, J.; Wang, J. MicroRNA miR-146a-5p inhibits the inflammatory response and injury of airway epithelial cells via targeting TNF receptor-associated factor 6. Bioengineered 2021, 12, 1916–1926. [Google Scholar] [CrossRef]
- Fussbroich, D.; Kohnle, C.; Schwenger, T.; Driessler, C.; Dücker, R.P.; Eickmeier, O.; Gottwald, G.; Jerkic, S.P.; Zielen, S.; Kreyenberg, H.; et al. A combination of LCPUFAs regulates the expression of miRNA-146a-5p in a murine asthma model and human alveolar cells. Prostaglandins Other Lipid Mediat. 2020, 147, 106378. [Google Scholar] [CrossRef]
- Wang, M.-C.; Hung, P.-S.; Tu, H.-F.; Shih, W.-Y.; Li, W.-C.; Chang, K.-W. Lipopolysaccharide Induces the Migration of Human Dental Pulp Cells by Up-regulating miR-146a. J. Endod. 2012, 38, 1598–1603. [Google Scholar] [CrossRef]
- Liu, L.; Shu, S.; Cheung, G.S.; Wei, X. Effect of miR-146a/bFGF/PEG-PEI Nanoparticles on Inflammation Response and Tissue Regeneration of Human Dental Pulp Cells. Biomed. Res. Int. 2016, 2016, 3892685. [Google Scholar] [CrossRef] [Green Version]
- Mo, Z.; Li, Q.; Cai, L.; Zhan, M.; Xu, Q. The effect of DNA methylation on the miRNA expression pattern in lipopolysaccharide-induced inflammatory responses in human dental pulp cells. Mol. Immunol. 2019, 111, 11–18. [Google Scholar] [CrossRef]
- Lin, Y.-C.; Kuo, M.-W.; Yu, J.; Kuo, H.-H.; Lin, R.-J.; Lo, W.-L.; Yu, A.L. c-Myb Is an Evolutionary Conserved miR-150 Target and miR-150/c-Myb Interaction Is Important for Embryonic Development. Mol. Biol. Evol. 2008, 25, 2189–2198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, B.; Wang, S.; Mayr, C.; Bartel, D.P.; Lodish, H.F. miR-150, a microRNA expressed in mature B and T cells, blocks early B cell development when expressed prematurely. Proc. Natl. Acad. Sci. USA 2007, 104, 7080–7085. [Google Scholar] [CrossRef] [Green Version]
- Xiao, C.; Calado, D.P.; Galler, G.; Thai, T.-H.; Patterson, H.C.; Wang, J.; Rajewsky, N.; Bender, T.P.; Rajewsky, K. MiR-150 Controls B Cell Differentiation by Targeting the Transcription Factor c-Myb. Cell 2007, 131, 146–159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moussa, F.M.; Cook, B.P.; Sondag, G.R.; DeSanto, M.; Obri, M.S.; McDermott, S.E.; Safadi, F.F. The role of miR-150 regulates bone cell differentiation and function. Bone 2021, 145, 115470. [Google Scholar] [CrossRef]
- Tsai, C.-L.; Hung, S.-L.; Lee, Y.-Y.; Ho, Y.-C.; Yang, S.-F. The role of fibroblasts in the modulation of dental pulp inflammation. J. Formos. Med. Assoc. 2021, in press. [Google Scholar] [CrossRef]
- Barkhordar, R.A.; Hayashi, C.; Hussain, M.Z. Detection of interleukin-6 in human dental pulp and periapical lesions. Dent. Traumatol. 1999, 15, 26–27. [Google Scholar] [CrossRef] [PubMed]
- Abd-Elmeguid, A.; Abdeldayem, M.; Kline, L.W.; Moqbel, R.; Vliagoftis, H.; Yu, D.C. Osteocalcin Expression in Pulp Inflammation. J. Endod. 2013, 39, 865–872. [Google Scholar] [CrossRef]
- ElSalhy, M.; Azizieh, F.; Raghupathy, R. Cytokines as diagnostic markers of pulpal inflammation. Int. Endod. J. 2013, 46, 573–580. [Google Scholar] [CrossRef]
- Zehnder, M.; Delaleu, N.; Du, Y.; Bickel, M. Cytokine gene expression—part of host defence in pulpitis. Cytokine 2003, 22, 84–88. [Google Scholar] [CrossRef]
- Hong, J.-H.; Kim, M.-R.; Lee, B.-N.; Oh, W.-M.; Min, K.-S.; Im, Y.-G.; Hwang, Y.-C. Anti-Inflammatory and Mineralization Effects of Bromelain on Lipopolysaccharide-Induced Inflammation of Human Dental Pulp Cells. Medicina 2021, 57, 591. [Google Scholar] [CrossRef]
- Chang, M.-C.; Lin, S.-I.; Pan, Y.-H.; Lin, L.-D.; Wang, Y.-L.; Yeung, S.-Y.; Chang, H.-H.; Jeng, J.-H. IL-1β-induced ICAM-1 and IL-8 expression/secretion of dental pulp cells is differentially regulated by IRAK and p38. J. Formos. Med. Assoc. 2019, 118, 1247–1254. [Google Scholar] [CrossRef]
- Pribadi, N.; Budiarti, D.; Kurniawan, H.J.; Widjiastuti, I. The NF-kB and Collagen Type 1 Expression in Dental Pulp after Treated Calcium Hydroxide Combined with Propolis. Eur. J. Dent. 2021, 15, 122–126. [Google Scholar]
- Wei, L.; Chen, Y.; Zhang, C.; Liu, M.; Xiong, H. Leptin induces IL-6 and IL-8 expression through leptin receptor Ob-Rb in human dental pulp fibroblasts. Acta Odontol. Scand. 2019, 77, 205–212. [Google Scholar] [CrossRef]
- Johnnidis, J.B.; Harris, M.H.; Wheeler, R.T.; Stehling-Sun, S.; Lam, M.H.; Kirak, O.; Brummelkamp, T.R.; Fleming, M.D.; Camargo, F.D. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature 2008, 451, 1125–1129. [Google Scholar] [CrossRef]
- Fazi, F.; Racanicchi, S.; Zardo, G.; Starnes, L.M.; Mancini, M.; Travaglini, L.; Diverio, D.; Ammatuna, E.; Cimino, G.; Lo-Coco, F.; et al. Epigenetic Silencing of the Myelopoiesis Regulator microRNA-223 by the AML1/ETO Oncoprotein. Cancer Cell 2007, 12, 457–466. [Google Scholar] [CrossRef]
- Chen, C.-Z.; Li, L.; Lodish, H.F.; Bartel, D.P. MicroRNAs modulate hematopoietic lineage differentiation. Science 2004, 303, 83–86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fazi, F.; Rosa, A.; Fatica, A.; Gelmetti, V.; De Marchis, M.L.; Nervi, C.; Bozzoni, I. A Minicircuitry Comprised of MicroRNA-223 and Transcription Factors NFI-A and C/EBPα Regulates Human Granulopoiesis. Cell 2005, 123, 819–831. [Google Scholar] [CrossRef] [Green Version]
- Bazzoni, F.; Rossato, M.; Fabbri, M.; Gaudiosi, D.; Mirolo, M.; Mori, L.; Tamassia, N.; Mantovani, A.; Cassatella, M.A.; Locati, M. Induction and regulatory function of miR-9 in human monocytes and neutrophils exposed to proinflammatory signals. Proc. Natl. Acad. Sci. USA 2009, 106, 5282–5287. [Google Scholar] [CrossRef] [Green Version]
- Muzio, M.; Bosisio, D.; Polentarutti, N.; D’amico, G.; Stoppacciaro, A.; Mancinelli, R.; van’t Veer, C.; Penton-Rol, G.; Ruco, L.P.; Allavena, P.; et al. Differential Expression and Regulation of Toll-Like Receptors (TLR) in Human Leukocytes: Selective Expression of TLR3 in Dendritic Cells. J. Immunol. 2000, 164, 5998–6004. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Bai, X.; Song, Q.; Fan, F.; Hu, Z.; Cheng, G.; Zhang, Y. miR-223 Inhibits Lipid Deposition and Inflammation by Suppressing Toll-Like Receptor 4 Signaling in Macrophages. Int. J. Mol. Sci. 2015, 16, 24965–24982. [Google Scholar] [CrossRef]
- Li, T.; Morgan, M.J.; Choksi, S.; Zhang, Y.; Kim, Y.-S.; Liu, Z. MicroRNAs modulate the noncanonical transcription factor NF-κB pathway by regulating expression of the kinase IKKα during macrophage differentiation. Nat. Immunol. 2010, 11, 799–805. [Google Scholar] [CrossRef] [Green Version]
- Haneklaus, M.; Gerlic, M.; O’Neill, L.A.J.; Masters, S.L. miR-223: Infection, inflammation and cancer. J. Intern. Med. 2013, 274, 215–226. [Google Scholar] [CrossRef]
- Matsui, S.; Ogata, Y. Effects of miR-223 on expression of IL-1β and IL-6 in human gingival fibroblasts. J. Oral Sci. 2016, 58, 101–108. [Google Scholar] [CrossRef] [Green Version]
- Huang, X.; Liu, F.; Hou, J.; Chen, K. Inflammation-induced overexpression of microRNA-223-3p regulates odontoblastic differentiation of human dental pulp stem cells by targeting SMAD3. Int. Endod. J. 2018, 52, 491–503. [Google Scholar] [CrossRef]
- Wang, D.; Sun, S.; Xue, Y.; Qiu, J.; Ye, T.; Zhang, R.; Song, B.; He, W.; Zhang, Y.; Jiang, W. MicroRNA-223 negatively regulates LPS-induced inflammatory responses by targeting NLRP3 in human dental pulp fibroblasts. Int. Endod. J. 2021, 54, 241–254. [Google Scholar] [CrossRef]
- Wang, Y.; Jiaqi, C.; Zhaoying, C.; Huimin, C. MicroRNA-506-3p regulates neural stem cell proliferation and differentiation through targeting TCF3. Gene 2016, 593, 193–200. [Google Scholar] [CrossRef]
- Erice, O.; Munoz-Garrido, P.; Vaquero, J.; Perugorria, M.J.; Fernandez-Barrena, M.G.; Saez, E.; Santos-Laso, A.; Arbelaiz, A.; Jimenez-Agüero, R.; Fernandez-Irigoyen, J.; et al. MicroRNA-506 promotes primary biliary cholangitis-like features in cholangiocytes and immune activation. Hepatology 2018, 67, 1420–1440. [Google Scholar] [CrossRef] [PubMed]
- Kempinska-Podhorodecka, A.; Adamowicz, M.; Ostrycharz, E.; Chmielarz, M.; Wójcicki, M.; Milkiewicz, P.; Milkiewicz, M. Role of miR-506 in ulcerative colitis associated with primary sclerosing cholangitis. Sci. Rep. 2021, 11, 10134. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Du, Y.; Deng, J.; Wang, X.; Long, F.; He, J. MicroRNA-506 Is Involved in Regulation of the Occurrence of Lipopolysaccharides (LPS)-Induced Pulpitis by Sirtuin 1 (SIRT1). Med. Sci. Monit. 2019, 25, 10008–10015. [Google Scholar] [CrossRef]
- Zhou, D.; Gan, L.; Peng, Y.; Zhou, Y.; Zhou, X.; Wan, M.; Fan, Y.; Xu, X.; Zhou, X.; Zheng, L.; et al. Epigenetic Regulation of Dental Pulp Stem Cell Fate. Stem Cells Int. 2020, 2020, 8876265. [Google Scholar] [CrossRef] [PubMed]
- Xiang, J.; Wu, Y.; Li, D.S.; Wang, Z.Y.; Shen, Q.; Sun, T.Q.; Guan, Q.; Wang, Y.J. miR-584 Suppresses Invasion and Cell Migration of Thyroid Carcinoma by Regulating the Target Oncogene ROCK1. Oncol. Res. Treat. 2015, 38, 436–440. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, Y.; Wang, J. MicroRNA-584 inhibits cell proliferation and invasion in non-small cell lung cancer by directly targeting MTDH. Exp. Ther. Med. 2018, 15, 2203–2211. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.; Hong, Y.; Liu, H.; Wang, Q.; Xu, J.; Zhang, Y.; Zhao, X.; Yao, Y.; Zhou, K.; Ding, X. miR-584 and miR-146 are candidate biomarkers for acute respiratory distress syndrome. Exp. Ther. Med. 2021, 21, 445. [Google Scholar] [CrossRef] [PubMed]
- Ouhara, K.; Savitri, I.J.; Fujita, T.; Kittaka, M.; Kajiya, M.; Iwata, T.; Miyagawa, T.; Yamakawa, M.; Shiba, H.; Kurihara, H. miR-584 expressed in human gingival epithelial cells is induced by Porphyromonas gingivalis stimulation and regulates interleukin-8 production via lactoferrin receptor. J. Periodontol. 2014, 85, 198–204. [Google Scholar] [CrossRef] [PubMed]
- Sehic, A.; Tulek, A.; Khuu, C.; Nirvani, M.; Sand, L.P.; Utheim, T.P. Regulatory roles of microRNAs in human dental tissues. Gene 2017, 596, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Tian, S.; Liu, Y.; Dong, F.; Dou, Y.; Li, W.; Wang, J. Knockdown of microRNA-584 promotes dental pulp stem cells proliferation by targeting TAZ. Cell Cycle. 2020, 19, 1048–1058. [Google Scholar] [CrossRef] [PubMed]
- Hayakawa, K.; Kawasaki, M.; Hirai, T.; Yoshida, Y.; Tsushima, H.; Fujishiro, M.; Ikeda, K.; Morimoto, S.; Takamori, K.; Sekigawa, I. MicroRNA-766-3p Contributes to Anti-Inflammatory Responses through the Indirect Inhibition of NF-κB Signaling. Int. J. Mol. Sci. 2019, 20, 809. [Google Scholar] [CrossRef] [Green Version]
- Tahamtan, A.; Teymoori-Rad, M.; Nakstad, B.; Salimi, V. Anti-Inflammatory MicroRNAs and Their Potential for Inflammatory Diseases Treatment. Front. Immunol. 2018, 9, 1377. [Google Scholar] [CrossRef] [Green Version]
- Feng, J.; Li, A.; Deng, J.; Yang, Y.; Dang, L.; Ye, Y.; Li, Y.; Zhang, W. miR-21 attenuates lipopolysaccharide-induced lipid accumulation and inflammatory response: Potential role in cerebrovascular disease. Lipids Health Dis. 2014, 13, 27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, T.X.; Hartner, J.; Lim, E.-J.; Fabry, V.; Mingler, M.K.; Cole, E.T.; Orkin, S.H.; Aronow, B.J.; Rothenberg, M.E. MicroRNA-21 Limits In Vivo Immune Response-Mediated Activation of the IL-12/IFN-γ Pathway, Th1 Polarization, and the Severity of Delayed-Type Hypersensitivity. J. Immunol. 2011, 187, 3362–3373. [Google Scholar] [CrossRef]
- Caescu, C.I.; Guo, X.; Tesfa, L.; Bhagat, T.D.; Verma, A.; Zheng, D.; Stanley, E.R. Colony stimulating factor-1 receptor signaling networks inhibit mouse macrophage inflammatory responses by induction of microRNA-21. Blood 2015, 125, e1–e13. [Google Scholar] [CrossRef] [PubMed]
- Dalbeth, N.; Pool, B.; Shaw, O.M.; Harper, J.L.; Tan, P.; Franklin, C.; House, M.E.; Cornish, J.; Naot, D. Role of miR-146a in regulation of the acute inflammatory response to monosodium urate crystals. Ann. Rheum. Dis. 2015, 74, 786–790. [Google Scholar] [CrossRef]
- Huang, Y.; Crawford, M.; Higuita-Castro, N.; Nana-Sinkam, P.; Ghadiali, S.N. miR-146a regulates mechanotransduction and pressure-induced inflammation in small airway epithelium. FASEB J. 2012, 26, 3351–3364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olivieri, F.; Prattichizzo, F.; Giuliani, A.; Matacchione, G.; Rippo, M.R.; Sabbatinelli, J.; Bonafè, M. miR-21 and miR-146a: The microRNAs of inflammaging and age-related diseases. Ageing Res. Rev. 2021, 70, 101374. [Google Scholar] [CrossRef] [PubMed]
- Taganov, K.D.; Boldin, M.P.; Baltimore, D. MicroRNAs and Immunity: Tiny Players in a Big Field. Immunity 2007, 26, 133–137. [Google Scholar] [CrossRef] [Green Version]
- Sipert, C.R.; Morandini, A.C.; Dionísio, T.J.; Trachtenberg, A.J.; Kuo, W.P.; Santos, C.F. MicroRNA-146a and microRNA-155 show tissue-dependent expression in dental pulp, gingival and periodontal ligament fibroblasts in vitro. J. Oral Sci. 2014, 56, 157–164. [Google Scholar] [CrossRef] [Green Version]
- Yao, Y.; Wang, H.; Xi, X.; Sun, W.; Ge, J.; Li, P. miR-150 and SRPK1 regulate AKT3 expression to participate in LPS-induced inflammatory response. Innate Immun. 2021, 27, 343–350. [Google Scholar] [CrossRef]
- Ma, Y.; Liu, Y.; Hou, H.; Yao, Y.; Meng, H. MiR-150 predicts survival in patients with sepsis and inhibits LPS-induced inflammatory factors and apoptosis by targeting NF-κB1 in human umbilical vein endothelial cells. Biochem. Biophys. Res. Commun. 2018, 500, 828–837. [Google Scholar] [CrossRef]
- Liu, L.; Yan, L.-N.; Sui, Z. MicroRNA-150 affects endoplasmic reticulum stress via MALAT1-miR-150 axis-mediated NF-κB pathway in LPS-challenged HUVECs and septic mice. Life Sci. 2021, 265, 118744. [Google Scholar] [CrossRef] [PubMed]
- Rangrez, A.Y.; Massy, Z.A.; Metzinger-Le Meuth, V.; Metzinger, L. miR-143 and miR-145. Circ. Cardiovasc. Genet. 2011, 4, 197–205. [Google Scholar] [CrossRef]
- Guan, X.; Gao, Y.; Zhou, J.; Wang, J.; Zheng, F.; Guo, F.; Chang, A.; Li, X.; Wang, B. miR-223 Regulates Adipogenic and Osteogenic Differentiation of Mesenchymal Stem Cells Through a C/EBPs/miR-223/FGFR2 Regulatory Feedback Loop. Stem Cells 2015, 33, 1589–1600. [Google Scholar] [CrossRef]
- Chuang, T.-Y.; Wu, H.-L.; Chen, C.-C.; Gamboa, G.M.; Layman, L.C.; Diamond, M.P.; Azziz, R.; Chen, Y.-H. MicroRNA-223 Expression Is Upregulated in Insulin Resistant Human Adipose Tissue. J. Diabetes Res. 2015, 2015, 943659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, D.; Zhou, Q.; Hu, G.; Wang, G. MiRNA-506 inhibits rheumatoid arthritis fibroblast-like synoviocytes proliferation and induces apoptosis by targetting TLR4. Biosci. Rep. 2019, 39, 20182500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zu, C.; Liu, T.; Zhang, G. MicroRNA-506 Inhibits Malignancy of Colorectal Carcinoma Cells by Targeting LAMC1. Ann. Clin. Lab. Sci. 2016, 46, 666–674. [Google Scholar] [PubMed]
- Cheng, R.-F.; Wang, J.; Zhang, J.-Y.; Sun, L.; Zhao, Y.-R.; Qiu, Z.-Q.; Sun, B.-C.; Sun, Y. MicroRNA-506 is up-regulated in the development of pancreatic ductal adenocarcinoma and is associated with attenuated disease progression. Chin. J. Cancer 2016, 35, 64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, W.-J.; Wang, Y.-L.; Lu, J.-G.; Guo, L.; Qi, B.; Chen, Z.-J. MicroRNA-506 inhibits esophageal cancer cell proliferation via targeting CREB1. Int. J. Clin. Exp. Pathol. 2015, 8, 10868–10874. [Google Scholar]
- Deng, J.; Lei, W.; Xiang, X.; Zhang, L.; Yu, F.; Chen, J.; Feng, M.; Xiong, J. MicroRNA-506 inhibits gastric cancer proliferation and invasion by directly targeting Yap1. Tumor. Biol. 2015, 36, 6823–6831. [Google Scholar] [CrossRef] [PubMed]
- Sand, M.; Skrygan, M.; Georgas, D.; Sand, D.; Hahn, S.A.; Gambichler, T.; Altmeyer, P.; Bechara, F.G. Microarray analysis of microRNA expression in cutaneous squamous cell carcinoma. J. Dermatol. Sci. 2012, 68, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.C.; Li, C.F.; Chen, L.B.; Li, D.D.; Yang, L.; Jin, J.P.; Zhang, B. MicroRNA-766 targeting regulation of SOX6 expression promoted cell proliferation of human colorectal cancer. Onco Targets Ther. 2015, 8, 2981–2988. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Xue, S.; Zhang, J.; Chen, W.; Gong, D.; Zheng, J.; Ma, J.; Xue, W.; Chen, Y.; Zhai, W.; et al. DNA-methylation-mediated repression of miR-766-3p promotes cell proliferation via targeting SF2 expression in renal cell carcinoma. Int. J. Cancer. 2017, 141, 1867–1878. [Google Scholar] [CrossRef]
- Johnson, S.M.; Grosshans, H.; Shingara, J.; Byrom, M.; Jarvis, R.; Cheng, A.; Labourier, E.; Reinert, K.L.; Brown, D.; Slack, F.J. RAS is regulated by the let-7 microRNA family. Cell 2005, 120, 635–647. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, Y.S.; Dutta, A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007, 21, 1025–1030. [Google Scholar] [CrossRef] [Green Version]
- Fu, X.; Mao, X.; Wang, Y.; Ding, X.; Li, Y. Let-7c-5p inhibits cell proliferation and induces cell apoptosis by targeting ERCC6 in breast cancer. Oncol. Rep. 2017, 38, 1851–1856. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, G.X.; Ma, S.; Li, Y.; Yu, Y.; Zhou, Y.X.; Lu, Y.D.; Jin, L.; Wang, Z.L.; Yu, J.H. Hsa-let-7c controls the committed differentiation of IGF-1-treated mesenchymal stem cells derived from dental pulps by targeting IGF-1R via the MAPK pathways. Exp. Mol. Med. 2018, 50, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, H.; Zhao, H.; Wang, J.; Zhang, H.; Hong, L.; Li, H.; Che, H.; Zhang, Z. MicroRNA let-7c-5p promotes osteo-genic differentiation of dental pulp stem cells by inhibiting lipopolysaccharide-induced inflammation via HMGA2/PI3K/Akt signal blockade. Clin. Exp. Pharmacol. Physiol. 2019, 46, 389–397. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Zhang, H.; Hong, L.; Zhao, H.; Wang, J.; Li, H.; Che, H.; Zhang, Z. MicroRNA let-7c-5p Suppressed Lip-opolysaccharide-Induced Dental Pulp Inflammation by Inhibiting Dentin Matrix Protein-1-Mediated Nuclear Factor kappa B (NF-κB) Pathway In Vitro and In Vivo. Med. Sci. Monit. 2018, 24, 6656–6665. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.M.; Lin, K.Y.; Chen, Y.Q. Diverse functions of miR-125 family in different cell contexts. J. Hematol. Oncol. 2013, 6, 6. [Google Scholar] [CrossRef] [Green Version]
- El Gazzar, M.; McCall, C.E. MicroRNAs distinguish translational from transcriptional silencing during endotoxin tolerance. J. Biol. Chem. 2010, 285, 20940–20951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tili, E.; Michaille, J.J.; Cimino, A.; Costinean, S.; Dumitru, C.D.; Adair, B.; Fabbri, M.; Alder, H.; Liu, C.G.; Calin, G.A.; et al. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock. J. Immunol. 2007, 179, 5082–5089. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Nerurkar, V.R. Integrated analysis of microRNAs and their disease related targets in the brain of mice infected with West Nile virus. Virology 2014, 452, 143–151. [Google Scholar] [CrossRef]
- Dong, Y.; Li, P.; Ni, Y.; Zhao, J.; Liu, Z. Decreased microRNA-125a-3p contributes to upregulation of p38 MAPK in rat trigeminal ganglions with orofacial inflammatory pain. PLoS ONE 2014, 9, e111594. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Zheng, Y.; Bai, B.; Song, Y.; Zheng, K.; Xiao, J.; Liang, Y.; Bao, L.; Zhou, Q.; Ji, L.; et al. MicroRNA-125a-3p participates in odontoblastic differentiation of dental pulp stem cells by targeting Fyn. Cytotechnology 2020, 72, 69–79. [Google Scholar] [CrossRef]
- Ouzounova, M.; Vuong, T.; Ancey, P.B.; Ferrand, M.; Durand, G.; Le-Calvez Kelm, F.; Croce, C.; Matar, C.; Herceg, Z.; Hernandez-Vargas, H. MicroRNA miR-30 family regulates non-attachment growth of breast cancer cells. BMC Genom. 2013, 14, 139. [Google Scholar] [CrossRef] [Green Version]
- Braun, J.; Hoang-Vu, C.; Dralle, H.; Hüttelmaier, S. Downregulation of microRNAs directs the EMT and invasive potential of anaplastic thyroid carcinomas. Oncogene 2010, 29, 4237–4244. [Google Scholar] [CrossRef] [Green Version]
- Yu, F.; Deng, H.; Yao, H.; Liu, Q.; Su, F.; Song, E. Mir-30 reduction maintains self-renewal and inhibits apoptosis in breast tumor-initiating cells. Oncogene 2010, 29, 4194–4204. [Google Scholar] [CrossRef] [Green Version]
- Gaziel-Sovran, A.; Segura, M.F.; Di, R.; Collins, M.K.; Hanniford, D.; Vega-Saenz de Miera, E.; Rakus, J.F.; Dankert, J.F.; Shang, S.; Kerbel, R.S.; et al. miR-30b/30d regulation of GalNAc transferases enhances invasion and immunosuppression during metastasis. Cancer Cell 2011, 20, 104–118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Donate, P.B.; Fornari, T.A.; Macedo, C.; Cunha, T.M.; Nascimento, D.C.; Sakamoto-Hojo, E.T.; Donadi, E.A.; Cunha, F.Q.; Passos, G.A. T cell post-transcriptional miRNA-mRNA interaction networks identify targets associated with susceptibility/resistance to collagen-induced arthritis. PLoS ONE 2013, 8, e54803. [Google Scholar] [CrossRef]
- Naqvi. A.R.; Fordham, J.B.; Nares, S. miR-24, miR-30b, and miR-142-3p regulate phagocytosis in myeloid inflammatory cells. J. Immunol. 2015, 194, 1916–1927. [Google Scholar] [CrossRef] [Green Version]
- Zhang, N.; Zhang, Q.; Yang, W.; Miao, L.; Wang, N.; Wei, S.; Ge, J.; Li, X.; Wu, J. Decreased expression of microRNA-30b promotes the development of pulpitis by upregulating the expression of interleukin-6 receptor. Exp. Ther. Med. 2019, 17, 3233–3238. [Google Scholar] [CrossRef] [Green Version]
- Friedrich, M.; Pracht, K.; Mashreghi, M.F.; Jäck, H.M.; Radbruch, A.; Seliger, B. The role of the miR-148/-152 family in physiology and disease. Eur. J. Immunol. 2017, 47, 2026–2038. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dougherty, U.; Mustafi, R.; Zhu, H.; Zhu, X.; Deb, D.; Meredith, S.C.; Ayaloglu-Butun, F.; Fletcher, M.; Sanchez, A.; Pekow, J.; et al. Upregulation of polycistronic microRNA-143 and microRNA-145 in colonocytes suppresses colitis and inflammation-associated colon cancer. Epigenetics 2020, 28, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhan, Z.; Xu, L.; Ma, F.; Li, D.; Guo, Z.; Li, N.; Cao, X. MicroRNA-148/152 impair innate response and antigen presentation of TLR-triggered dendritic cells by targeting CaMKIIα. J. Immunol. 2010, 185, 7244–7251. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Du, J.; Fei, D.; Xing, J.; Liu, J.; Lu, H. miR-152 inhibits rheumatoid arthritis synovial fibroblast prolifera-tion and induces apoptosis by targeting ADAM10. Int. J. Mol. Med. 2018, 42, 643–650. [Google Scholar]
- Hu, L.; Zhang, H.; Wang, B.; Ao, Q.; He, Z. MicroRNA-152 attenuates neuroinflammation in intracerebral hem-orrhage by inhibiting thioredoxin interacting protein (TXNIP)-mediated NLRP3 inflammasome activation. Int. Immunopharmacol. 2020, 80, 106141. [Google Scholar] [CrossRef]
- Ma, P.; Zhang, C.; Huo, P.; Li, Y.; Yang, H. A novel role of the miR-152-3p/ERRFI1/STAT3 pathway modulates the apoptosis and inflammatory response after acute kidney injury. J. Biochem. Mol. Toxicol. 2020, 25, e22540. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.B.; Lai, X.; Guo, X.F. Activation of Nrf2 by miR-152 Inhibits Doxorubicin-Induced Cardiotoxicity via Attenuation of Oxidative Stress, Inflammation, and Apoptosis. Oxid. Med. Cell Longev. 2021, 2021, 8860883. [Google Scholar] [PubMed]
- Gu, S.; Ran, S.; Liu, B.; Liang, J. miR-152 induces human dental pulp stem cell senescence by inhibiting SIRT7 expression. FEBS Lett. 2016, 590, 1123–1131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marisetty, A.; Wei, J.; Kong, L.Y.; Ott, M.; Fang, D.; Sabbagh, A.; Heimberger, A.B. MiR-181 Family Modulates Osteopontin in Glioblastoma Multiforme. Cancers 2020, 12, 3813. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Sit, A.; Feinberg, M.W. Role of miR-181 family in regulating vascular inflammation and immunity. Trends Cardiovasc. Med. 2014, 24, 105–112. [Google Scholar] [CrossRef] [Green Version]
- Sun, X.; Icli, B.; Wara, A.K.; Belkin, N.; He, S.; Kobzik, L.; Hunninghake, G.M.; Vera, M.P.; MICU Registry; Blackwell, T.S.; et al. MicroRNA-181b regulates NF-κB-mediated vascular inflammation. J. Clin. Investig. 2012, 122, 1973–1990. [Google Scholar]
- Hutchison, E.R.; Kawamoto, E.M.; Taub, D.D.; Lal, A.; Abdelmohsen, K.; Zhang, Y.; Wood, W.H., III; Lehrmann, E.; Camandola, S.; Becker, K.G.; et al. Evidence for miR-181 involvement in neuroinflammatory responses of astrocytes. Glia 2013, 61, 1018–1028. [Google Scholar] [CrossRef] [PubMed]
- Xie, W.; Li, M.; Xu, N.; Lv, Q.; Huang, N.; He, J.; Zhang, Y. MiR-181a regulates inflammation responses in monocytes and macrophages. PLoS ONE 2013, 8, e58639. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pichiorri, F.; Suh, S.S.; Ladetto, M.; Kuehl, M.; Palumbo, T.; Drandi, D.; Taccioli, C.; Zanesi, N.; Alder, H.; Hagan, J.P.; et al. MicroRNAs regulate critical genes associated with multiple myeloma pathogenesis. Proc. Natl. Acad. Sci. USA 2008, 105, 12885–12890. [Google Scholar] [CrossRef] [Green Version]
- Dave, R.S.; Khalili, K. Morphine treatment of human monocyte-derived macrophages induces differential miRNA and protein expression: Impact on inflammation and oxidative stress in the central nervous system. J. Cell. Biochem. 2010, 110, 834–845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xue, Q.; Guo, Z.Y.; Li, W.; Wen, W.H.; Meng, Y.L.; Jia, L.T.; Wang, J.; Yao, L.B.; Jin, B.Q.; Wang, T.; et al. Human activated CD4(+) T lymphocytes increase IL-2 expression by downregulating microRNA-181c. Mol. Immunol. 2011, 48, 592–599. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Hsu, S.H.; Majumder, S.; Kutay, H.; Huang, W.; Jacob, S.T.; Ghoshal, K. TGFbeta-mediated upregulation of hepatic miR-181b promotes hepatocarcinogenesis by targeting TIMP3. Oncogene 2010, 29, 1787–1797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galicia, J.C.; Naqvi, A.R.; Ko, C.C.; Nares, S.; Khan, A.A. MiRNA-181a regulates Toll-like receptor agonist-induced inflammatory response in human fibroblasts. Genes Immun. 2014, 15, 333–337. [Google Scholar] [CrossRef] [Green Version]
- Li, T.; Pan, H.; Li, R. The dual regulatory role of miR-204 in cancer. Tumour. Biol. 2016, 37, 11667–11677. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Qiu, J.; Chen, B.; Lin, Y.; Chen, Y.; Xie, G.; Qiu, J.; Tong, H.; Jiang, D. Long non-coding RNA NEAT1 plays an important role in sepsis-induced acute kidney injury by targeting miR-204 and modulating the NF-κB pathway. Int. Immunopharmacol. 2018, 59, 252–260. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Xu, J. Downregulation of microRNA-204 increases the expression of matrix metallopeptidase 9 in pediatric patients with pulpitis and Helicobacter pylori infection in the stomach. Exp. Ther. Med. 2019, 18, 253–259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Howe, E.N.; Cochrane, D.R.; Richer, J.K. The miR-200 and miR-221/222 microRNA families: Opposing effects on epithelial identity. J. Mammary Gland. Biol. Neoplasia 2012, 17, 65–77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Urbich, C.; Kuehbacher, A.; Dimmeler, S. Role of microRNAs in vascular diseases, inflammation, and angio-genesis. Cardiovasc. Res. 2008, 79, 581–588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gramantieri, L.; Fornari, F.; Ferracin, M.; Veronese, A.; Sabbioni, S.; Calin, G.A.; Grazi, G.L.; Croce, C.M.; Bolondi, L.; Negrini, M. MicroRNA-221 targets Bmf in hepatocellular carcinoma and correlates with tumor multifocality. Clin. Cancer Res. 2009, 15, 5073–5081. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Huang, Y.; Zhou, C.; Wu, H.; Zhao, J.; Wu, L.; Zhao, M.; Zhang, F.; Liu, H. The Role of Autophagy and Related MicroRNAs in Inflammatory Bowel Disease. Gastroenterol. Res. Pract. 2018, 2018, 7565076. [Google Scholar] [CrossRef]
- Fang, K.; Sideri, A.; Law, I.K.; Bakirtzi, K.; Polytarchou, C.; Iliopoulos, D.; Pothoulakis, C. Identification of a novel substance P (SP)-neurokinin-1 receptor (NK-1R) microRNA-221-5p inflammatory network in human colonic epithelial cells. Cell Mol. Gastroenterol. Hepatol. 2015, 1, 503–515. [Google Scholar] [CrossRef] [Green Version]
- Kong, Q.; Liu, L.; Huang, Y.; Zhang, F.; Wei, X.; Ling, J. The effect of octamer-binding transcription factor 4B1 on microRNA signals in human dental pulp cells with inflammatory response. J. Endod. 2014, 40, 101–108. [Google Scholar] [CrossRef]
- Korkmaz, B.; Ozveren, E.; Buharalioglu, C.K.; Tunctan, B. Extracellular signal-regulated kinase (ERK1/2) contributes to endotoxin-induced hyporeactivity via nitric oxide and prostacyclin production in rat aorta. Pharmacology 2006, 78, 123–128. [Google Scholar] [CrossRef]
- Felaco, M.; Di Maio, F.D.; De Fazio, P.; D’Arcangelo, C.; De Lutiis, M.A.; Varvara, G.; Grilli, A.; Barbacane, R.C.; Reale, M.; Conti, P. Localization of the e-NOS enzyme in endothelial cells and odontoblasts of healthy human dental pulp. Life Sci. 2000, 68, 297–306. [Google Scholar] [CrossRef]
- Korkmaz, Y.; Bloch, W.; Steinritz, D.; Baumann, M.A.; Addicks, K.; Schneider, K.; Raab, W.H. Bradykinin mediates phosphorylation of eNOS in odontoblasts. J. Dent. Res. 2006, 85, 536–541. [Google Scholar] [CrossRef]
- Zhang, Y.F.; Yu, Y.; Song, W.Z.; Zhang, R.M.; Jin, S.; Bai, J.W.; Kang, H.B.; Wang, X.; Cao, X.C. miR-410-3p suppresses breast cancer progression by targeting Snail. Oncol. Rep. 2016, 36, 480–486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, J.; Zhou, H.; Zhao, H.; Zhao, Y.; Chang, C. Hesperetin ameliorates lipopolysaccharide-induced acute lung injury via the miR-410/SOX18 axis. J. Biochem. Mol. Toxicol. 2020, 34, 22588. [Google Scholar] [CrossRef] [PubMed]
- Pan, H.; Dai, H.; Wang, L.; Lin, S.; Tao, Y.; Zheng, Y.; Jiang, R.; Fang, F.; Wu, Y. MicroRNA-410-3p modulates chondrocyte apoptosis and inflammation by targeting high mobility group box 1 (HMGB1) in an osteoarthritis mouse model. BMC Musculoskelet. Disord. 2020, 21, 486. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, N.; Zhao, S.; Jiao, T.; Fu, W.; Yang, L.; Zhang, N. miR-410-3p Suppresses Cytokine Release from Fi-broblast-Like Synoviocytes by Regulating NF-κB Signaling in Rheumatoid Arthritis. Inflammation 2019, 42, 331–341. [Google Scholar] [CrossRef] [Green Version]
- Brodzikowska, A.; Gondek, A.; Rak, B.; Paskal, W.; Pełka, K.; Cudnoch-Jędrzejewska, A.; Włodarski, P. Metalloproteinase 14 (MMP-14) and hsa-miR-410-3p expression in human inflamed dental pulp and odontoblasts. Histochem. Cell Biol. 2019, 152, 345–353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duncan, H.F.; Galler, K.M.; Tomson, P.L.; Simon, S.; El-Karim, I.; Kundzina, R.; Krastl, G.; Dammaschke, T.; Fransson, H.; Markvart, M.; et al. European Society of Endodontology position statement: Management of deep caries and the exposed pulp. Int. Endod. J. 2019, 52, 923–934. [Google Scholar]
- Lin, L.M.; Ricucci, D.; Saoud, T.M.; Sigurdsson, A.; Kahler, B. Vital pulp therapy of mature permanent teeth with irreversible pulpitis from the perspective of pulp biology. Aust. Endod. J. 2020, 46, 154–166. [Google Scholar] [CrossRef]

| miRNA | Target Gene | MiRNA Function | Ref |
|---|---|---|---|
| miR-21 | TLR-4, TRAF6, PDCD | Anti-inflammatory activity Inhibition NF-κB signal pathway | [53,54,55,56,57] |
| miR-146a | IRAK1, TRAF6, COX-2, 5-LO | Anti-inflammatory activity Suppressing MAPK pathway Cell proliferation and odontogenic differentiation of hDPC | [58,59,60,61,62,63,64] |
| miR-150 | IL-6, NF-κB1, JAK2, IRAK2, c-Myb, Osteoactivin/GPNMB | Differentiation of B cells Differentiation and function of osteoblasts, and osteoclasts Regulator of inflammatory responses | [51,65,66,67,68,69,70,71,72,73,74,75,76,77] |
| miR-223 | TLR-3, TLR-4, IKK-α, MKP-5 | Granulopoiesis and monocyte activation Cell differentiation Pro-inflammatory Pulpitis repair | [78,79,80,81,82,83,84,85,86,87,88,89] |
| miR-506 | SIRT1 | Cell differentiation and proliferation Activating the immune system Pro-inflammatory | [90,91,92,93,94] |
| miR-584 | MAPK8 | Suppress cell proliferation, migration, and invasion Pulp tissue homeostasis Favoring the inflammatory response | [51,95,96,97,98,99,100] |
| miR-766 | HSF1 | Inflammatory gene suppressor Temperature stress response | [51,52,101] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muñoz-Carrillo, J.L.; Vázquez-Alcaraz, S.J.; Vargas-Barbosa, J.M.; Ramos-Gracia, L.G.; Alvarez-Barreto, I.; Medina-Quiroz, A.; Díaz-Huerta, K.K. The Role of microRNAs in Pulp Inflammation. Cells 2021, 10, 2142. https://doi.org/10.3390/cells10082142
Muñoz-Carrillo JL, Vázquez-Alcaraz SJ, Vargas-Barbosa JM, Ramos-Gracia LG, Alvarez-Barreto I, Medina-Quiroz A, Díaz-Huerta KK. The Role of microRNAs in Pulp Inflammation. Cells. 2021; 10(8):2142. https://doi.org/10.3390/cells10082142
Chicago/Turabian StyleMuñoz-Carrillo, José Luis, Silverio Jafet Vázquez-Alcaraz, Jazmín Monserrat Vargas-Barbosa, Luis Guillermo Ramos-Gracia, Israel Alvarez-Barreto, Alejandro Medina-Quiroz, and Karla Karina Díaz-Huerta. 2021. "The Role of microRNAs in Pulp Inflammation" Cells 10, no. 8: 2142. https://doi.org/10.3390/cells10082142
APA StyleMuñoz-Carrillo, J. L., Vázquez-Alcaraz, S. J., Vargas-Barbosa, J. M., Ramos-Gracia, L. G., Alvarez-Barreto, I., Medina-Quiroz, A., & Díaz-Huerta, K. K. (2021). The Role of microRNAs in Pulp Inflammation. Cells, 10(8), 2142. https://doi.org/10.3390/cells10082142







