Mechanisms of Smoothened Regulation in Hedgehog Signaling
Abstract
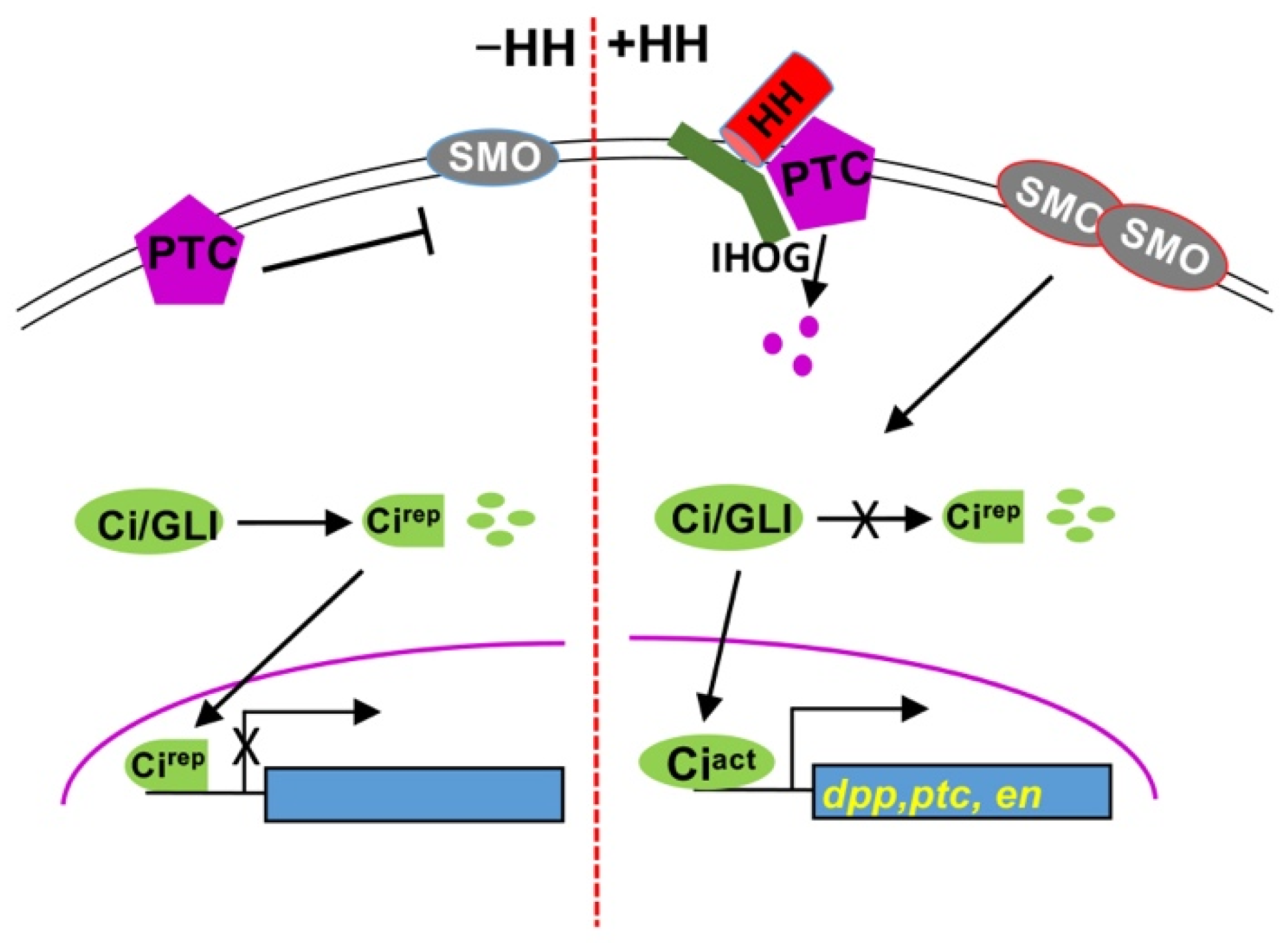
1. Introduction
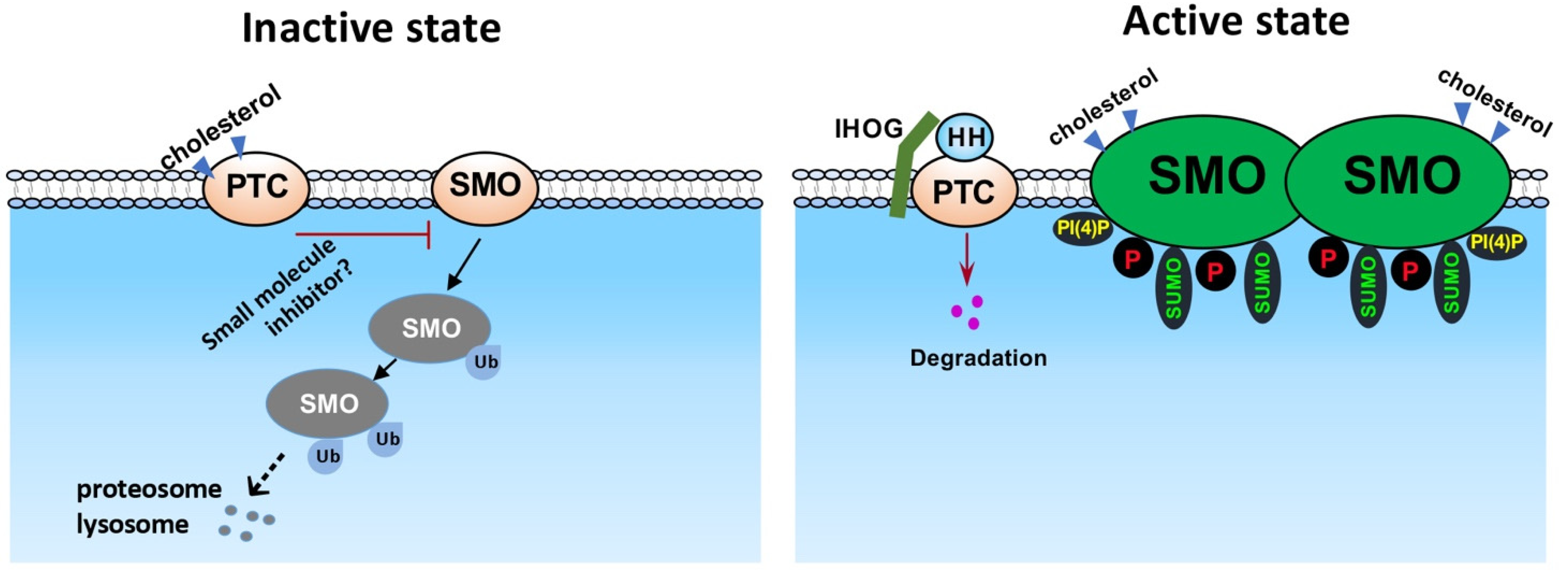
2. SMO Phosphorylation, Dimerization, and Conformational Change
3. SMO Ubiquitination, Sumoylation, and Stability Control
4. SMO Regulation by Lipid-Based Modulators
5. Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ingham, P.W.; McMahon, A.P. Hedgehog signaling in animal development: Paradigms and principles. Genes Dev. 2001, 15, 3059–3087. [Google Scholar] [CrossRef] [PubMed]
- Jia, J.; Jiang, J. Decoding the Hedgehog signal in animal development. Cell Mol. Life Sci. 2006, 63, 1249–1265. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Hui, C.C. Hedgehog signaling in development and cancer. Dev. Cell 2008, 15, 801–812. [Google Scholar] [CrossRef] [PubMed]
- Villavicencio, E.H.; Walterhouse, D.O.; Iannaccone, P.M. The sonic hedgehog-patched-gli pathway in human development and disease. Am. J. Hum. Genet. 2000, 67, 1047–1054. [Google Scholar] [CrossRef]
- Taipale, J.; Beachy, P.A. The Hedgehog and Wnt signalling pathways in cancer. Nature 2001, 411, 349–354. [Google Scholar] [CrossRef]
- Pasca di Magliano, M.; Hebrok, M. Hedgehog signalling in cancer formation and maintenance. Nat. Rev. Cancer 2003, 3, 903–911. [Google Scholar] [CrossRef]
- Pak, E.; Segal, R.A. Hedgehog Signal Transduction: Key Players, Oncogenic Drivers, and Cancer Therapy. Dev. Cell 2016, 38, 333–344. [Google Scholar] [CrossRef]
- Zheng, X.; Mann, R.K.; Sever, N.; Beachy, P.A. Genetic and biochemical definition of the Hedgehog receptor. Genes Dev. 2010, 24, 57–71. [Google Scholar] [CrossRef]
- Lum, L.; Beachy, P.A. The Hedgehog response network: Sensors, switches, and routers. Science 2004, 304, 1755–1759. [Google Scholar] [CrossRef]
- Camp, D.; Currie, K.; Labbe, A.; van Meyel, D.J.; Charron, F. Ihog and Boi are essential for Hedgehog signaling in Drosophila. Neural Dev. 2010, 5, 28. [Google Scholar] [CrossRef]
- Hooper, J.E.; Scott, M.P. Communicating with Hedgehogs. Nat. Rev. Mol. Cell Biol. 2005, 6, 306–317. [Google Scholar] [CrossRef]
- Stone, D.M.; Hynes, M.; Armanini, M.; Swanson, T.A.; Gu, Q.; Johnson, R.L.; Scott, M.P.; Pennica, D.; Goddard, A.; Phillips, H.; et al. The tumour-suppressor gene patched encodes a candidate receptor for Sonic hedgehog. Nature 1996, 384, 129–134. [Google Scholar] [CrossRef] [PubMed]
- Nakano, Y.; Nystedt, S.; Shivdasani, A.A.; Strutt, H.; Thomas, C.; Ingham, P.W. Functional domains and sub-cellular distribution of the Hedgehog transducing protein Smoothened in Drosophila. Mech. Dev. 2004, 121, 507–518. [Google Scholar] [CrossRef] [PubMed]
- Aanstad, P.; Santos, N.; Corbit, K.C.; Scherz, P.J.; Trinh, L.A.; Salvenmoser, W.; Huisken, J.; Reiter, J.F.; Stainier, D.Y. The extracellular domain of Smoothened regulates ciliary localization and is required for high-level Hh signaling. Curr. Biol. 2009, 19, 1034–1039. [Google Scholar] [CrossRef]
- Jiang, J. Hedgehog signaling mechanism and role in cancer. Semin. Cancer Biol. 2021. [Google Scholar] [CrossRef] [PubMed]
- Denef, N.; Neubuser, D.; Perez, L.; Cohen, S.M. Hedgehog induces opposite changes in turnover and subcellular localization of patched and smoothened. Cell 2000, 102, 521–531. [Google Scholar] [CrossRef]
- Taipale, J.; Cooper, M.K.; Maiti, T.; Beachy, P.A. Patched acts catalytically to suppress the activity of Smoothened. Nature 2002, 418, 892–897. [Google Scholar] [CrossRef] [PubMed]
- Casali, A.; Struhl, G. Reading the Hedgehog morphogen gradient by measuring the ratio of bound to unbound Patched protein. Nature 2004, 431, 76–80. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Murone, M.; Luoh, S.M.; Ryan, A.; Gu, Q.; Zhang, C.; Bonifas, J.M.; Lam, C.W.; Hynes, M.; Goddard, A.; et al. Activating Smoothened mutations in sporadic basal-cell carcinoma. Nature 1998, 391, 90–92. [Google Scholar] [CrossRef]
- Yang, L.; Xie, G.; Fan, Q.; Xie, J. Activation of the hedgehog-signaling pathway in human cancer and the clinical implications. Oncogene 2010, 29, 469–481. [Google Scholar] [CrossRef]
- Guha, M. Hedgehog inhibitor gets landmark skin cancer approval, but questions remain for wider potential. Nat. Rev. Drug Discov. 2012, 11, 257–258. [Google Scholar] [CrossRef] [PubMed]
- Sekulic, A.; Von Hoff, D. Hedgehog Pathway Inhibition. Cell 2016, 164, 831. [Google Scholar] [CrossRef] [PubMed]
- Bohl, S.R.; Bullinger, L.; Rucker, F.G. New Targeted Agents in Acute Myeloid Leukemia: New Hope on the Rise. Int. J. Mol. Sci. 2019, 20, 1983. [Google Scholar] [CrossRef] [PubMed]
- Pietrobono, S.; Stecca, B. Targeting the Oncoprotein Smoothened by Small Molecules: Focus on Novel Acylguanidine Derivatives as Potent Smoothened Inhibitors. Cells 2018, 7, 272. [Google Scholar] [CrossRef]
- Yauch, R.L.; Dijkgraaf, G.J.; Alicke, B.; Januario, T.; Ahn, C.P.; Holcomb, T.; Pujara, K.; Stinson, J.; Callahan, C.A.; Tang, T.; et al. Smoothened mutation confers resistance to a Hedgehog pathway inhibitor in medulloblastoma. Science 2009, 326, 572–574. [Google Scholar] [CrossRef]
- Rudin, C.M.; Hann, C.L.; Laterra, J.; Yauch, R.L.; Callahan, C.A.; Fu, L.; Holcomb, T.; Stinson, J.; Gould, S.E.; Coleman, B.; et al. Treatment of medulloblastoma with hedgehog pathway inhibitor GDC-0449. N. Engl. J. Med. 2009, 361, 1173–1178. [Google Scholar] [CrossRef]
- Ogden, S.K.; Fei, D.L.; Schilling, N.S.; Ahmed, Y.F.; Hwa, J.; Robbins, D.J. G protein Galphai functions immediately downstream of Smoothened in Hedgehog signalling. Nature 2008, 456, 967–970. [Google Scholar] [CrossRef]
- Apionishev, S.; Katanayeva, N.M.; Marks, S.A.; Kalderon, D.; Tomlinson, A. Drosophila Smoothened phosphorylation sites essential for Hedgehog signal transduction. Nat. Cell Biol. 2005, 7, 86–92. [Google Scholar] [CrossRef]
- Jia, J.; Tong, C.; Wang, B.; Luo, L.; Jiang, J. Hedgehog signalling activity of Smoothened requires phosphorylation by protein kinase A and casein kinase I. Nature 2004, 432, 1045–1050. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Williams, E.H.; Guo, Y.; Lum, L.; Beachy, P.A. Extensive phosphorylation of Smoothened in Hedgehog pathway activation. Proc. Natl. Acad. Sci. USA 2004, 101, 17900–17907. [Google Scholar] [CrossRef]
- Jia, H.; Liu, Y.; Xia, R.; Tong, C.; Yue, T.; Jiang, J.; Jia, J. Casein kinase 2 promotes Hedgehog signaling by regulating both smoothened and Cubitus interruptus. J. Biol. Chem. 2010, 285, 37218–37226. [Google Scholar] [CrossRef]
- Chen, Y.; Li, S.; Tong, C.; Zhao, Y.; Wang, B.; Liu, Y.; Jia, J.; Jiang, J. G protein-coupled receptor kinase 2 promotes high-level Hedgehog signaling by regulating the active state of Smo through kinase-dependent and kinase-independent mechanisms in Drosophila. Genes Dev. 2010, 24, 2054–2067. [Google Scholar] [CrossRef]
- Jiang, K.; Liu, Y.; Fan, J.; Epperly, G.; Gao, T.; Jiang, J.; Jia, J. Hedgehog-regulated atypical PKC promotes phosphorylation and activation of Smoothened and Cubitus interruptus in Drosophila. Proc. Natl. Acad. Sci. USA 2014, 111, E4842–E4850. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Liu, Y.; Jia, J. Hh-induced Smoothened conformational switch is mediated by differential phosphorylation at its C-terminal tail in a dose- and position-dependent manner. Dev. Biol. 2012, 366, 172–184. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Tong, C.; Jiang, J. Hedgehog regulates smoothened activity by inducing a conformational switch. Nature 2007, 450, 252–258. [Google Scholar] [CrossRef]
- Rohatgi, R.; Milenkovic, L.; Scott, M.P. Patched1 regulates hedgehog signaling at the primary cilium. Science 2007, 317, 372–376. [Google Scholar] [CrossRef] [PubMed]
- Rohatgi, R.; Milenkovic, L.; Corcoran, R.B.; Scott, M.P. Hedgehog signal transduction by Smoothened: Pharmacologic evidence for a 2-step activation process. Proc. Natl. Acad. Sci. USA 2009, 106, 3196–3201. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhou, Z.; Walsh, C.T.; McMahon, A.P. Selective translocation of intracellular Smoothened to the primary cilium in response to Hedgehog pathway modulation. Proc. Natl. Acad. Sci. USA 2009, 106, 2623–2628. [Google Scholar] [CrossRef]
- Corbit, K.C.; Aanstad, P.; Singla, V.; Norman, A.R.; Stainier, D.Y.; Reiter, J.F. Vertebrate Smoothened functions at the primary cilium. Nature 2005, 437, 1018–1021. [Google Scholar] [CrossRef]
- Nusslein-Volhard, C.; Wieschaus, E. Mutations affecting segment number and polarity in Drosophila. Nature 1980, 287, 795–801. [Google Scholar] [CrossRef]
- Chen, Y.; Jiang, J. Decoding the phosphorylation code in Hedgehog signal transduction. Cell Res. 2013, 23, 186–200. [Google Scholar] [CrossRef]
- Jia, J. Phosphorylation regulation of Hedgehog signaling. Vitam. Horm. 2012, 88, 253–272. [Google Scholar] [CrossRef]
- Li, S.; Ma, G.; Wang, B.; Jiang, J. Hedgehog induces formation of PKA-Smoothened complexes to promote Smoothened phosphorylation and pathway activation. Sci. Signal. 2014, 7, ra62. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Li, S.; Han, Y.; Tong, C.; Wang, B.; Chen, Y.; Jiang, J. Regulation of Smoothened Phosphorylation and High-Level Hedgehog Signaling Activity by a Plasma Membrane Associated Kinase. PLoS Biol. 2016, 14, e1002481. [Google Scholar] [CrossRef] [PubMed]
- Nybakken, K.; Vokes, S.A.; Lin, T.Y.; McMahon, A.P.; Perrimon, N. A genome-wide RNA interference screen in Drosophila melanogaster cells for new components of the Hh signaling pathway. Nat. Genet. 2005, 37, 1323–1332. [Google Scholar] [CrossRef] [PubMed]
- Casso, D.J.; Liu, S.; Iwaki, D.D.; Ogden, S.K.; Kornberg, T.B. A screen for modifiers of hedgehog signaling in Drosophila melanogaster identifies swm and mts. Genetics 2008, 178, 1399–1413. [Google Scholar] [CrossRef][Green Version]
- Rorick, A.M.; Mei, W.; Liette, N.L.; Phiel, C.; El-Hodiri, H.M.; Yang, J. PP2A:B56epsilon is required for eye induction and eye field separation. Dev. Biol. 2007, 302, 477–493. [Google Scholar] [CrossRef]
- Jia, H.; Liu, Y.; Yan, W.; Jia, J. PP4 and PP2A regulate Hedgehog signaling by controlling Smo and Ci phosphorylation. Development 2009, 136, 307–316. [Google Scholar] [CrossRef]
- Su, Y.; Ospina, J.K.; Zhang, J.; Michelson, A.P.; Schoen, A.M.; Zhu, A.J. Sequential phosphorylation of smoothened transduces graded hedgehog signaling. Sci. Signal. 2011, 4, ra43. [Google Scholar] [CrossRef]
- Liu, M.; Liu, A.; Wang, J.; Zhang, Y.; Li, Y.; Su, Y.; Zhu, A.J. Competition between two phosphatases fine-tunes Hedgehog signaling. J. Cell Biol. 2021, 220, e202010078. [Google Scholar] [CrossRef]
- Li, S.; Chen, Y.; Shi, Q.; Yue, T.; Wang, B.; Jiang, J. Hedgehog-regulated ubiquitination controls smoothened trafficking and cell surface expression in Drosophila. PLoS Biol. 2012, 10, e1001239. [Google Scholar] [CrossRef]
- Xia, R.; Jia, H.; Fan, J.; Liu, Y.; Jia, J. USP8 promotes smoothened signaling by preventing its ubiquitination and changing its subcellular localization. PLoS Biol. 2012, 10, e1001238. [Google Scholar] [CrossRef]
- Li, S.; Li, S.; Wang, B.; Jiang, J. Hedgehog reciprocally controls trafficking of Smo and Ptc through the Smurf family of E3 ubiquitin ligases. Sci. Signal. 2018, 11, eaan8660. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Cho, Y.S.; Wang, B.; Li, S.; Jiang, J. Regulation of Smoothened ubiquitylation and cell surface expression through a Cul4-DDB1-Gbeta E3 ubiquitin ligase complex. J. Cell Sci. 2018, 131, jcs218016. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Yao, X.; Shan, Z.; Li, W.; Gao, Y.; Zhang, Q. E3 ligase Herc4 regulates Hedgehog signalling through promoting Smoothened degradation. J. Mol. Cell Biol. 2019, 11, 791–803. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Yao, X.; Pang, S.; Chen, P.; Jiang, W.; Shan, Z.; Zhang, Q. The deubiquitinase UCHL5/UCH37 positively regulates Hedgehog signaling by deubiquitinating Smoothened. J. Mol. Cell Biol. 2018, 10, 243–257. [Google Scholar] [CrossRef] [PubMed]
- Kong, J.H.; Young, C.B.; Pusapati, G.V.; Patel, C.B.; Ho, S.; Krishnan, A.; Lin, J.I.; Devine, W.; Moreau de Bellaing, A.; Athni, T.S.; et al. A Membrane-Tethered Ubiquitination Pathway Regulates Hedgehog Signaling and Heart Development. Dev. Cell 2020, 55, 432–449.e412. [Google Scholar] [CrossRef] [PubMed]
- Lv, B.; Stuck, M.W.; Desai, P.B.; Cabrera, O.A.; Pazour, G.J. E3 ubiquitin ligase Wwp1 regulates ciliary dynamics of the Hedgehog receptor Smoothened. J. Cell Biol. 2021, 220, e202010177. [Google Scholar] [CrossRef]
- Ma, G.; Li, S.; Han, Y.; Li, S.; Yue, T.; Wang, B.; Jiang, J. Regulation of Smoothened Trafficking and Hedgehog Signaling by the SUMO Pathway. Dev. Cell 2016, 39, 438–451. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, Y.; Jiang, K.; Jia, J. SUMO regulates the activity of Smoothened and Costal-2 in Drosophila Hedgehog signaling. Sci. Rep. 2017, 7, 42749. [Google Scholar] [CrossRef]
- Fan, J.; Jiang, K.; Liu, Y.; Jia, J. Hrs promotes ubiquitination and mediates endosomal trafficking of smoothened in Drosophila hedgehog signaling. PLoS ONE 2013, 8, e79021. [Google Scholar] [CrossRef]
- Jiang, K.; Liu, Y.; Zhang, J.; Jia, J. An intracellular activation of Smoothened that is independent of Hedgehog stimulation in Drosophila. J. Cell Sci. 2018, 131, jcs211367. [Google Scholar] [CrossRef]
- Matusek, T.; Wendler, F.; Poles, S.; Pizette, S.; D’Angelo, G.; Furthauer, M.; Therond, P.P. The ESCRT machinery regulates the secretion and long-range activity of Hedgehog. Nature 2014, 516, 99–103. [Google Scholar] [CrossRef]
- Jiang, K.; Liu, Y.; Fan, J.; Zhang, J.; Li, X.A.; Evers, B.M.; Zhu, H.; Jia, J. PI(4)P Promotes Phosphorylation and Conformational Change of Smoothened through Interaction with Its C-terminal Tail. PLoS Biol. 2016, 14, e1002375. [Google Scholar] [CrossRef] [PubMed]
- Yavari, A.; Nagaraj, R.; Owusu-Ansah, E.; Folick, A.; Ngo, K.; Hillman, T.; Call, G.; Rohatgi, R.; Scott, M.P.; Banerjee, U. Role of lipid metabolism in smoothened derepression in hedgehog signaling. Dev. Cell 2010, 19, 54–65. [Google Scholar] [CrossRef] [PubMed]
- Hedger, G.; Koldso, H.; Chavent, M.; Siebold, C.; Rohatgi, R.; Sansom, M.S.P. Cholesterol Interaction Sites on the Transmembrane Domain of the Hedgehog Signal Transducer and Class F G Protein-Coupled Receptor Smoothened. Structure 2019, 27, 549–559.e2. [Google Scholar] [CrossRef] [PubMed]
- Arensdorf, A.M.; Dillard, M.E.; Menke, J.M.; Frank, M.W.; Rock, C.O.; Ogden, S.K. Sonic Hedgehog Activates Phospholipase A2 to Enhance Smoothened Ciliary Translocation. Cell Rep. 2017, 19, 2074–2087. [Google Scholar] [CrossRef] [PubMed]
- Chavez, M.; Ena, S.; Van Sande, J.; de Kerchove d’Exaerde, A.; Schurmans, S.; Schiffmann, S.N. Modulation of Ciliary Phosphoinositide Content Regulates Trafficking and Sonic Hedgehog Signaling Output. Dev. Cell 2015, 34, 338–350. [Google Scholar] [CrossRef]
- Garcia-Gonzalo, F.R.; Phua, S.C.; Roberson, E.C.; Garcia, G., 3rd; Abedin, M.; Schurmans, S.; Inoue, T.; Reiter, J.F. Phosphoinositides Regulate Ciliary Protein Trafficking to Modulate Hedgehog Signaling. Dev. Cell 2015, 34, 400–409. [Google Scholar] [CrossRef]
- Mukhopadhyay, S.; Wen, X.; Ratti, N.; Loktev, A.; Rangell, L.; Scales, S.J.; Jackson, P.K. The ciliary G-protein-coupled receptor Gpr161 negatively regulates the Sonic hedgehog pathway via cAMP signaling. Cell 2013, 152, 210–223. [Google Scholar] [CrossRef]
- Constable, S.; Long, A.B.; Floyd, K.A.; Schurmans, S.; Caspary, T. The ciliary phosphatidylinositol phosphatase Inpp5e plays positive and negative regulatory roles in Shh signaling. Development 2020, 147, dev183301. [Google Scholar] [CrossRef] [PubMed]
- Cooper, M.K.; Porter, J.A.; Young, K.E.; Beachy, P.A. Teratogen-mediated inhibition of target tissue response to Shh signaling. Science 1998, 280, 1603–1607. [Google Scholar] [CrossRef] [PubMed]
- Corcoran, R.B.; Scott, M.P. Oxysterols stimulate Sonic hedgehog signal transduction and proliferation of medulloblastoma cells. Proc. Natl. Acad. Sci. USA 2006, 103, 8408–8413. [Google Scholar] [CrossRef]
- Myers, B.R.; Sever, N.; Chong, Y.C.; Kim, J.; Belani, J.D.; Rychnovsky, S.; Bazan, J.F.; Beachy, P.A. Hedgehog pathway modulation by multiple lipid binding sites on the smoothened effector of signal response. Dev. Cell 2013, 26, 346–357. [Google Scholar] [CrossRef]
- Rana, R.; Carroll, C.E.; Lee, H.J.; Bao, J.; Marada, S.; Grace, C.R.; Guibao, C.D.; Ogden, S.K.; Zheng, J.J. Structural insights into the role of the Smoothened cysteine-rich domain in Hedgehog signalling. Nat. Commun. 2013, 4, 2965. [Google Scholar] [CrossRef] [PubMed]
- Byrne, E.F.; Sircar, R.; Miller, P.S.; Hedger, G.; Luchetti, G.; Nachtergaele, S.; Tully, M.D.; Mydock-McGrane, L.; Covey, D.F.; Rambo, R.P.; et al. Structural basis of Smoothened regulation by its extracellular domains. Nature 2016, 535, 517–522. [Google Scholar] [CrossRef]
- Nachtergaele, S.; Mydock, L.K.; Krishnan, K.; Rammohan, J.; Schlesinger, P.H.; Covey, D.F.; Rohatgi, R. Oxysterols are allosteric activators of the oncoprotein Smoothened. Nat. Chem. Biol. 2012, 8, 211–220. [Google Scholar] [CrossRef]
- Nedelcu, D.; Liu, J.; Xu, Y.; Jao, C.; Salic, A. Oxysterol binding to the extracellular domain of Smoothened in Hedgehog signaling. Nat. Chem. Biol. 2013, 9, 557–564. [Google Scholar] [CrossRef] [PubMed]
- Dwyer, J.R.; Sever, N.; Carlson, M.; Nelson, S.F.; Beachy, P.A.; Parhami, F. Oxysterols are novel activators of the hedgehog signaling pathway in pluripotent mesenchymal cells. J. Biol. Chem. 2007, 282, 8959–8968. [Google Scholar] [CrossRef]
- Nachtergaele, S.; Whalen, D.M.; Mydock, L.K.; Zhao, Z.; Malinauskas, T.; Krishnan, K.; Ingham, P.W.; Covey, D.F.; Siebold, C.; Rohatgi, R. Structure and function of the Smoothened extracellular domain in vertebrate Hedgehog signaling. eLife 2013, 2, e01340. [Google Scholar] [CrossRef]
- Byrne, E.F.; Luchetti, G.; Rohatgi, R.; Siebold, C. Multiple ligand binding sites regulate the Hedgehog signal transducer Smoothened in vertebrates. Curr. Opin. Cell Biol. 2017, 51, 81–88. [Google Scholar] [CrossRef]
- Xiao, X.; Tang, J.J.; Peng, C.; Wang, Y.; Fu, L.; Qiu, Z.P.; Xiong, Y.; Yang, L.F.; Cui, H.W.; He, X.L.; et al. Cholesterol Modification of Smoothened Is Required for Hedgehog Signaling. Mol. Cell 2017, 66, 154–162.e110. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.; Nedelcu, D.; Watanabe, M.; Jao, C.; Kim, Y.; Liu, J.; Salic, A. Cellular Cholesterol Directly Activates Smoothened in Hedgehog Signaling. Cell 2016, 166, 1176–1187.e1114. [Google Scholar] [CrossRef] [PubMed]
- Kinnebrew, M.; Iverson, E.J.; Patel, B.B.; Pusapati, G.V.; Kong, J.H.; Johnson, K.A.; Luchetti, G.; Eckert, K.M.; McDonald, J.G.; Covey, D.F.; et al. Cholesterol accessibility at the ciliary membrane controls hedgehog signaling. eLife 2019, 8, e50051. [Google Scholar] [CrossRef] [PubMed]
- Myers, B.R.; Neahring, L.; Zhang, Y.; Roberts, K.J.; Beachy, P.A. Rapid, direct activity assays for Smoothened reveal Hedgehog pathway regulation by membrane cholesterol and extracellular sodium. Proc. Natl. Acad. Sci. USA 2017, 114, E11141–E11150. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.; Zheng, S.; Wierbowski, B.M.; Kim, Y.; Nedelcu, D.; Aravena, L.; Liu, J.; Kruse, A.C.; Salic, A. Structural Basis of Smoothened Activation in Hedgehog Signaling. Cell 2018, 174, 312–324. [Google Scholar] [CrossRef]
- Raleigh, D.R.; Sever, N.; Choksi, P.K.; Sigg, M.A.; Hines, K.M.; Thompson, B.M.; Elnatan, D.; Jaishankar, P.; Bisignano, P.; Garcia-Gonzalo, F.R.; et al. Cilia-Associated Oxysterols Activate Smoothened. Mol. Cell 2018, 72, 316–327.e315. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.; Schmiege, P.; Coutavas, E.; Li, X. Two Patched molecules engage distinct sites on Hedgehog yielding a signaling-competent complex. Science 2018, 362, eaas8843. [Google Scholar] [CrossRef]
- Qi, X.; Schmiege, P.; Coutavas, E.; Wang, J.; Li, X. Structures of human Patched and its complex with native palmitoylated sonic hedgehog. Nature 2018, 560, 128–132. [Google Scholar] [CrossRef]
- Qi, X.; Liu, H.; Thompson, B.; McDonald, J.; Zhang, C.; Li, X. Cryo-EM structure of oxysterol-bound human Smoothened coupled to a heterotrimeric Gi. Nature 2019, 571, 279–283. [Google Scholar] [CrossRef]
- Findakly, S.; Daggubati, V.; Garcia, G.; LaStella, S.A.; Choudhury, A.; Tran, C.; Li, A.; Tong, P.; Garcia, J.Q.; Puri, N.; et al. Sterol and oxysterol synthases near the ciliary base activate the Hedgehog pathway. J. Cell Biol. 2021, 220, e202002026. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 1–11. [Google Scholar] [CrossRef]
- Kong, J.H.; Siebold, C.; Rohatgi, R. Biochemical mechanisms of vertebrate hedgehog signaling. Development 2019, 146, dev166892. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Liu, Z.; Jia, J. Mechanisms of Smoothened Regulation in Hedgehog Signaling. Cells 2021, 10, 2138. https://doi.org/10.3390/cells10082138
Zhang J, Liu Z, Jia J. Mechanisms of Smoothened Regulation in Hedgehog Signaling. Cells. 2021; 10(8):2138. https://doi.org/10.3390/cells10082138
Chicago/Turabian StyleZhang, Jie, Zulong Liu, and Jianhang Jia. 2021. "Mechanisms of Smoothened Regulation in Hedgehog Signaling" Cells 10, no. 8: 2138. https://doi.org/10.3390/cells10082138
APA StyleZhang, J., Liu, Z., & Jia, J. (2021). Mechanisms of Smoothened Regulation in Hedgehog Signaling. Cells, 10(8), 2138. https://doi.org/10.3390/cells10082138






