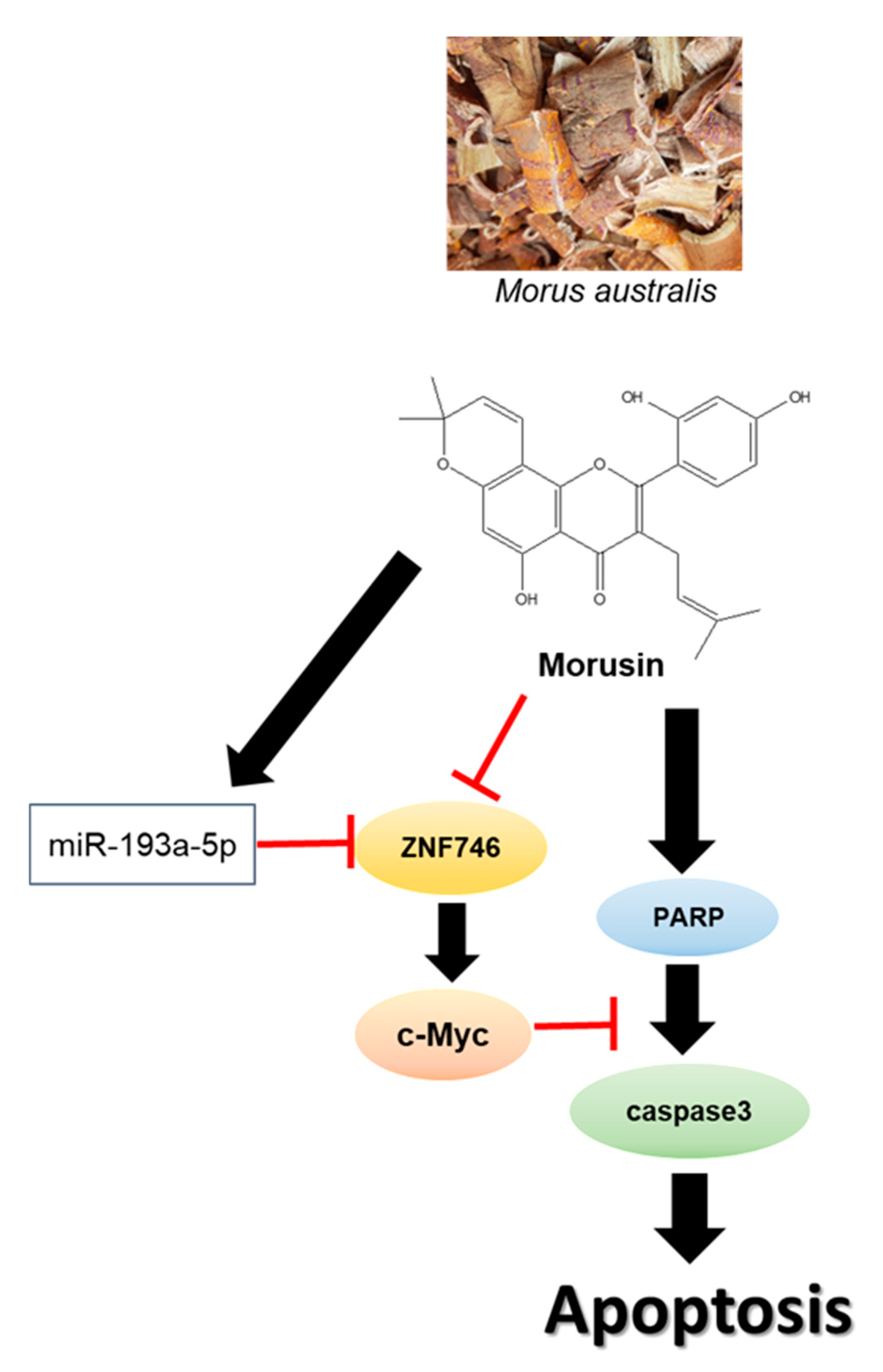
miR193a-5p Mediated ZNF746 and c-Myc Signaling Axis Is Critically Involved in Morusin Induced Apoptosis in Colorectal Cancer Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Cell Viability Assay
2.3. Colony Formation Assay
2.4. Cell Cycle Analysis
2.5. Cycloheximide Assay
2.6. Western Blotting
2.7. TUNEL Assay
2.8. RT-qPCR Analysis
2.9. RNA Interference
2.10. Co-Immunoprecipitation
2.11. Immunofluorescence
2.12. Statistical Analysis
3. Results
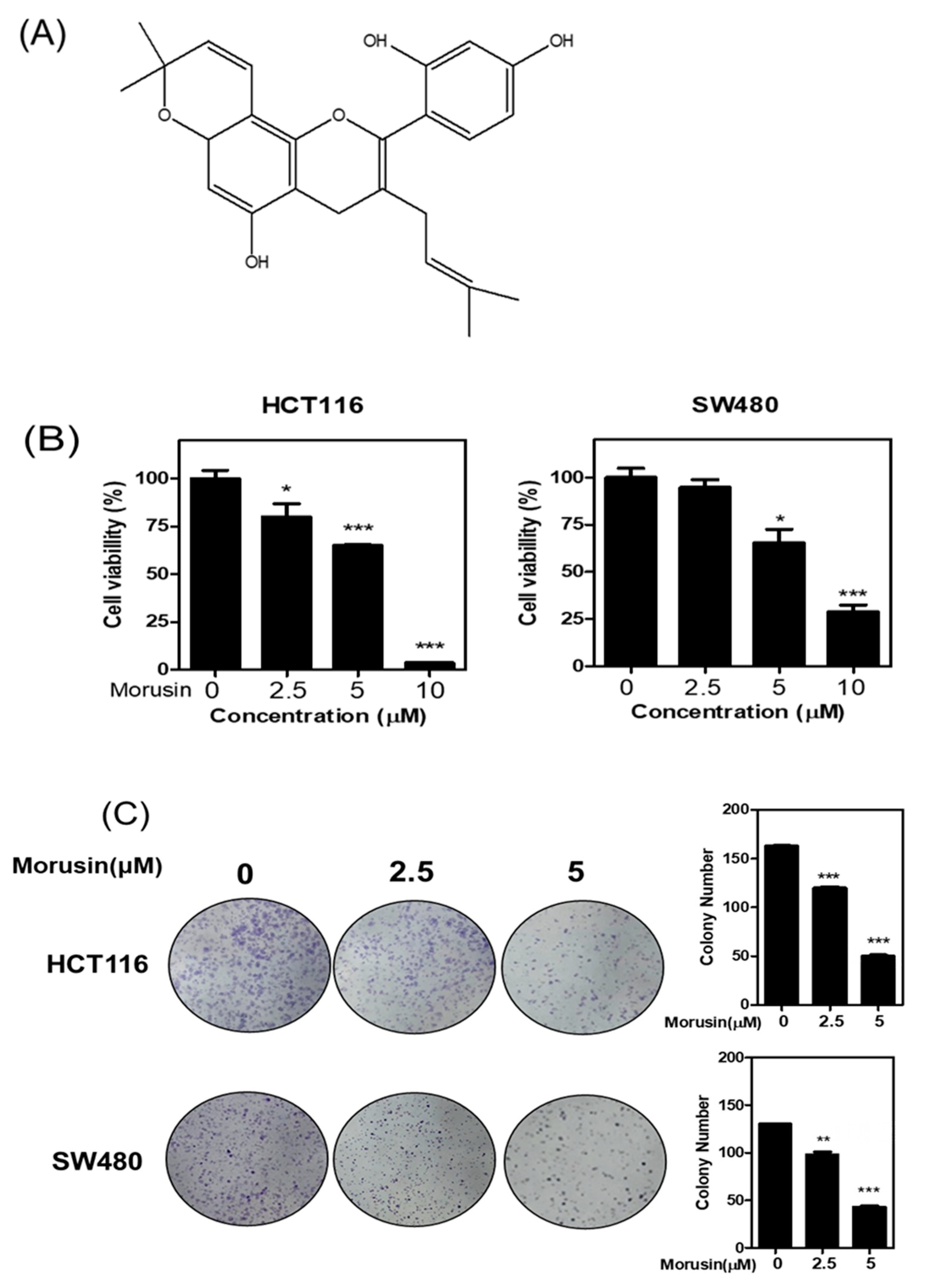
3.1. Cytotoxic Effect of Morusin in Colorectal Cancer Cells
3.2. Morusin Induced Apoptosis in HCT116 and SW480 Cells
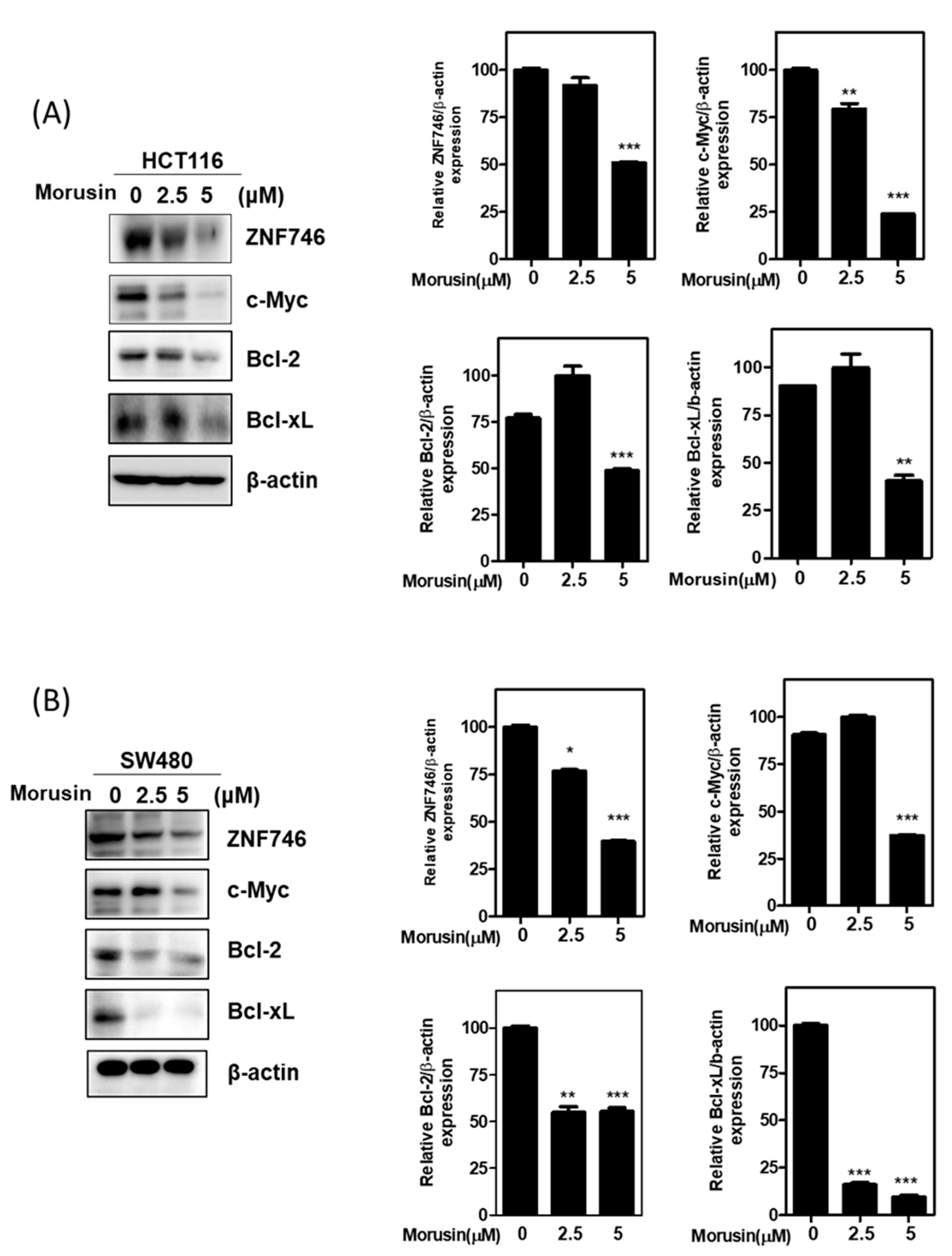
3.3. Morusin Attenuated the Expression of ZNF746 and c-Myc in HCT116 and SW480 Cells
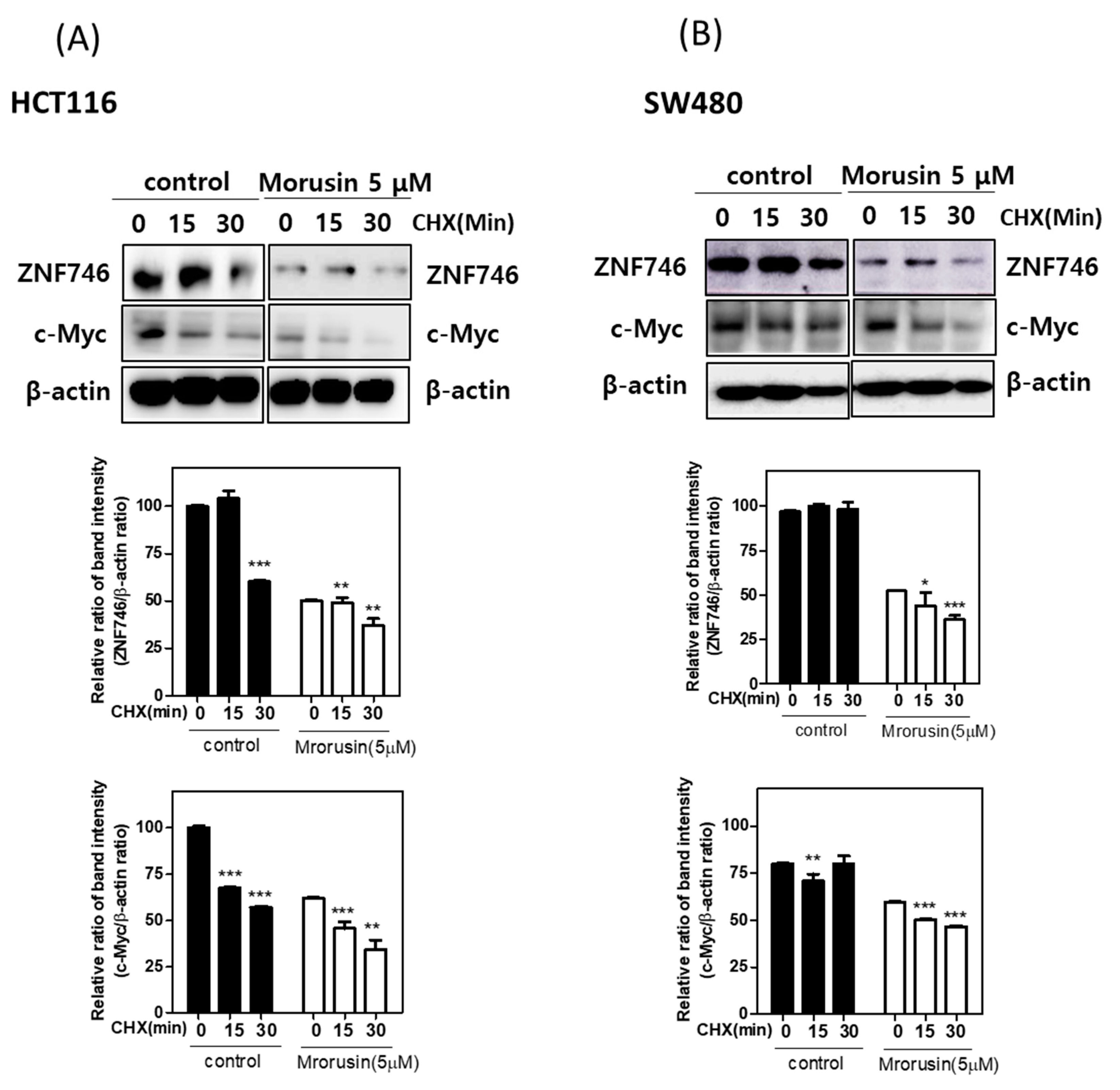
3.4. Morusin Reduced the Stability of ZNF746 and c-Myc in HCT116 and SW480 Cells in the Presence of Cycloheximide
3.5. Ectopic Expression of ZNF746 Reduces Apoptotic Effect of Morusin in HCT116 Cells
3.6. Morusin Disrupted the Binding of c-Myc and ZNF746 in HCT116 Cells
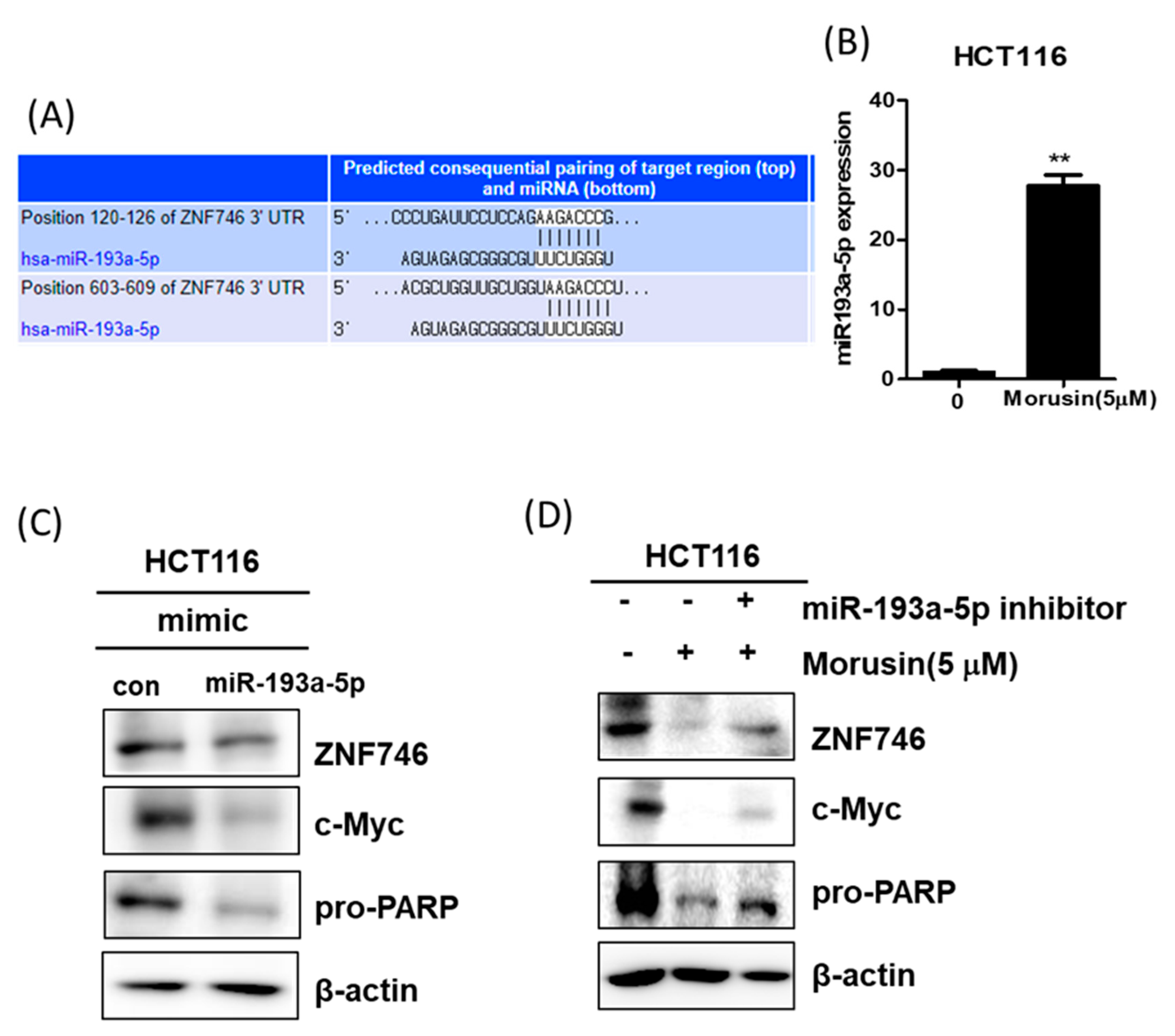
3.7. miR193a-5p Plays a Pivotal Role in Morusin-Induced Apoptosis in HCT116 Cells
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Jung, J.H.; Jung, D.-B.; Kim, H.; Lee, H.; Kang, S.-E.; Srivastava, S.K.; Yun, M.; Kim, S.-H. Zinc finger protein 746 promotes colorectal cancer progression via c-Myc stability mediated by glycogen synthase kinase 3β and F-box and WD repeat domain-containing 7. Oncogene 2018, 37, 3715–3728. [Google Scholar] [CrossRef]
- Thanikachalam, K.; Khan, G. Colorectal Cancer and Nutrition. Nutrients 2019, 11, 164. [Google Scholar] [CrossRef]
- Zhou, Y.; Li, X.; Ye, M. Morusin inhibits the growth of human colorectal cancer HCT116-derived sphere-forming cells via the inactivation of Akt pathway. Int. J. Mol. Med. 2021, 47, 1. [Google Scholar] [CrossRef]
- Piawah, S.; Venook, A.P. Targeted therapy for colorectal cancer metastases: A review of current methods of molecularly targeted therapy and the use of tumor biomarkers in the treatment of metastatic colorectal cancer. Cancer 2019, 125, 4139–4147. [Google Scholar] [CrossRef]
- Yiu, A.J.; Yiu, C.Y. Biomarkers in Colorectal Cancer. Anticancer Res. 2016, 36, 1093–1102. [Google Scholar]
- Park, J.E.; Jung, J.H.; Lee, H.; Sim, D.Y.; Im, E.; Park, W.Y.; Shim, B.S.; Ko, S.; Kim, S. Ribosomal protein L5 mediated inhibition of c-Myc is critically involved in sanggenon G induced apoptosis in non-small lung cancer cells. Phytother. Res. 2021, 35, 1080–1088. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Ren, P.; Xu, D.; Liu, X.; Liu, Z.; Zhang, C.; Li, Y.; Wang, L.; Du, X.; Xing, B. Human UTP14a promotes colorectal cancer progression by forming a positive regulation loop with c-Myc. Cancer Lett. 2019, 440–441, 106–115. [Google Scholar] [CrossRef]
- Huang, J.; Wang, J.-F.; Zhang, H.-S. Structure and function of plant C2H2 zinc finger protein. Yi Chuan 2004, 26, 414–418. [Google Scholar] [PubMed]
- Han, G.; Lu, C.; Guo, J.; Qiao, Z.; Sui, N.; Qiu, N.; Wang, B. C2H2 Zinc Finger Proteins: Master Regulators of Abiotic Stress Responses in Plants. Front. Plant Sci. 2020, 11, 115. [Google Scholar] [CrossRef]
- Kamitani, S.; Togi, S.; Ikeda, O.; Nakasuji, M.; Sakauchi, A.; Sekine, Y.; Muromoto, R.; Oritani, K.; Matsuda, T. Krüppel-Associated Box-Associated Protein 1 Negatively Regulates TNF-α–Induced NF-κB Transcriptional Activity by Influencing the Interactions among STAT3, p300, and NF-κB/p65. J. Immunol. 2011, 187, 2476–2483. [Google Scholar] [CrossRef]
- Cesaro, E.; Sodaro, G.; Montano, G.; Grosso, M.; Lupo, A.; Costanzo, P. The Complex Role of the ZNF224 Transcription Factor in Cancer. Protein-Protein Interact. Hum. Dis. Part A 2017, 107, 191–222. [Google Scholar] [CrossRef]
- Kim, S.; Sohn, E.J.; Jung, J.H.; Shin, E.A.; You, O.H.; Im, J. Inhibition of ZNF746 suppresses invasion and epithelial to mesenchymal transition in H460 non-small cell lung cancer cells. Oncol. Rep. 2013, 31, 73–78. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kane, L.A.; Lazarou, M.; Fogel, A.I.; Li, Y.; Yamano, K.; Sarraf, S.; Banerjee, S.; Youle, R.J. PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J. Cell Biol. 2014, 205, 143–153. [Google Scholar] [CrossRef]
- Lee, Y.S.; Dutta, A. MicroRNAs in Cancer. Annu. Rev. Pathol. Mech. Dis. 2009, 4, 199–227. [Google Scholar] [CrossRef]
- Goodall, G.J.; Wickramasinghe, V.O. RNA in cancer. Nat. Rev. Cancer 2021, 21, 22–36. [Google Scholar] [CrossRef]
- Toden, S.; Zumwalt, T.J.; Goel, A. Non-coding RNAs and potential therapeutic targeting in cancer. Biochim. Biophys. Acta (BBA)-Bioenerg. 2021, 1875, 188491. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, T.; Chen, D.; Ma, Q.; Zheng, Y.; Liao, S.; Wang, Y.; Zhang, J. Quercetin preferentially induces apoptosis in KRAS-mutant colorectal cancer cells via JNK signaling pathways. Cell Biol. Int. 2019, 43, 117–124. [Google Scholar] [CrossRef]
- Shan, J.; Xuan, Y.; Zhang, Q.; Zhu, C.; Liu, Z.; Zhang, S. Ursolic acid synergistically enhances the therapeutic effects of oxaliplatin in colorectal cancer. Protein Cell 2016, 7, 571–585. [Google Scholar] [CrossRef][Green Version]
- Pricci, M.; Girardi, B.; Giorgio, F.; Losurdo, G.; Ierardi, E.; Di Leo, A. Curcumin and Colorectal Cancer: From Basic to Clinical Evidences. Int. J. Mol. Sci. 2020, 21, 2364. [Google Scholar] [CrossRef]
- Hazafa, A.; Rehman, K.-U.; Jahan, N.; Jabeen, Z. The Role of Polyphenol (Flavonoids) Compounds in the Treatment of Cancer Cells. Nutr. Cancer 2020, 72, 386–397. [Google Scholar] [CrossRef]
- Venturelli, S.; Burkard, M.; Biendl, M.; Lauer, U.M.; Frank, J.; Busch, C. Prenylated chalcones and flavonoids for the prevention and treatment of cancer. Nutrition 2016, 32, 1171–1178. [Google Scholar] [CrossRef]
- Dat, N.T.; Binh, P.T.X.; Quynh, L.T.P.; Van Minh, C.; Huong, H.T.; Lee, J.J. Cytotoxic prenylated flavonoids from Morus alba. Fitoterapia 2010, 81, 1224–1227. [Google Scholar] [CrossRef]
- Wu, S.-C.; Han, F.; Song, M.-R.; Chen, S.; Li, Q.; Zhang, Q.; Zhu, K.; Shen, J.-Z. Natural Flavones fromMorus albaagainst Methicillin-ResistantStaphylococcus aureusvia Targeting the Proton Motive Force and Membrane Permeability. J. Agric. Food Chem. 2019, 67, 10222–10234. [Google Scholar] [CrossRef]
- Jia, Y.; He, W.; Zhang, H.; He, L.; Wang, Y.; Zhang, T.; Peng, J.; Sun, P.; Qian, Y. Morusin Ameliorates IL-1β-Induced Chondrocyte Inflammation and Osteoarthritis via NF-κB Signal Pathway. Drug Des. Dev. Ther. 2020, 14, 1227–1240. [Google Scholar] [CrossRef]
- Yang, C.; Luo, J.; Luo, X.; Jia, W.; Fang, Z.; Yi, S.; Li, L. Morusin exerts anti-cancer activity in renal cell carcinoma by disturbing MAPK signaling pathways. Ann. Transl. Med. 2020, 8, 327. [Google Scholar] [CrossRef]
- Huang, C.-C.; Wang, P.-H.; Lu, Y.-T.; Yang, J.-S.; Yang, S.-F.; Ho, Y.-T.; Lin, C.-W.; Hsin, C.-H. Morusin Suppresses Cancer Cell Invasion and MMP-2 Expression through ERK Signaling in Human Nasopharyngeal Carcinoma. Molecules 2020, 25, 4851. [Google Scholar] [CrossRef]
- Costea, T.; Hudiță, A.; Ciolac, O.-A.; Gălățeanu, B.; Ginghină, O.; Costache, M.; Ganea, C.; Mocanu, M.-M. Chemoprevention of Colorectal Cancer by Dietary Compounds. Int. J. Mol. Sci. 2018, 19, 3787. [Google Scholar] [CrossRef] [PubMed]
- Rejhová, A.; Opattová, A.; Čumová, A.; Slíva, D.; Vodička, P. Natural compounds and combination therapy in colorectal cancer treatment. Eur. J. Med. Chem. 2018, 144, 582–594. [Google Scholar] [CrossRef]
- Lee, J.-C.; Won, S.-J.; Chao, C.-L.; Wu, F.-L.; Liu, H.-S.; Ling, P.; Lin, C.-N.; Su, C.-L. Morusin induces apoptosis and suppresses NF-κB activity in human colorectal cancer HT-29 cells. Biochem. Biophys. Res. Commun. 2008, 372, 236–242. [Google Scholar] [CrossRef]
- Kajstura, M.; Halicka, H.D.; Pryjma, J.; Darzynkiewicz, Z. Discontinuous fragmentation of nuclear DNA during apoptosis revealed by discrete “sub-G1” peaks on DNA content histograms. Cytom. Part A 2007, 71, 125–131. [Google Scholar] [CrossRef]
- Porter, A.G.; Jänicke, R.U. Emerging roles of caspase-3 in apoptosis. Cell Death Differ. 1999, 6, 99–104. [Google Scholar] [CrossRef] [PubMed]
- Larsen, B.; Sørensen, C.S. The caspase-activated DN ase: Apoptosis and beyond. FEBS J. 2017, 284, 1160–1170. [Google Scholar] [CrossRef] [PubMed]
- Van Opdenbosch, N.; Lamkanfi, M. Caspases in Cell Death, Inflammation, and Disease. Immunity 2019, 50, 1352–1364. [Google Scholar] [CrossRef]
- Elbadawy, M.; Usui, T.; Yamawaki, H.; Sasaki, K. Emerging Roles of C-Myc in Cancer Stem Cell-Related Signaling and Resistance to Cancer Chemotherapy: A Potential Therapeutic Target Against Colorectal Cancer. Int. J. Mol. Sci. 2019, 20, 2340. [Google Scholar] [CrossRef]
- Pérez-Sayáns, M.; Suárez-Peñaranda, J.M.; Pilar, G.-D.; Barros, F.; Gándara-Rey, J.M.; Garcia-Garcia, A. What real influence does the proto-oncogene c-Myc have in OSCC behavior? Oral Oncol. 2011, 47, 688–692. [Google Scholar] [CrossRef] [PubMed]
- Dang, C.V. MYC on the Path to Cancer. Cell 2012, 149, 22–35. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Yang, T.; Li, Y.; Ye, W.; Liu, F.; He, X.; Wang, J.; Gan, W.; Li, X.; Zhang, S.; et al. Tumor Necrosis Factor Receptor–Associated Factor 6 Promotes Hepatocarcinogenesis by Interacting With Histone Deacetylase 3 to Enhance c-Myc Gene Expression and Protein Stability. Hepatology 2020, 71, 148–163. [Google Scholar] [CrossRef]
- Chen, Y.-T.; Yang, C.-C.; Shao, P.-L.; Huang, C.-R.; Yip, H.-K. Melatonin-mediated downregulation of ZNF746 suppresses bladder tumorigenesis mainly through inhibiting the AKT-MMP-9 signaling pathway. J. Pineal Res. 2018, 66, e12536. [Google Scholar] [CrossRef]
- Tutar, Y. Editorial (Thematic Issue: “miRNA and Cancer; Computational and Experimental Approaches”). Curr. Pharm. Biotechnol. 2014, 15, 429. [Google Scholar] [CrossRef] [PubMed]
- Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017, 16, 203–222. [Google Scholar] [CrossRef]
- Roy, S.; Hooiveld, G.; Seehawer, M.; Caruso, S.; Heinzmann, F.; Schneider, A.T.; Frank, A.K.; Cardenas, D.V.; Sonntag, R.; Luedde, M.; et al. microRNA 193a-5p Regulates Levels of Nucleolar- and Spindle-Associated Protein 1 to Suppress Hepatocarcinogenesis. Gastroenterology 2018, 155, 1951–1966.e26. [Google Scholar] [CrossRef]
- Noorolyai, S.; Baghbani, E.; Maleki, L.A.; Kojabad, A.B.; Shanehbandi, D.; Shahgoli, V.K.; Mokhtarzadeh, A.; Baradaran, B. Restoration of miR-193a-5p and miR-146 a-5p Expression Induces G1 Arrest in Colorectal Cancer through Targeting of MDM2/p53. Adv. Pharm. Bull. 2019, 10, 130–134. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, W.-Y.; Lee, H.-J.; Sim, D.-Y.; Im, E.; Park, J.-E.; Ahn, C.-H.; Shim, B.-S.; Kim, S.-H. miR193a-5p Mediated ZNF746 and c-Myc Signaling Axis Is Critically Involved in Morusin Induced Apoptosis in Colorectal Cancer Cells. Cells 2021, 10, 2065. https://doi.org/10.3390/cells10082065
Park W-Y, Lee H-J, Sim D-Y, Im E, Park J-E, Ahn C-H, Shim B-S, Kim S-H. miR193a-5p Mediated ZNF746 and c-Myc Signaling Axis Is Critically Involved in Morusin Induced Apoptosis in Colorectal Cancer Cells. Cells. 2021; 10(8):2065. https://doi.org/10.3390/cells10082065
Chicago/Turabian StylePark, Woon-Yi, Hyo-Jung Lee, Deok-Yong Sim, Eunji Im, Ji-Eon Park, Chi-Hoon Ahn, Bum-Sang Shim, and Sung-Hoon Kim. 2021. "miR193a-5p Mediated ZNF746 and c-Myc Signaling Axis Is Critically Involved in Morusin Induced Apoptosis in Colorectal Cancer Cells" Cells 10, no. 8: 2065. https://doi.org/10.3390/cells10082065
APA StylePark, W.-Y., Lee, H.-J., Sim, D.-Y., Im, E., Park, J.-E., Ahn, C.-H., Shim, B.-S., & Kim, S.-H. (2021). miR193a-5p Mediated ZNF746 and c-Myc Signaling Axis Is Critically Involved in Morusin Induced Apoptosis in Colorectal Cancer Cells. Cells, 10(8), 2065. https://doi.org/10.3390/cells10082065






