Transcriptomic Analysis of Human Brain -Microvascular Endothelial Cell Driven Changes in -Vascular Pericytes
Abstract
1. Introduction
2. Materials and Methods
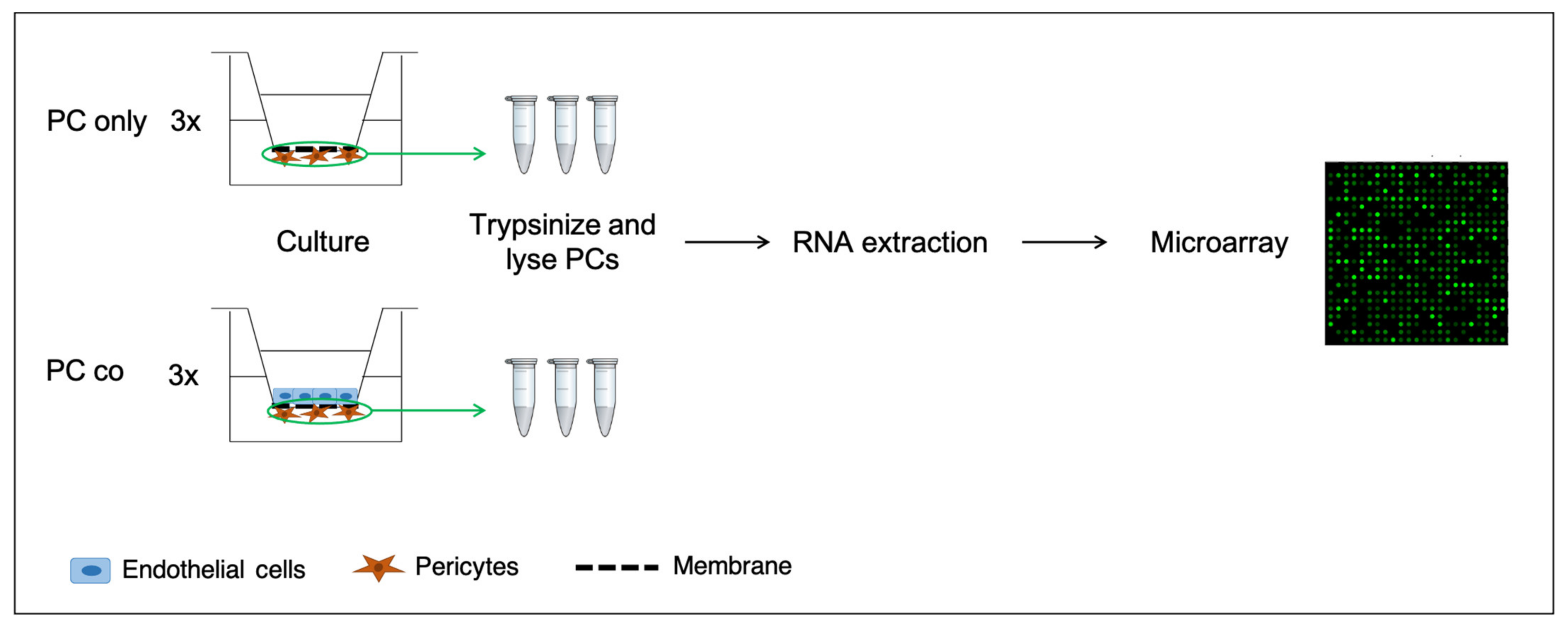
2.1. Cell Culture
2.2. Immuno-Fluorescence
2.3. Microarray Analysis
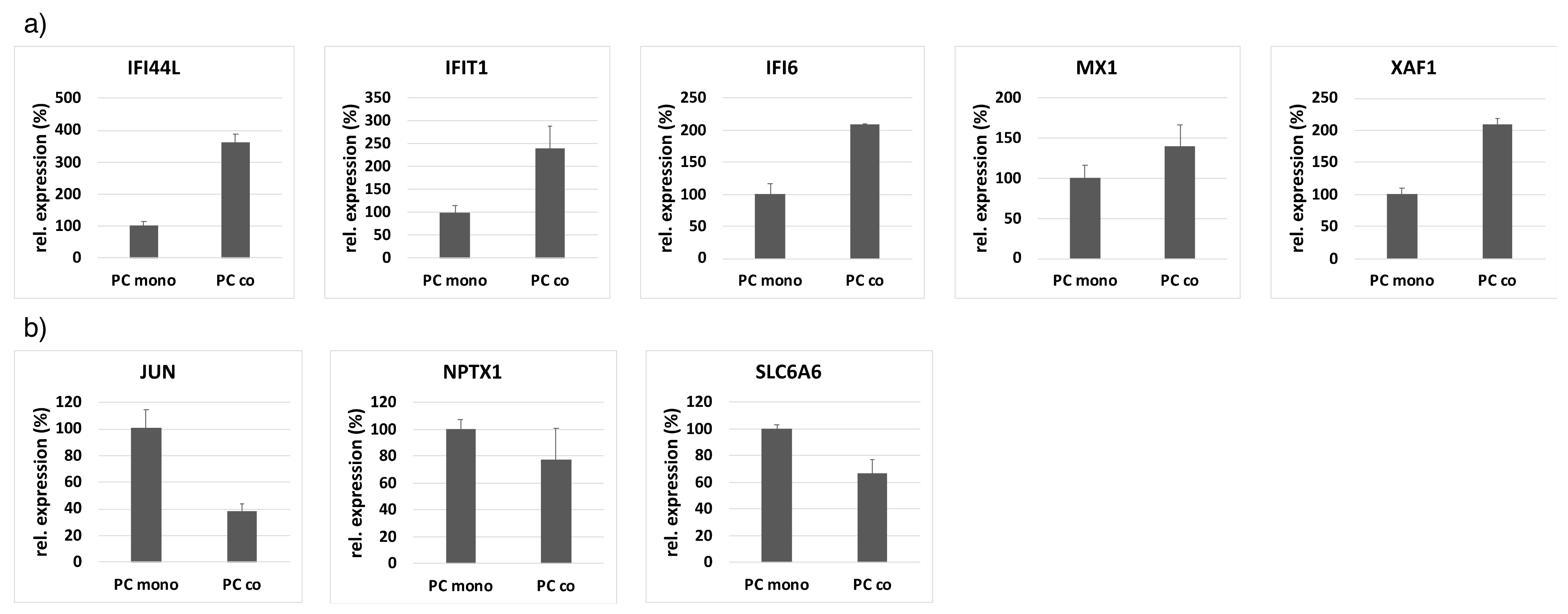
2.4. Quantitative RT-PCR
2.5. Cytokine Proteome Array
2.6. Western Blot Analysis
2.7. Statistical Analysis
3. Results
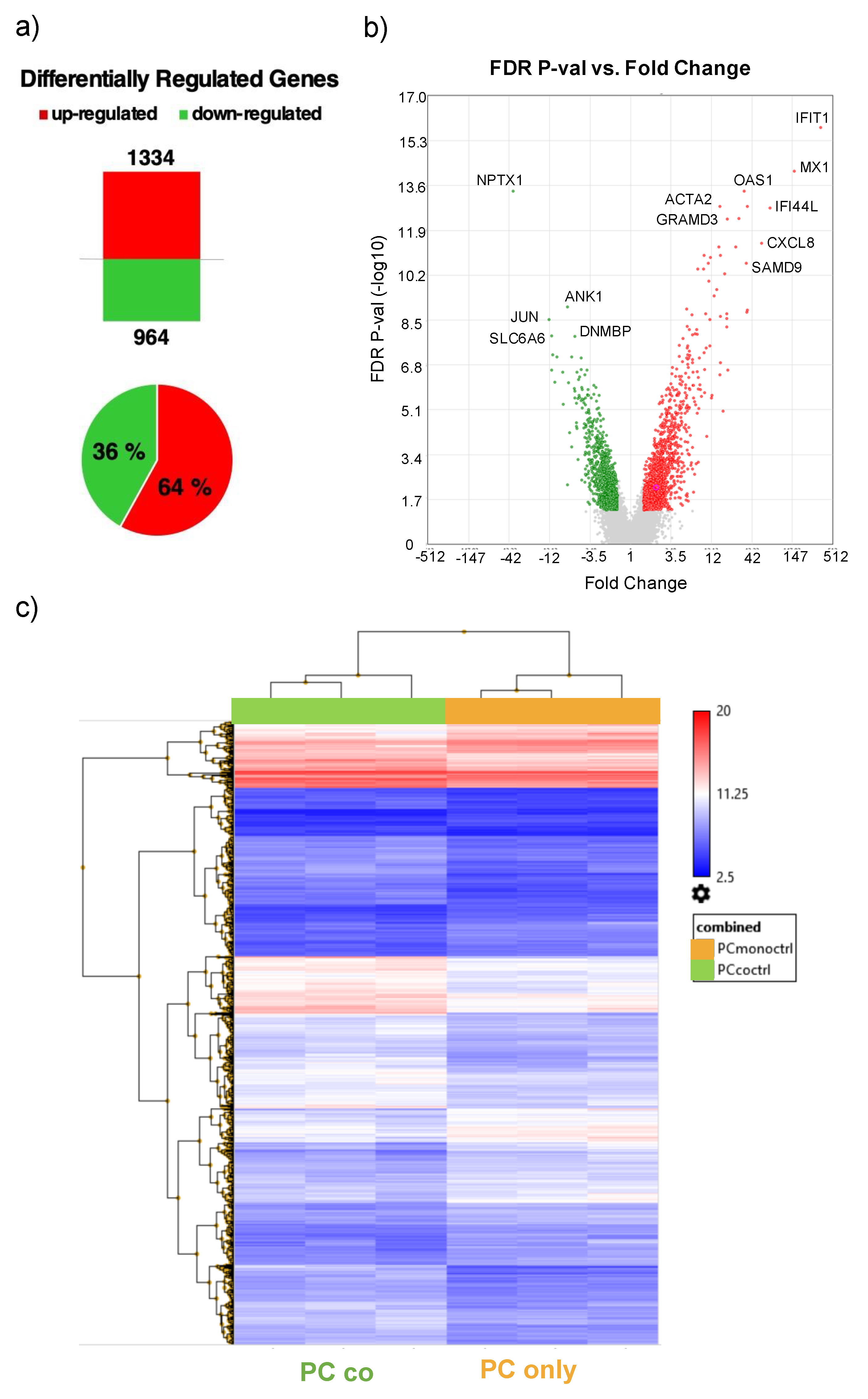
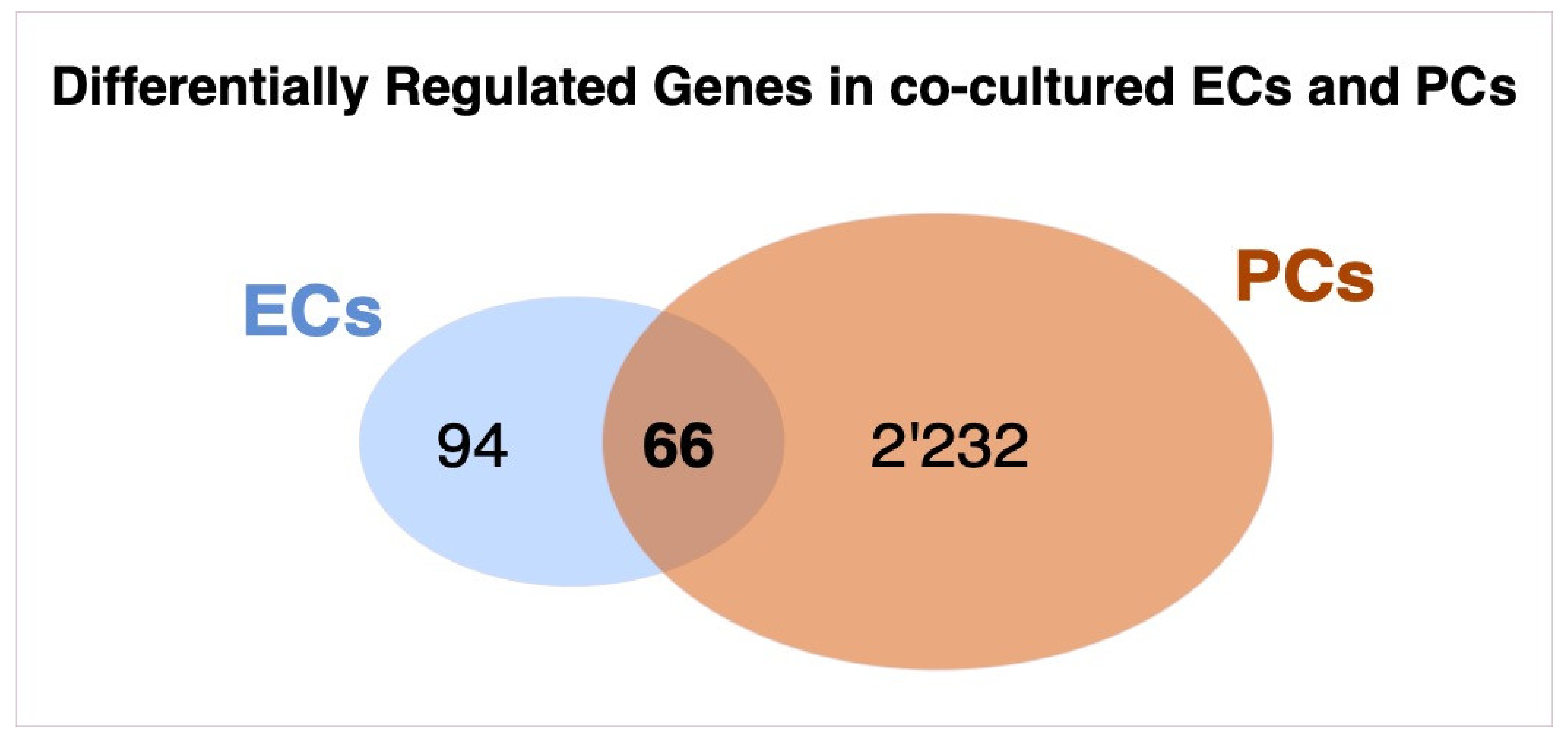
3.1. Differentially Regulated Genes
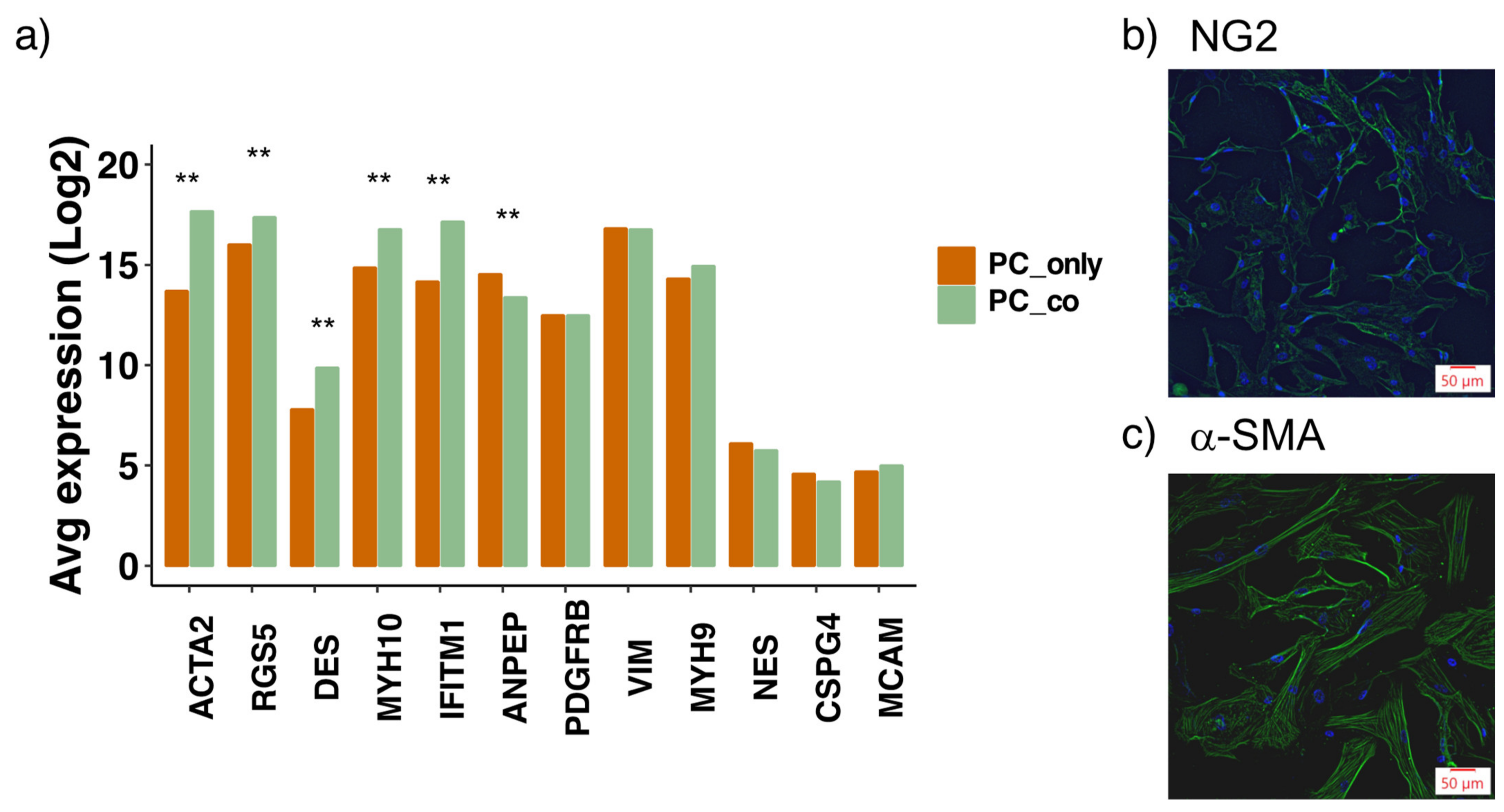
3.2. Assessment of Pericyte Marker Expression
3.3. Effect of Co-Culture on Proteins of the Extracellular Matrix
3.4. Effect of Co-Culture on Inflammatory Profile
3.5. Effect of Co-Culture on Secreted Soluble Proteins
3.6. Effect of Co-Culture on Transporters, Carriers, Ion Pumps and G Protein-Coupled Receptors
3.7. Effect of Co-Culture on Other Proteins
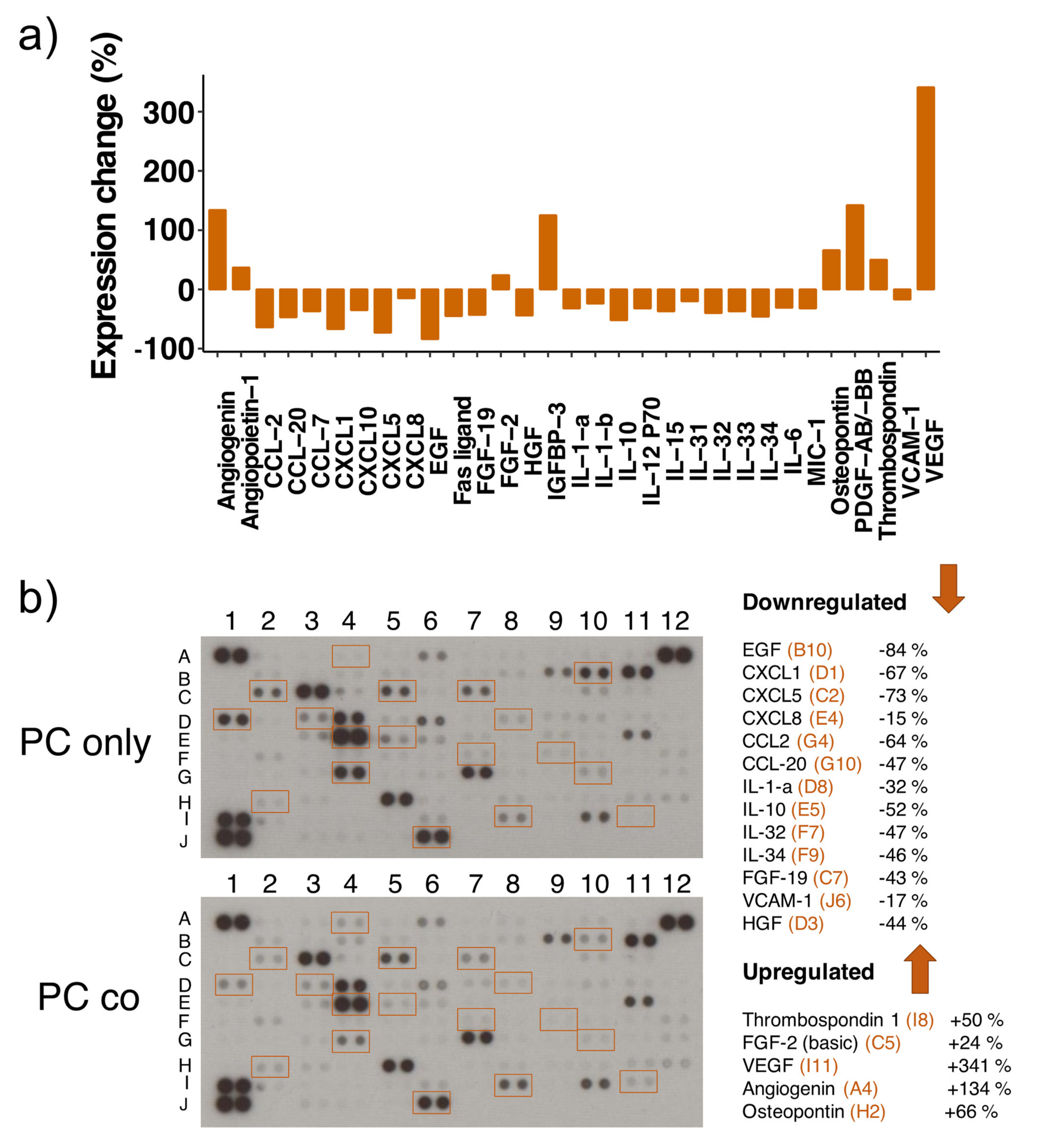
3.8. Cytokine Proteome Profiler
3.9. Overlap of Transcriptomic Changes in Co-Cultured Pericytes and Endothelial Cells
4. Discussion
4.1. PC Markers
4.2. Inflammatory Profile
4.3. ECM Proteins
4.4. Soluble Factors
4.5. Solute Carriers, Transporters, Ion Channels and G Protein-Coupled Receptors (GPCRs)
4.6. Other Proteins
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sweeney, M.D.; Ayyadurai, S.; Zlokovic, B.V. Pericytes of the neurovascular unit: Key functions and signaling pathways. Nat. Neurosci. 2016, 19, 771–783. [Google Scholar] [CrossRef]
- Daneman, R.; Zhou, L.; Kebede, A.A.; Barres, B.A. Pericytes are required for blood-brain barrier integrity during embryogenesis. Nature 2010, 468, 562–566. [Google Scholar] [CrossRef]
- Armulik, A.; Genove, G.; Betsholtz, C. Pericytes: Developmental, Physiological, and Pathological Perspectives, Problems, and Promises. Dev. Cell 2011, 21, 193–215. [Google Scholar] [CrossRef]
- Sweeney, M.; Foldes, G. Takes Two: Endothelial-Perivascular Cell Cross-Talk in Vascular Development and Disease. Front. Cardiovasc. Med. 2018, 5. [Google Scholar] [CrossRef]
- Lendahl, U.; Nilsson, P.; Betsholtz, C. Emerging links between cerebrovascular and neurodegenerative diseases-a special role for pericytes. EMBO Rep. 2019, 20, e48070. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, A.L.; Okamoto, O.K. Combined Effects of Pericytes in the Tumor Microenvironment. Stem Cells Int. 2015, 2015. [Google Scholar] [CrossRef]
- Armulik, A.; Abramsson, A.; Betsholtz, C. Endothelial/pericyte interactions. Circ. Res. 2005, 97, 512–523. [Google Scholar] [CrossRef]
- Zhang, Z.-S.; Zhou, H.-N.; He, S.-S.; Xue, M.-Y.; Li, T.; Liu, L.-M. Research advances in pericyte function and their roles in diseases. Chin. J. Traumatol. 2020, 23, 89–95. [Google Scholar] [CrossRef]
- Stratman, A.N.; Davis, G.E. Endothelial Cell-Pericyte Interactions Stimulate Basement Membrane Matrix Assembly: Influence on Vascular Tube Remodeling, Maturation, and Stabilization. Microsc. Microanal. 2012, 18, 68–80. [Google Scholar] [CrossRef]
- Baeten, K.M.; Akassoglou, K. Extracellular Matrix and Matrix Receptors in Blood-Brain Barrier Formation and Stroke. Dev. Neurobiol. 2011, 71, 1018–1039. [Google Scholar] [CrossRef]
- Bhattacharya, A.; Kaushik, D.K.; Lozinski, B.M.; Yong, V.W. Beyond barrier functions: Roles of pericytes in homeostasis and regulation of neuroinflammation. J. Neurosci. Res. 2020, 98, 2390–2405. [Google Scholar] [CrossRef]
- Pieper, C.; Marek, J.J.; Unterberg, M.; Schwerdtle, T.; Galla, H.-J. Brain capillary pericytes contribute to the immune defense in response to cytokines or LPS in vitro. Brain Res. 2014, 1550, 1–8. [Google Scholar] [CrossRef]
- Ferland-McCollough, D.; Slater, S.; Richard, J.; Reni, C.; Mangialardi, G. Pericytes, an overlooked player in vascular pathobiology. Pharmacol. Ther. 2017, 171, 30–42. [Google Scholar] [CrossRef]
- Winkler, E.A.; Sagare, A.P.; Zlokovic, B.V. The Pericyte: A Forgotten Cell Type with Important Implications for Alzheimer’s Disease? Brain Pathol. 2014, 24, 371–386. [Google Scholar] [CrossRef]
- Navarro, R.; Compte, M.; Alvarez-Vallina, L.; Sanz, L. Immune Regulation by Pericytes: Modulating Innate and Adaptive Immunity. Front. Immunol. 2016, 7. [Google Scholar] [CrossRef]
- Kang, T.-Y.; Bocci, F.; Jolly, M.K.; Levin, H.; Onuchic, J.N.; Levchenko, A. Pericytes enable effective angiogenesis in the presence of proinflammatory signals. Proc. Natl. Acad. Sci. USA 2019, 116, 23551–23561. [Google Scholar] [CrossRef] [PubMed]
- Smyth, L.C.D.; Rustenhoven, J.; Park, T.I.H.; Schweder, P.; Jansson, D.; Heppner, P.A.; O’Carroll, S.J.; Mee, E.W.; Faull, R.L.M.; Curtis, M.; et al. Unique and shared inflammatory profiles o human brain endothelia and pericytes. J. Neuroinflamm. 2018, 15. [Google Scholar] [CrossRef]
- Kurmann, L.; Okoniewski, M.; Ogunshola, O.O.; Leeners, B.; Imthurn, B.; Dubey, R.K. Transcryptomic Analysis of Human Brain-Microvascular Endothelial Response to -Pericytes: Cell Orientation Defines Barrier Function. Cells 2021, 10, 963. [Google Scholar] [CrossRef]
- Weksler, B.; Subileau, E.; Perriere, N.; Charneau, P.; Holloway, K.; Leveque, M.; Tricoire-Leignel, H.; Nicotra, A.; Bourdoulous, S.; Turowski, P.; et al. Blood-brain barrier-specific properties of a human adult brain endothelial cell line. FASEB J. 2005, 19, 1872–1874. [Google Scholar] [CrossRef]
- Hamad, S.; Derichsweiler, D.; Papadopoulos, S.; Nguemo, F.; Saric, T.; Sachinidis, A.; Brockmeier, K.; Hescheler, J.; Boukens, B.J.; Pfannkuche, K. Generation of human induced pluripotent stem cell-derived cardiomyocytes in 2D monolayer and scalable 3D suspension bioreactor cultures with reduced batch-to-batch variations. Theranostics 2019, 9, 7222–7238. [Google Scholar] [CrossRef]
- Huang, R.; Grishagin, I.; Wang, Y.; Zhao, T.; Greene, J.; Obenauer, J.C.; Ngan, D.; Dac-Trung, N.; Guha, R.; Jadhav, A.; et al. The NCATS BioPlanet—An Integrated Platform for Exploring the Universe of Cellular Signaling Pathways for Toxicology, Systems Biology, and Chemical Genomics. Front. Pharmacol. 2019, 10. [Google Scholar] [CrossRef]
- Brandt, M.M.; van Dijk, C.G.M.; Maringanti, R.; Chrifi, I.; Kramann, R.; Verhaar, M.C.; Duncker, D.J.; Mokry, M.; Cheng, C. Transcriptome analysis reveals microvascular endothelial cell-dependent pericyte differentiation. Sci. Rep. 2019, 9, 15586. [Google Scholar] [CrossRef]
- He, L.; Vanlandewijck, M.; Raschperger, E.; Mae, M.A.; Jung, B.; Lebouvier, T.; Ando, K.; Hofmann, J.; Keller, A.; Betsholtz, C. Analysis of the brain mural cell transcriptome. Sci. Rep. 2016, 6, 35108. [Google Scholar] [CrossRef]
- Winkler, E.A.; Bell, R.D.; Zlokovic, B.V. Central nervous system pericytes in health and disease. Nat. Neurosci. 2011, 14, 1398–1405. [Google Scholar] [CrossRef]
- Heymans, M.; Figueiredo, R.; Dehouck, L.; Francisco, D.; Sano, Y.; Shimizu, F.; Kanda, T.; Bruggmann, R.; Engelhardt, B.; Winter, P.; et al. Contribution of brain pericytes in blood-brain barrier formation and maintenance: A transcriptomic study of cocultured human endothelial cells derived from hematopoietic stem cells. Fluids Barriers CNS 2020, 17. [Google Scholar] [CrossRef] [PubMed]
- Jamieson, J.J.; Linville, R.M.; Ding, Y.Y.; Gerecht, S.; Searson, P.C. Role of iPSC-derived pericytes on barrier function of iPSC-derived brain microvascular endothelial cells in 2D and 3D. Fluids Barriers CNS 2019, 16. [Google Scholar] [CrossRef]
- Murfee, W.L.; Skalak, T.C.; Peirce, S.M. Differential arterial/venous expression of NG2 proteoglycan in perivascular cells along microvessels: Identifying a venule-specific phenotype. Microcirculation 2005, 12, 151–160. [Google Scholar] [CrossRef]
- Jansson, D.; Rustenhoven, J.; Feng, S.; Hurley, D.; Oldfield, R.L.; Bergin, P.S.; Mee, E.W.; Faull, R.L.M.; Dragunow, M. A role for human brain pericytes in neuroinflammation (vol 11, 104, 2014). J. NeuroInflamm. 2015, 12. [Google Scholar] [CrossRef]
- Zlokovic, B.V. The blood-brain barrier in health and chronic neurodegenerative disorders. Neuron 2008, 57, 178–201. [Google Scholar] [CrossRef]
- Guijarro-Munoz, I.; Compte, M.; Alvarez-Cienfuegos, A.; Alvarez-Vallina, L.; Sanz, L. Lipopolysaccharide Activates Toll-like Receptor 4 (TLR4)-mediated NF-kappa B Signaling Pathway and Proinflammatory Response in Human Pericytes. J. Biol. Chem. 2014, 289, 2457–2468. [Google Scholar] [CrossRef]
- Zeuke, S.; Ulmer, A.J.; Kusumoto, S.; Katus, H.A.; Heine, H. TLR4-mediated inflammatory activation of human coronary artery endothelial cells by LPS. Cardiovasc. Res. 2002, 56, 126–134. [Google Scholar] [CrossRef]
- Duan, L.; Zhang, X.-D.; Miao, W.-Y.; Sun, Y.-J.; Xiong, G.; Wu, Q.; Li, G.; Yang, P.; Yu, H.; Li, H.; et al. PDGFR beta Cells Rapidly Relay Inflammatory Signal from the Circulatory System to Neurons via Chemokine CCL2. Neuron 2018, 100, 183–200. [Google Scholar] [CrossRef] [PubMed]
- Anderson, P. Post-transcriptional regulons coordinate the initiation and resolution of inflammation. Nat. Rev. Immunol. 2010, 10, 24–35. [Google Scholar] [CrossRef] [PubMed]
- Anderson, P. Post-transcriptional control of cytokine production. Nat. Immunol. 2008, 9, 353–359. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, T.A.; Novotny, M.; Datta, S.; Mandal, P.; Hartupee, J.; Tebo, J.; Li, X. Chemokine and chemoattractant receptor expression: Post-transcriptional regulation. J. Leukoc. Biol. 2007, 82, 213–219. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wilczynska, A.; Bushell, M. The complexity of miRNA-mediated repression. Cell Death Differ. 2015, 22, 22–33. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, P.; Kedersha, N.; Anderson, P. Stress Granules and Processing Bodies in Translational Control. Cold Spring Harb. Perspect. Biol. 2019, 11, 18. [Google Scholar] [CrossRef]
- Leone, P.; Shin, E.-C.; Perosa, F.; Vacca, A.; Dammacco, F.; Racanelli, V. MHC Class I Antigen Processing and Presenting Machinery: Organization, Function, and Defects in Tumor Cells. J. Natl. Cancer Inst. 2013, 105, 1172–1187. [Google Scholar] [CrossRef]
- Pichlmair, A.; Lassnig, C.; Eberle, C.-A.; Gorna, M.W.; Baumann, C.L.; Burkard, T.R.; Buerckstuemmer, T.; Stefanovic, A.; Krieger, S.; Bennett, K.L.; et al. IFIT1 is an antiviral protein that recognizes 5′-triphosphate RNA. Nat. Immunol. 2011, 12, 624–630. [Google Scholar] [CrossRef]
- Busse, D.C.; Habgood-Coote, D.; Clare, S.; Brandt, C.; Bassano, I.; Kaforou, M.; Herberg, J.; Levin, M.; Eleouet, J.F.; Kellam, P.; et al. Interferon-Induced Protein 44 and Interferon-Induced Protein 44-Like Restrict Replication of Respiratory Syncytial Virus. J. Virol. 2020, 94. [Google Scholar] [CrossRef]
- Daniels, B.P.; Holman, D.W.; Cruz-Orengo, L.; Jujjavarapu, H.; Durrant, D.M.; Klein, R.S. Viral Pathogen-Associated Molecular Patterns Regulate Blood-Brain Barrier Integrity via Competing Innate Cytokine Signals. Mbio 2014, 5. [Google Scholar] [CrossRef]
- Bian, P.; Ye, C.; Zheng, X.; Luo, C.; Yang, J.; Li, M.; Wang, Y.; Yang, J.; Zhou, Y.; Zhang, F.; et al. RIPK3 Promotes JEV Replication in Neurons via Downregulation of IFI44L. Front. Microbiol. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Richardson, R.B.; Ohlson, M.B.; Eitson, J.L.; Kumar, A.; McDougal, M.B.; Boys, I.N.; Mar, K.B.; De La Cruz-Rivera, P.C.; Douglas, C.; Konopka, G.; et al. A CRISPR screen identifies IFI6 as an ER-resident interferon effector that blocks flavivirus replication. Nat. Microbiol. 2018, 3, 1214–1223. [Google Scholar] [CrossRef] [PubMed]
- Spindler, K.R.; Hsu, T.-H. Viral disruption of the blood brain barrier. Trends Microbiol. 2012, 20, 282–290. [Google Scholar] [CrossRef] [PubMed]
- Klein, R.S.; Hunter, C.A. Protective and Pathological Immunity during Central Nervous System Infections. Immunity 2017, 46, 891–909. [Google Scholar] [CrossRef] [PubMed]
- Ni, C.; Wang, C.; Zhang, J.; Qu, L.; Liu, X.; Lu, Y.; Yang, W.; Deng, J.; Lorenz, D.; Gao, P.; et al. Interferon-gamma Safeguards Blood-Brain Barrier during Experimental Autoimmune Encephalomyelitis. Am. J. Pathol. 2014, 184, 3308–3320. [Google Scholar] [CrossRef] [PubMed]
- Bonney, S.; Seitz, S.; Ryan, C.A.; Jones, K.L.; Clarke, P.; Tyler, K.L.; Siegenthaler, J.A. Gamma Interferon Alters Junctional Integrity via Rho Kinase, Resulting in Blood-Brain Barrier Leakage in Experimental Viral Encephalitis. Mbio 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Daniels, B.P.; Jujjavarapu, H.; Durrant, D.M.; Williams, J.L.; Green, R.R.; White, J.P.; Lazear, H.M.; Gale, M., Jr.; Diamond, M.S.; Klein, R.S. Regional astrocyte IFN signaling restricts pathogenesis during neurotropic viral infection. J. Clin. Investig. 2017, 127, 843–856. [Google Scholar] [CrossRef] [PubMed]
- Daniels, B.P.; Klein, R.S. Knocking on Closed Doors: Host Interferons Dynamically Regulate Blood-Brain Barrier Function during Viral Infections of the Central Nervous System. PLoS Pathog. 2015, 11. [Google Scholar] [CrossRef]
- Witjas, F.M.R.; van den Berg, B.M.; van den Berg, C.W.; Engelse, M.A.; Rabelink, T.J. Concise Review: The Endothelial Cell Extracellular Matrix Regulates Tissue Homeostasis and Repair. Stem Cells Transl. Med. 2019, 8, 375–382. [Google Scholar] [CrossRef]
- Hartmann, C.; Zozulya, A.; Wegener, J.; Galla, H.-J. The impact of glia-derived extracellular matrices on the barrier function of cerebral endothelial cells: An in vitro study. Exp. Cell Res. 2007, 313, 1318–1325. [Google Scholar] [CrossRef]
- Sato, Y.; Tsuboi, R.; Lyons, R.; Moses, H.; Rifkin, D.B. Characterization of the Activation of Latent Tgf-Beta by Cocultures of Endothelial-Cells and Pericytes or Smooth-Muscle Cells—A Self-Regulating System. J. Cell Biol. 1990, 111, 757–763. [Google Scholar] [CrossRef] [PubMed]
- Antonelliorlidge, A.; Saunders, K.B.; Smith, S.R.; Damore, P.A. An Activated Form of Transforming Growth Factor-Beta Is Produced by Cocultures of Endothelial-Cells and Pericytes. Proc. Natl. Acad. Sci. USA 1989, 86, 4544–4548. [Google Scholar] [CrossRef]
- Weiss, A.; Attisano, L. The TGFbeta Superfamily Signaling Pathway. Wiley Interdiscip. Rev. Dev. Biol. 2013, 2, 47–63. [Google Scholar] [CrossRef]
- Horiguchi, M.; Ota, M.; Rifkin, D.B. Matrix control of transforming growth factor-beta function. J. Biochem. 2012, 152, 321–329. [Google Scholar] [CrossRef] [PubMed]
- Moon, S.; Lee, S.; Caesar, J.A.; Pruchenko, S.; Leask, A.; Knowles, J.A.; Sinon, J.; Chaqour, B. A CTGF-YAP Regulatory Pathway Is Essential for Angiogenesis and Barriergenesis in the Retina. Iscience 2020, 23. [Google Scholar] [CrossRef]
- Gautam, J.; Zhang, X.; Yao, Y. The role of pericytic laminin in blood brain barrier integrity maintenance. Sci. Rep. 2016, 6, 36450. [Google Scholar] [CrossRef] [PubMed]
- McCarty, J.H.; Lacy-Hulbert, A.; Charest, A.; Bronson, R.T.; Crowley, D.; Housman, D.; Savill, J.; Roes, J.; Hynes, R.O. Selective ablation of alpha v integrins in the central nervous system leads to cerebral hemorrhage, seizures, axonal degeneration and premature death. Development 2005, 132, 165–176. [Google Scholar] [CrossRef] [PubMed]
- Dieguez-Hurtado, R.; Kato, K.; Giaimo, B.D.; Nieminen-Kelhae, M.; Arf, H.; Ferrante, F.; Bartkuhn, M.; Zimmermann, T.; Bixel, M.G.; Borggrefe, T.; et al. Loss of the transcription factor RBPJ induces disease-promoting properties in brain pericytes. Acta Physiol. 2019, 227, 2817. [Google Scholar] [CrossRef]
- Lemos, D.R.; Marsh, G.; Huang, A.; Campanholle, G.; Aburatani, T.; Dang, L.; Gomez, I.; Fisher, K.; Ligresti, G.; Peti-Peterdi, J.; et al. Maintenance of vascular integrity by pericytes is essential for normal kidney function. Am. J. Physiol. Ren. Physiol. 2016, 311, F1230–F1242. [Google Scholar] [CrossRef] [PubMed]
- Marek, I.; Hilgers, K.F.; Rascher, W.; Woelfle, J.; Hartner, A. A role for the alpha-8 integrin chain (itga8) in glomerular homeostasis of the kidney. Mol. Cell. Pediatrics 2020, 7, 13. [Google Scholar] [CrossRef]
- Huang, B.; Krafft, P.R.; Ma, Q.; Rolland, W.B.; Caner, B.; Lekic, T.; Manaenko, A.; Le, M.; Tang, J.; Zhang, J.H. Fibroblast growth factors preserve blood-brain barrier integrity through RhoA inhibition after intracerebral hemorrhage in mice. Neurobiol. Dis. 2012, 46, 204–214. [Google Scholar] [CrossRef] [PubMed]
- Hori, S.; Ohtsuki, S.; Hosoya, K.; Nakashima, E.; Terasaki, T. A pericyte-derived angiopoietin-1 multimeric complex induces occludin gene expression in brain capillary endothelial cells through Tie-2 activation in vitro. J. Neurochem. 2004, 89, 503–513. [Google Scholar] [CrossRef]
- Mirzapoiazova, T.; Kolosova, I.; Usatyuk, P.V.; Natarajan, V.; Verin, A.D. Diverse effects of vascular endothelial growth factor on human pulmonary endothelial barrier and migration. Am. J. Physiol. Lung Cell. Mol. Physiol. 2006, 291, L718–L724. [Google Scholar] [CrossRef]
- Yamagishi, S.; Yonekura, H.; Yamamoto, Y.; Fujimori, H.; Sakurai, S.; Tanaka, N.; Yamamoto, H. Vascular endothelial growth factor acts as a pericyte mitogen under hypoxic conditions. Lab. Investig. 1999, 79, 501–509. [Google Scholar]
- Stratman, A.N.; Davis, M.J.; Davis, G.E. VEGF and FGF prime vascular tube morphogenesis and sprouting directed by hematopoietic stem cell cytokines. Blood 2011, 117, 3709–3719. [Google Scholar] [CrossRef] [PubMed]
- Darland, D.C.; Massingham, L.J.; Smith, S.R.; Piek, E.; St-Geniez, M.; D’Amore, P.A. Pericyte production of cell-associated VEGF is differentiation-dependent and is associated with endothelial survival. Dev. Biol. 2003, 264, 275–288. [Google Scholar] [CrossRef] [PubMed]
- Rahimi, N. Defenders and Challengers of Endothelial Barrier Function. Front. Immunol. 2017, 8. [Google Scholar] [CrossRef]
- Tratwal, J.; Mathiasen, A.B.; Juhl, M.; Brorsen, S.K.; Kastrup, J.; Ekblond, A. Influence of vascular endothelial growth factor stimulation and serum deprivation on gene activation patterns of human adipose tissue-derived stromal cells. Stem Cell Res. Ther. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Dumic, J.; Dabelic, S.; Flogel, M. Galectin-3: An open-ended story. Biochim. Biophys. Acta Gen. Subj. 2006, 1760, 616–635. [Google Scholar] [CrossRef]
- Fukushi, J.; Makagiansar, I.T.; Stallcup, W.B. NG2 proteoglycan promotes endothelial cell motility and angiogenesis via engagement of galectin-3 and alpha 3 beta 1 integrin. Mol. Biol. Cell 2004, 15, 3580–3590. [Google Scholar] [CrossRef]
- Cheng, J.; Korte, N.; Nortley, R.; Sethi, H.; Tang, Y.; Attwell, D. Targeting pericytes for therapeutic approaches to neurological disorders. Acta Neuropathol. 2018, 136, 507–523. [Google Scholar] [CrossRef]
- Longden, T.A.; Hill-Eubanks, D.C.; Nelson, M.T. Ion channel networks in the control of cerebral blood flow. J. Cereb. Blood Flow Metab. 2016, 36, 492–512. [Google Scholar] [CrossRef]
- Hariharan, A.; Weir, N.; Robertson, C.; He, L.; Betsholtz, C.; Longden, T.A. The Ion Channel and GPCR Toolkit of Brain Capillary Pericytes. Front. Cell. Neurosci. 2020, 14. [Google Scholar] [CrossRef]
- Hu, C.; Tao, L.; Cao, X.; Chen, L. The solute carrier transporters and the brain: Physiological and pharmacological implications. Asian J. Pharm. Sci. 2020, 15, 131–144. [Google Scholar] [CrossRef]
- Perrot, C.Y.; Herrera, J.L.; Fournier-Goss, A.E.; Komatsu, M. Prostaglandin E2 breaks down pericyte-endothelial cell interaction via EP1 and EP4-dependent downregulation of pericyte N-cadherin, connexin-43, and R-Ras. Sci. Rep. 2020, 10, 11186. [Google Scholar] [CrossRef] [PubMed]
- Ezan, P.; Andre, P.; Cisternino, S.; Saubamea, B.; Boulay, A.-C.; Doutremer, S.; Thomas, M.-A.; Quenech’du, N.; Giaume, C.; Cohen-Salmon, M. Deletion of astroglial connexins weakens the blood-brain barrier. J. Cereb. Blood Flow Metab. 2012, 32, 1457–1467. [Google Scholar] [CrossRef]
- Ivanova, E.; Kovacs-Oller, T.; Sagdullaev, B.T. Vascular Pericyte Impairment and Connexin43 Gap Junction Deficit Contribute to Vasomotor Decline in Diabetic Retinopathy. J. Neurosci. 2017, 37, 7580–7594. [Google Scholar] [CrossRef]
- Hirschi, K.K.; Burt, J.M.; Hirschi, K.D.; Dai, C.P. Gap junction communication mediates transforming growth factor-beta activation and endothelial-induced mural cell differentiation. Circ. Res. 2003, 93, 429–437. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Quan, Y.; Ganesh, T.; Pouliot, W.A.; Dudek, F.E.; Dingledine, R. Inhibition of the prostaglandin receptor EP2 following status epilepticus reduces delayed mortality and brain inflammation. Proc. Natl. Acad. Sci. USA 2013, 110, 3591–3596. [Google Scholar] [CrossRef] [PubMed]
- Higashiyama, S.; Nanba, D.; Nakayama, H.; Inoue, H.; Fukuda, S. Ectodomain shedding and remnant peptide signalling of EGFRs and their ligands. J. Biochem. 2011, 150, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Shin, W.K. Epidermal Growth-Factor Receptor at Endothelial-Cell and Pericyte Interdigitation in Human Granulation-Tissue. Microvasc. Res. 1992, 44, 255–262. [Google Scholar] [CrossRef]
- Kielczewski, J.L.; Hu, P.; Shaw, L.C.; Calzi, S.L.; Mames, R.N.; Gardiner, T.A.; McFarland, E.; Chan-Ling, T.; Grant, M.B. Novel Protective Properties of IGFBP-3 Result in Enhanced Pericyte Ensheathment, Reduced Microglial Activation, Increased Microglial Apoptosis, and Neuronal Protection after Ischemic Retinal Injury. Am. J. Pathol. 2011, 178, 1517–1528. [Google Scholar] [CrossRef] [PubMed]
- Kielczewski, J.L.; Jarajapu, Y.P.R.; McFarland, E.L.; Cai, J.; Afzal, A.; Calzi, S.L.; Chang, K.H.; Lydic, T.; Shaw, L.C.; Busik, J.; et al. Insulin-Like Growth Factor Binding Protein-3 Mediates Vascular Repair by Enhancing Nitric Oxide Generation. Circ. Res. 2009, 105, 897–905. [Google Scholar] [CrossRef]
- Lofqvist, C.; Chen, J.; Connor, K.M.; Smith, A.C.H.; Aderman, C.M.; Liu, N.; Pintar, J.E.; Ludwig, T.; Hellstrom, A.; Smith, L.E.H. IGFBP3 suppresses retinopathy through suppression of oxygen-induced vessel loss and promotion of vascular regrowth. Proc. Natl. Acad. Sci. USA 2007, 104, 10589–10594. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Jiang, Y.; Miller, M.J.; Peng, B.; Liu, L.; Soderland, C.; Tang, J.; Kern, T.S.; Pintar, J.; Steinle, J.J. IGFBP-3 and TNF-alpha Regulate Retinal Endothelial Cell Apoptosis. Investig. Ophthalmol. Vis. Sci. 2013, 54, 5376–5384. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Touyz, R.M.; Briones, A.M. Reactive oxygen species and vascular biology: Implications in human hypertension. Hypertens. Res. 2011, 34, 5–14. [Google Scholar] [CrossRef]
- DiSabato, D.J.; Quan, N.; Godbout, J.P. Neuroinflammation: The devil is in the details. J. Neurochem. 2016, 139, 136–153. [Google Scholar] [CrossRef]
- Norden, D.M.; Trojanowski, P.J.; Villanueva, E.; Navarro, E.; Godbout, J.P. Sequential Activation of Microglia and Astrocyte Cytokine Expression Precedes Increased Iba-1 or GFAP Immunoreactivity Following Systemic Immune Challenge. Glia 2016, 64, 300–316. [Google Scholar] [CrossRef]
- Galani, I.-E.; Rovina, N.; Lampropoulou, V.; Triantafyllia, V.; Manioudaki, M.; Pavlos, E.; Koukaki, E.; Fragkou, P.C.; Panou, V.; Rapti, V.; et al. Untuned antiviral immunity in COVID-19 revealed by temporal type I/III interferon patterns and flu comparison. Nat. Immunol. 2021, 22, 32–40. [Google Scholar] [CrossRef]
- Cardot-Leccia, N.; Hubiche, T.; Dellamonica, J.; Burel-Vandenbos, F.; Passeron, T. Pericyte alteration sheds light on micro-vasculopathy in COVID-19 infection. Intensive Care Med. 2020, 46, 1777–1778. [Google Scholar] [CrossRef] [PubMed]
| Gene | Gene Description | Log2 FC (Co- Vs. Mono-Culture) | FDR p-Value |
|---|---|---|---|
| IFIT1 | Interferon-induced protein with tetratricopeptide repeats 1 | 8.5 | 1.59 × 10−16 |
| MX1 | MX dynamin-like GTPase 1 (interferon-induced) | 7.3 | 7.05 × 10−15 |
| IFI44L | Interferon-induced protein 44-like | 6.2 | 1.80 × 10−13 |
| CXCL8 | Chemokine (C-X-C motif) ligand 8 | 5.8 | 3.89 × 10−12 |
| IFI6 | Interferon, alpha-inducible protein 6 | 5.2 | 1.30 × 10−9 |
| XAF1 | XIAP associated factor 1 | 5.2 | 1.58 × 10−13 |
| SAMD9L | Sterile alpha motif domain containing 9-like | 5.2 | 1.61 × 10−9 |
| SAMD9 | Sterile alpha motif domain containing 9 | 5.2 | 2.18 × 10−11 |
| OAS1 | 2-5-oligoadenylate synthetase 1 | 5.1 | 4.12 × 10−14 |
| RCAN1 | Regulator of calcineurin 1 | 4.8 | 4.36 × 10−13 |
| Gene | Gene Description | Log2 FC (Co- Vs. Mono-Culture) | FDR p-Value |
|---|---|---|---|
| NPTX1 | Neuronal pentraxin I | −5.2 | 4.12 × 10−14 |
| JUN | Jun proto-oncogene | −3.6 | 2.96 × 10−9 |
| SLC6A6 | Solute carrier family 6 (neurotransmitter transporter), member 6 | −3.5 | 1.28 × 10−8 |
| PTGER2 | Prostaglandin E receptor 2 | −3.5 | 2.50 × 10−7 |
| MN1 | Meningioma (disrupted in balanced translocation) 1 | −3.5 | 6.56 × 10−8 |
| F2RL2 | Coagulation factor II (thrombin) receptor-like 2 | −3.4 | 7.00 × 10−7 |
| SLC14A1 | Solute carrier family 14 (urea transporter), member 1 (Kidd blood group) | −3.3 | 8.03 × 10−8 |
| STEAP3 | STEAP family member 3, metalloreductase | −3.0 | 3.05 × 10−7 |
| SCARA3 | Scavenger receptor class A, member 3 | −3.0 | 4.89 × 10−6 |
| SHISA2 | Shisa family member 2 | −2.8 | 0.0055 |
| Pathway | Overlap | Adj. p-Value |
|---|---|---|
| TGF-beta regulation of extracellular matrix | 132/565 | 1.09 × 10−11 |
| Epidermal growth factor receptor 1 (EGFR1) pathway | 45/152 | 2.32 × 10−6 |
| Brain-derived neurotrophic factor (BDNF) signaling pathway | 63/261 | 8.33 × 10−6 |
| T cell receptor regulation of apoptosis | 118/603 | 8.33 × 10−6 |
| Follicle-stimulating hormone (FSH) regulation of apoptosis | 63/263 | 8.77 × 10−6 |
| Messenger RNA processing | 52/203 | 1.14 × 10−5 |
| Interleukin-1 regulation of extracellular matrix | 36/120 | 1.64 × 10−5 |
| Interferon alpha/beta signaling | 24/64 | 2.03 × 10−5 |
| Smooth muscle contraction | 13/22 | 2.41 × 10−5 |
| Interferon signaling | 44/168 | 3.61 × 10−5 |
| Interleukin-2 signaling pathway | 149/847 | 5.46 × 10−5 |
| Ataxia telangiectasia mutated (ATM) pathway | 19/48 | 1.05 × 10−4 |
| Interleukin-4 regulation of apoptosis | 58/267 | 3.54 × 10−4 |
| Lipid metabolism regulation by peroxisome proliferator-activated receptor alpha (PPAR-alpha) | 31/112 | 4.19 × 10−4 |
| Lipid and lipoprotein metabolism | 92/489 | 4.28 × 10−4 |
| Fatty acid, triacylglycerol, and ketone body metabolism | 41/173 | 8.60 × 10−4 |
| Activator protein 1 (AP-1) transcription factor network | 22/70 | 9.30 × 10−4 |
| Integrated breast cancer pathway | 37/152 | 1.07 × 10−3 |
| Retinoic acid receptor-mediated signaling | 13/30 | 1.09 × 10−3 |
| Gene expression | 158/968 | 1.17 × 10−3 |
| Angiogenesis | 11/23 | 1.34 × 10−3 |
| Senescence and autophagy | 27/99 | 1.34 × 10−3 |
| Uridine diphosphate (UDP)-N-acetyl-glucosamine biosynthesis | 5/5 | 1.34 × 10−3 |
| Capped intron-containing pre-mRNA processing | 34/138 | 1.34 × 10−3 |
| Type II interferon signaling (interferon-gamma) | 17/50 | 1.91 × 10−3 |
| Protein | Array Coordinates | Protein Regulation | mRNA Regulation |
|---|---|---|---|
| CCL2 | G4 | −64% | up |
| CCL7 | G5 | −37% | down (ns) |
| CCL20 | G10 | −47% | up |
| CXCL1 | D1 | −67% | up |
| CXCL5 | C2 | −73% | up |
| CXCL8 | E4 | −15% | up |
| CXCL10 | F10 | −35% | up |
| EGF | B10 | −84% | nr |
| Fas ligand | C4 | −45% | nr |
| FGF-19 | C7 | −43% | nr |
| HGF | D3 | −44% | nr |
| IL-1-a | D8 | −32% | nr |
| IL-1-b | D9 | −24% | up (ns) |
| IL-6 | E3 | −31% | up |
| IL-10 | E5 | −52% | nr |
| IL-15 | E9 | −37% | nr |
| IL-31 | F6 | −20% | nr |
| IL-32 | F7 | −40% | nr |
| IL-33 | F8 | −37% | nr |
| IL-34 | F9 | −46% | down (ns) |
| IL-12 P70 | E7 | −32% | nr |
| MIC-1 | C10 | −32% | nr |
| VCAM1 | J6 | −17% | up |
| Angiogenin | A4 | +134% | nr |
| Angiopoietin 1 | A5 | +37% | up |
| FGF-2 | C5 | +24% | up |
| IGFBP 3 | D7 | +125% | down |
| Osteopontin | H2 | +66% | nr |
| PDGF-AB/-BB | H4 | +142% | nr |
| Thrombospondin-1 | I8 | +50% | nr |
| VEGF-A | I11 | +341% | down |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kurmann, L.; Okoniewski, M.; Dubey, R.K. Transcriptomic Analysis of Human Brain -Microvascular Endothelial Cell Driven Changes in -Vascular Pericytes. Cells 2021, 10, 1784. https://doi.org/10.3390/cells10071784
Kurmann L, Okoniewski M, Dubey RK. Transcriptomic Analysis of Human Brain -Microvascular Endothelial Cell Driven Changes in -Vascular Pericytes. Cells. 2021; 10(7):1784. https://doi.org/10.3390/cells10071784
Chicago/Turabian StyleKurmann, Lisa, Michal Okoniewski, and Raghvendra K. Dubey. 2021. "Transcriptomic Analysis of Human Brain -Microvascular Endothelial Cell Driven Changes in -Vascular Pericytes" Cells 10, no. 7: 1784. https://doi.org/10.3390/cells10071784
APA StyleKurmann, L., Okoniewski, M., & Dubey, R. K. (2021). Transcriptomic Analysis of Human Brain -Microvascular Endothelial Cell Driven Changes in -Vascular Pericytes. Cells, 10(7), 1784. https://doi.org/10.3390/cells10071784






