Role of Alternative Splicing in Regulating Host Response to Viral Infection
Abstract
1. Introduction
2. Alternative RNA Splicing and Its Isoforms in Type I and III IFN Responses
3. Other Innate Immune Pathways Impacted by Alternative RNA Splicing
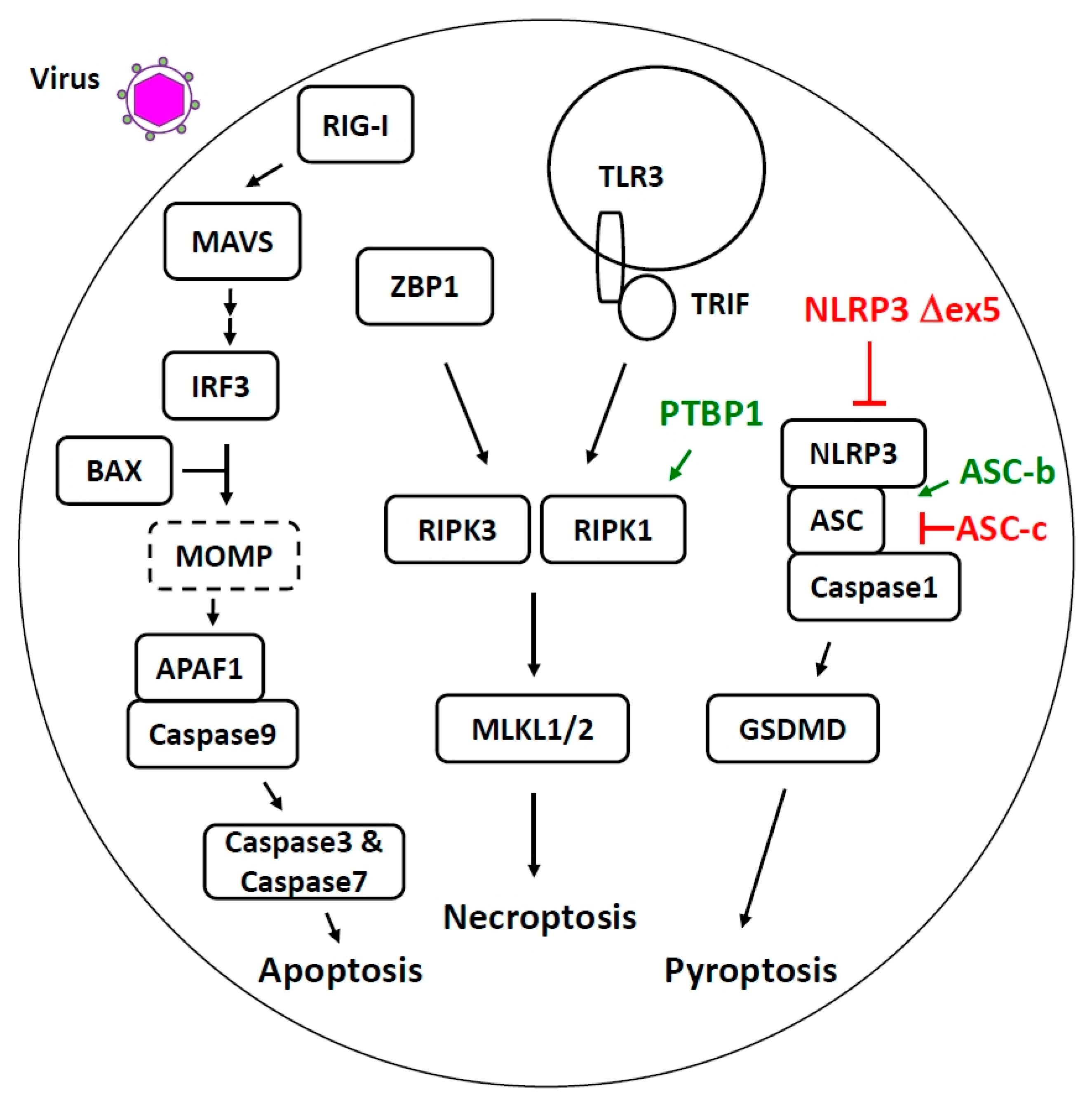
4. Alternative Splicing Regulates Host Cell Death Pathways Activated during Viral Infection
5. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wilkinson, M.E.; Charenton, C.; Nagai, K. RNA Splicing by the Spliceosome. Annu. Rev. Biochem. 2020, 89, 359–388. [Google Scholar] [CrossRef] [PubMed]
- Wang, E.T.; Sandberg, R.; Luo, S.; Khrebtukova, I.; Zhang, L.; Mayr, C.; Kingsmore, S.F.; Schroth, G.P.; Burge, C.B. Alternative isoform regulation in human tissue transcriptomes. Nature 2008, 456, 470–476. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.D.; Ares, M., Jr. Context-dependent control of alternative splicing by RNA-binding proteins. Nat. Rev. Genet. 2014, 15, 689–701. [Google Scholar] [CrossRef] [PubMed]
- Evsyukova, I.; Somarelli, J.A.; Gregory, S.G.; Garcia-Blanco, M.A. Alternative splicing in multiple sclerosis and other autoimmune diseases. RNA Biol. 2010, 7, 462–473. [Google Scholar] [CrossRef]
- Tazi, J.; Bakkour, N.; Stamm, S. Alternative splicing and disease. Biochim. Biophys. Acta 2009, 1792, 14–26. [Google Scholar] [CrossRef]
- Hopfner, K.P.; Hornung, V. Molecular mechanisms and cellular functions of cGAS-STING signalling. Nat. Rev. Mol. Cell Biol. 2020, 21, 501–521. [Google Scholar] [CrossRef]
- Sun, B.; Sundstrom, K.B.; Chew, J.J.; Bist, P.; Gan, E.S.; Tan, H.C.; Goh, K.C.; Chawla, T.; Tang, C.K.; Ooi, E.E. Dengue virus activates cGAS through the release of mitochondrial DNA. Sci. Rep. 2017, 7, 3594. [Google Scholar] [CrossRef]
- Lazear, H.M.; Schoggins, J.W.; Diamond, M.S. Shared and Distinct Functions of Type I and Type III Interferons. Immunity 2019, 50, 907–923. [Google Scholar] [CrossRef]
- Gack, M.U.; Kirchhofer, A.; Shin, Y.C.; Inn, K.S.; Liang, C.; Cui, S.; Myong, S.; Ha, T.; Hopfner, K.P.; Jung, J.U. Roles of RIG-I N-terminal tandem CARD and splice variant in TRIM25-mediated antiviral signal transduction. Proc. Natl. Acad. Sci. USA 2008, 105, 16743–16748. [Google Scholar] [CrossRef]
- Seo, J.W.; Yang, E.J.; Kim, S.H.; Choi, I.H. An inhibitory alternative splice isoform of Toll-like receptor 3 is induced by type I interferons in human astrocyte cell lines. BMB Rep. 2015, 48, 696–701. [Google Scholar] [CrossRef]
- Yang, E.; Shin, J.S.; Kim, H.; Park, H.W.; Kim, M.H.; Kim, S.J.; Choi, I.H. Cloning of TLR3 isoform. Yonsei Med. J. 2004, 45, 359–361. [Google Scholar] [CrossRef] [PubMed]
- Lad, S.P.; Yang, G.; Scott, D.A.; Chao, T.H.; Correia Jda, S.; de la Torre, J.C.; Li, E. Identification of MAVS splicing variants that interfere with RIGI/MAVS pathway signaling. Mol. Immunol. 2008, 45, 2277–2287. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Pei, R.; Zhu, W.; Zeng, R.; Wang, Y.; Wang, Y.; Lu, M.; Chen, X. An alternative splicing isoform of MITA antagonizes MITA-mediated induction of type I IFNs. J. Immunol. 2014, 192, 1162–1170. [Google Scholar] [CrossRef]
- Han, K.J.; Yang, Y.; Xu, L.G.; Shu, H.B. Analysis of a TIR-less splice variant of TRIF reveals an unexpected mechanism of TLR3-mediated signaling. J. Biol. Chem. 2010, 285, 12543–12550. [Google Scholar] [CrossRef] [PubMed]
- Oshiumi, H.; Matsumoto, M.; Funami, K.; Akazawa, T.; Seya, T. TICAM-1, an adaptor molecule that participates in Toll-like receptor 3-mediated interferon-beta induction. Nat. Immunol. 2003, 4, 161–167. [Google Scholar] [CrossRef]
- Michel, M.; Wilhelmi, I.; Schultz, A.S.; Preussner, M.; Heyd, F. Activation-induced tumor necrosis factor receptor-associated factor 3 (Traf3) alternative splicing controls the noncanonical nuclear factor kappaB pathway and chemokine expression in human T cells. J. Biol. Chem. 2014, 289, 13651–13660. [Google Scholar] [CrossRef]
- Schultz, A.S.; Preussner, M.; Bunse, M.; Karni, R.; Heyd, F. Activation-Dependent TRAF3 Exon 8 Alternative Splicing Is Controlled by CELF2 and hnRNP C Binding to an Upstream Intronic Element. Mol. Cell Biol. 2017, 37, e00488-16. [Google Scholar] [CrossRef]
- Deng, W.; Shi, M.; Han, M.; Zhong, J.; Li, Z.; Li, W.; Hu, Y.; Yan, L.; Wang, J.; He, Y.; et al. Negative regulation of virus-triggered IFN-beta signaling pathway by alternative splicing of TBK1. J. Biol. Chem. 2008, 283, 35590–35597. [Google Scholar] [CrossRef]
- Fabozzi, G.; Oler, A.J.; Liu, P.; Chen, Y.; Mindaye, S.; Dolan, M.A.; Kenney, H.; Gucek, M.; Zhu, J.; Rabin, R.L.; et al. Strand-Specific Dual RNA Sequencing of Bronchial Epithelial Cells Infected with Influenza A/H3N2 Viruses Reveals Splicing of Gene Segment 6 and Novel Host-Virus Interactions. J. Virol. 2018, 92, e00518-18. [Google Scholar] [CrossRef]
- Koop, A.; Lepenies, I.; Braum, O.; Davarnia, P.; Scherer, G.; Fickenscher, H.; Kabelitz, D.; Adam-Klages, S. Novel splice variants of human IKKepsilon negatively regulate IKKepsilon-induced IRF3 and NF-kB activation. Eur. J. Immunol. 2011, 41, 224–234. [Google Scholar] [CrossRef]
- De Maio, F.A.; Risso, G.; Iglesias, N.G.; Shah, P.; Pozzi, B.; Gebhard, L.G.; Mammi, P.; Mancini, E.; Yanovsky, M.J.; Andino, R.; et al. The Dengue Virus NS5 Protein Intrudes in the Cellular Spliceosome and Modulates Splicing. PLoS Pathog. 2016, 12, e1005841. [Google Scholar] [CrossRef] [PubMed]
- Karpova, A.Y.; Ronco, L.V.; Howley, P.M. Functional characterization of interferon regulatory factor 3a (IRF-3a), an alternative splice isoform of IRF-3. Mol. Cell Biol. 2001, 21, 4169–4176. [Google Scholar] [CrossRef]
- Karpova, A.Y.; Howley, P.M.; Ronco, L.V. Dual utilization of an acceptor/donor splice site governs the alternative splicing of the IRF-3 gene. Genes Dev. 2000, 14, 2813–2818. [Google Scholar] [CrossRef]
- Li, C.; Ma, L.; Chen, X. Interferon regulatory factor 3-CL, an isoform of IRF3, antagonizes activity of IRF3. Cell Mol. Immunol. 2011, 8, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Marozin, S.; Altomonte, J.; Stadler, F.; Thasler, W.E.; Schmid, R.M.; Ebert, O. Inhibition of the IFN-beta response in hepatocellular carcinoma by alternative spliced isoform of IFN regulatory factor-3. Mol. Ther. 2008, 16, 1789–1797. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Hu, X.; Song, Y.; Lu, Z.; Ning, T.; Cai, H.; Ke, Y. Identification of novel alternative splicing variants of interferon regulatory factor 3. Biochim. Biophys. Acta 2011, 1809, 166–175. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.; Li, Y.; Ning, J.; Sun, D.; Lin, L.; Liu, X. HnRNP A1/A2 and SF2/ASF regulate alternative splicing of interferon regulatory factor-3 and affect immunomodulatory functions in human non-small cell lung cancer cells. PLoS ONE 2013, 8, e62729. [Google Scholar] [CrossRef]
- Frankiw, L.; Majumdar, D.; Burns, C.; Vlach, L.; Moradian, A.; Sweredoski, M.J.; Baltimore, D. BUD13 Promotes a Type I Interferon Response by Countering Intron Retention in Irf7. Mol. Cell 2019, 73, 803–814. [Google Scholar] [CrossRef]
- Xu, X.; Mann, M.; Qiao, D.; Brasier, A.R. Alternative mRNA Processing of Innate Response Pathways in Respiratory Syncytial Virus (RSV) Infection. Viruses 2021, 13, 218. [Google Scholar] [CrossRef]
- Zhang, L.; Pagano, J.S. Structure and function of IRF-7. J. Interferon Cytokine Res. 2002, 22, 95–101. [Google Scholar] [CrossRef]
- Prokunina-Olsson, L.; Muchmore, B.; Tang, W.; Pfeiffer, R.M.; Park, H.; Dickensheets, H.; Hergott, D.; Porter-Gill, P.; Mumy, A.; Kohaar, I.; et al. A variant upstream of IFNL3 (IL28B) creating a new interferon gene IFNL4 is associated with impaired clearance of hepatitis C virus. Nat. Genet. 2013, 45, 164–171. [Google Scholar] [CrossRef]
- Hong, M.; Schwerk, J.; Lim, C.; Kell, A.; Jarret, A.; Pangallo, J.; Loo, Y.M.; Liu, S.; Hagedorn, C.H.; Gale, M., Jr.; et al. Interferon lambda 4 expression is suppressed by the host during viral infection. J. Exp. Med. 2016, 213, 2539–2552. [Google Scholar] [CrossRef]
- Fang, M.Z.; Jackson, S.S.; O’Brien, T.R. IFNL4: Notable variants and associated phenotypes. Gene 2020, 730, 144289. [Google Scholar] [CrossRef] [PubMed]
- Lutfalla, G.; Holland, S.J.; Cinato, E.; Monneron, D.; Reboul, J.; Rogers, N.C.; Smith, J.M.; Stark, G.R.; Gardiner, K.; Mogensen, K.E.; et al. Mutant U5A cells are complemented by an interferon-alpha beta receptor subunit generated by alternative processing of a new member of a cytokine receptor gene cluster. EMBO J. 1995, 14, 5100–5108. [Google Scholar] [CrossRef]
- Cohen, B.; Novick, D.; Barak, S.; Rubinstein, M. Ligand-induced association of the type I interferon receptor components. Mol. Cell Biol. 1995, 15, 4208–4214. [Google Scholar] [CrossRef] [PubMed]
- Gazziola, C.; Cordani, N.; Carta, S.; De Lorenzo, E.; Colombatti, A.; Perris, R. The relative endogenous expression levels of the IFNAR2 isoforms influence the cytostatic and pro-apoptotic effect of IFNalpha on pleomorphic sarcoma cells. Int. J. Oncol. 2005, 26, 129–140. [Google Scholar]
- Sheppard, P.; Kindsvogel, W.; Xu, W.; Henderson, K.; Schlutsmeyer, S.; Whitmore, T.E.; Kuestner, R.; Garrigues, U.; Birks, C.; Roraback, J.; et al. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat. Immunol. 2003, 4, 63–68. [Google Scholar] [CrossRef] [PubMed]
- Dumoutier, L.; Lejeune, D.; Hor, S.; Fickenscher, H.; Renauld, J.C. Cloning of a new type II cytokine receptor activating signal transducer and activator of transcription (STAT)1, STAT2 and STAT3. Biochem. J. 2003, 370, 391–396. [Google Scholar] [CrossRef] [PubMed]
- Witte, K.; Gruetz, G.; Volk, H.D.; Looman, A.C.; Asadullah, K.; Sterry, W.; Sabat, R.; Wolk, K. Despite IFN-lambda receptor expression, blood immune cells, but not keratinocytes or melanocytes, have an impaired response to type III interferons: Implications for therapeutic applications of these cytokines. Genes Immun. 2009, 10, 702–714. [Google Scholar] [CrossRef]
- Santer, D.M.; Minty, G.E.S.; Golec, D.P.; Lu, J.; May, J.; Namdar, A.; Shah, J.; Elahi, S.; Proud, D.; Joyce, M.; et al. Differential expression of interferon-lambda receptor 1 splice variants determines the magnitude of the antiviral response induced by interferon-lambda 3 in human immune cells. PLoS Pathog. 2020, 16, e1008515. [Google Scholar] [CrossRef]
- Schindler, C.; Fu, X.Y.; Improta, T.; Aebersold, R.; Darnell, J.E., Jr. Proteins of transcription factor ISGF-3: One gene encodes the 91-and 84-kDa ISGF-3 proteins that are activated by interferon alpha. Proc. Natl. Acad. Sci. USA 1992, 89, 7836–7839. [Google Scholar] [CrossRef] [PubMed]
- Baran-Marszak, F.; Feuillard, J.; Najjar, I.; Le Clorennec, C.; Bechet, J.M.; Dusanter-Fourt, I.; Bornkamm, G.W.; Raphael, M.; Fagard, R. Differential roles of STAT1alpha and STAT1beta in fludarabine-induced cell cycle arrest and apoptosis in human B cells. Blood 2004, 104, 2475–2483. [Google Scholar] [CrossRef] [PubMed]
- Walter, M.J.; Look, D.C.; Tidwell, R.M.; Roswit, W.T.; Holtzman, M.J. Targeted inhibition of interferon-gamma-dependent intercellular adhesion molecule-1 (ICAM-1) expression using dominant-negative Stat1. J. Biol. Chem. 1997, 272, 28582–28589. [Google Scholar] [CrossRef]
- Semper, C.; Leitner, N.R.; Lassnig, C.; Parrini, M.; Mahlakoiv, T.; Rammerstorfer, M.; Lorenz, K.; Rigler, D.; Muller, S.; Kolbe, T.; et al. STAT1beta is not dominant negative and is capable of contributing to gamma interferon-dependent innate immunity. Mol. Cell Biol. 2014, 34, 2235–2248. [Google Scholar] [CrossRef] [PubMed]
- Verma, D.; Swaminathan, S. Epstein-Barr virus SM protein functions as an alternative splicing factor. J. Virol. 2008, 82, 7180–7188. [Google Scholar] [CrossRef] [PubMed]
- Verma, D.; Bais, S.; Gaillard, M.; Swaminathan, S. Epstein-Barr Virus SM protein utilizes cellular splicing factor SRp20 to mediate alternative splicing. J. Virol. 2010, 84, 11781–11789. [Google Scholar] [CrossRef]
- Du, Z.; Fan, M.; Kim, J.G.; Eckerle, D.; Lothstein, L.; Wei, L.; Pfeffer, L.M. Interferon-resistant Daudi cell line with a Stat2 defect is resistant to apoptosis induced by chemotherapeutic agents. J. Biol. Chem. 2009, 284, 27808–27815. [Google Scholar] [CrossRef]
- Hambleton, S.; Goodbourn, S.; Young, D.F.; Dickinson, P.; Mohamad, S.M.; Valappil, M.; McGovern, N.; Cant, A.J.; Hackett, S.J.; Ghazal, P.; et al. STAT2 deficiency and susceptibility to viral illness in humans. Proc. Natl. Acad. Sci. USA 2013, 110, 3053–3058. [Google Scholar] [CrossRef]
- Vairo, D.; Tassone, L.; Tabellini, G.; Tamassia, N.; Gasperini, S.; Bazzoni, F.; Plebani, A.; Porta, F.; Notarangelo, L.D.; Parolini, S.; et al. Severe impairment of IFN-gamma and IFN-alpha responses in cells of a patient with a novel STAT1 splicing mutation. Blood 2011, 118, 1806–1817. [Google Scholar] [CrossRef]
- Frankiw, L.; Mann, M.; Li, G.; Joglekar, A.; Baltimore, D. Alternative splicing coupled with transcript degradation modulates OAS1g antiviral activity. RNA 2020, 26, 126–136. [Google Scholar] [CrossRef]
- Ku, C.C.; Che, X.B.; Reichelt, M.; Rajamani, J.; Schaap-Nutt, A.; Huang, K.J.; Sommer, M.H.; Chen, Y.S.; Chen, Y.Y.; Arvin, A.M. Herpes simplex virus-1 induces expression of a novel MxA isoform that enhances viral replication. Immunol. Cell Biol. 2011, 89, 173–182. [Google Scholar] [CrossRef]
- Scherer, M.; Stamminger, T. Emerging Role of PML Nuclear Bodies in Innate Immune Signaling. J. Virol. 2016, 90, 5850–5854. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.E.; Ahn, J.H. Positive role of promyelocytic leukemia protein in type I interferon response and its regulation by human cytomegalovirus. PLoS Pathog. 2015, 11, e1004785. [Google Scholar] [CrossRef] [PubMed]
- Jensen, K.; Shiels, C.; Freemont, P.S. PML protein isoforms and the RBCC/TRIM motif. Oncogene 2001, 20, 7223–7233. [Google Scholar] [CrossRef]
- El Asmi, F.; Maroui, M.A.; Dutrieux, J.; Blondel, D.; Nisole, S.; Chelbi-Alix, M.K. Implication of PMLIV in both intrinsic and innate immunity. PLoS Pathog. 2014, 10, e1003975. [Google Scholar] [CrossRef]
- Giovannoni, F.; Ladelfa, M.F.; Monte, M.; Jans, D.A.; Hemmerich, P.; Garcia, C. Dengue Non-structural Protein 5 Polymerase Complexes With Promyelocytic Leukemia Protein (PML) Isoforms III and IV to Disrupt PML-Nuclear Bodies in Infected Cells. Front. Cell Infect. Microbiol. 2019, 9, 284. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wright, J.; Meng, X.; Leppard, K.N. Promyelocytic Leukemia Protein Isoform II Promotes Transcription Factor Recruitment to Activate Interferon Beta and Interferon-Responsive Gene Expression. Mol. Cell Biol. 2015, 35, 1660–1672. [Google Scholar] [CrossRef]
- Nojima, T.; Oshiro-Ideue, T.; Nakanoya, H.; Kawamura, H.; Morimoto, T.; Kawaguchi, Y.; Kataoka, N.; Hagiwara, M. Herpesvirus protein ICP27 switches PML isoform by altering mRNA splicing. Nucleic Acids Res. 2009, 37, 6515–6527. [Google Scholar] [CrossRef]
- Haque, N.; Ouda, R.; Chen, C.; Ozato, K.; Hogg, J.R. ZFR coordinates crosstalk between RNA decay and transcription in innate immunity. Nat. Commun. 2018, 9, 1145. [Google Scholar] [CrossRef]
- Cao, P.; Luo, W.W.; Li, C.; Tong, Z.; Zheng, Z.Q.; Zhou, L.; Xiong, Y.; Li, S. The heterogeneous nuclear ribonucleoprotein hnRNPM inhibits RNA virus-triggered innate immunity by antagonizing RNA sensing of RIG-I-like receptors. PLoS Pathog. 2019, 15, e1007983. [Google Scholar] [CrossRef]
- West, K.O.; Scott, H.M.; Torres-Odio, S.; West, A.P.; Patrick, K.L.; Watson, R.O. The Splicing Factor hnRNP M Is a Critical Regulator of Innate Immune Gene Expression in Macrophages. Cell Rep. 2019, 29, 1594–1609. [Google Scholar] [CrossRef]
- Galluzzi, L.; Vitale, I.; Aaronson, S.A.; Abrams, J.M.; Adam, D.; Agostinis, P.; Alnemri, E.S.; Altucci, L.; Amelio, I.; Andrews, D.W.; et al. Molecular mechanisms of cell death: Recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 2018, 25, 486–541. [Google Scholar] [CrossRef]
- Zhou, X.; Jiang, W.; Liu, Z.; Liu, S.; Liang, X. Virus Infection and Death Receptor-Mediated Apoptosis. Viruses 2017, 9, 316. [Google Scholar] [CrossRef]
- Galluzzi, L.; Brenner, C.; Morselli, E.; Touat, Z.; Kroemer, G. Viral control of mitochondrial apoptosis. PLoS Pathog. 2008, 4, e1000018. [Google Scholar] [CrossRef]
- Paronetto, M.P.; Passacantilli, I.; Sette, C. Alternative splicing and cell survival: From tissue homeostasis to disease. Cell Death Differ. 2016, 23, 1919–1929. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.C.; Tsao, M.F.; Lin, Y.J. Differential Impacts of Alternative Splicing Networks on Apoptosis. Int. J. Mol. Sci. 2016, 17, 2097. [Google Scholar] [CrossRef] [PubMed]
- Chattopadhyay, S.; Yamashita, M.; Zhang, Y.; Sen, G.C. The IRF-3/Bax-mediated apoptotic pathway, activated by viral cytoplasmic RNA and DNA, inhibits virus replication. J. Virol. 2011, 85, 3708–3716. [Google Scholar] [CrossRef] [PubMed]
- Chattopadhyay, S.; Marques, J.T.; Yamashita, M.; Peters, K.L.; Smith, K.; Desai, A.; Williams, B.R.; Sen, G.C. Viral apoptosis is induced by IRF-3-mediated activation of Bax. EMBO J. 2010, 29, 1762–1773. [Google Scholar] [CrossRef]
- Djerbi, M.; Darreh-Shori, T.; Zhivotovsky, B.; Grandien, A. Characterization of the human FLICE-inhibitory protein locus and comparison of the anti-apoptotic activity of four different flip isoforms. Scand. J. Immunol. 2001, 54, 180–189. [Google Scholar] [CrossRef]
- Feoktistova, M.; Geserick, P.; Kellert, B.; Dimitrova, D.P.; Langlais, C.; Hupe, M.; Cain, K.; MacFarlane, M.; Hacker, G.; Leverkus, M. cIAPs block Ripoptosome formation, a RIP1/caspase-8 containing intracellular cell death complex differentially regulated by cFLIP isoforms. Mol. Cell 2011, 43, 449–463. [Google Scholar] [CrossRef]
- Krueger, A.; Schmitz, I.; Baumann, S.; Krammer, P.H.; Kirchhoff, S. Cellular FLICE-inhibitory protein splice variants inhibit different steps of caspase-8 activation at the CD95 death-inducing signaling complex. J. Biol. Chem. 2001, 276, 20633–20640. [Google Scholar] [CrossRef] [PubMed]
- Hughes, M.A.; Powley, I.R.; Jukes-Jones, R.; Horn, S.; Feoktistova, M.; Fairall, L.; Schwabe, J.W.; Leverkus, M.; Cain, K.; MacFarlane, M. Co-operative and Hierarchical Binding of c-FLIP and Caspase-8: A Unified Model Defines How c-FLIP Isoforms Differentially Control Cell Fate. Mol. Cell 2016, 61, 834–849. [Google Scholar] [CrossRef] [PubMed]
- Gates, L.T.; Shisler, J.L. cFLIPL Interrupts IRF3-CBP-DNA Interactions to Inhibit IRF3-Driven Transcription. J. Immunol. 2016, 197, 923–933. [Google Scholar] [CrossRef] [PubMed]
- Gates-Tanzer, L.T.; Shisler, J.L. Cellular FLIP long isoform (cFLIPL)-IKKalpha interactions inhibit IRF7 activation, representing a new cellular strategy to inhibit IFNalpha expression. J. Biol. Chem. 2018, 293, 1745–1755. [Google Scholar] [CrossRef]
- Ram, D.R.; Ilyukha, V.; Volkova, T.; Buzdin, A.; Tai, A.; Smirnova, I.; Poltorak, A. Balance between short and long isoforms of cFLIP regulates Fas-mediated apoptosis in vivo. Proc. Natl. Acad. Sci. USA 2016, 113, 1606–1611. [Google Scholar] [CrossRef]
- Ueffing, N.; Singh, K.K.; Christians, A.; Thorns, C.; Feller, A.C.; Nagl, F.; Fend, F.; Heikaus, S.; Marx, A.; Zotz, R.B.; et al. A single nucleotide polymorphism determines protein isoform production of the human c-FLIP protein. Blood 2009, 114, 572–579. [Google Scholar] [CrossRef]
- Pasparakis, M.; Vandenabeele, P. Necroptosis and its role in inflammation. Nature 2015, 517, 311–320. [Google Scholar] [CrossRef]
- Kaiser, W.J.; Sridharan, H.; Huang, C.; Mandal, P.; Upton, J.W.; Gough, P.J.; Sehon, C.A.; Marquis, R.W.; Bertin, J.; Mocarski, E.S. Toll-like receptor 3-mediated necrosis via TRIF, RIP3, and MLKL. J. Biol. Chem. 2013, 288, 31268–31279. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Liang, Y.; Shao, F.; Wang, X. Toll-like receptors activate programmed necrosis in macrophages through a receptor-interacting kinase-3-mediated pathway. Proc. Natl. Acad. Sci. USA 2011, 108, 20054–20059. [Google Scholar] [CrossRef]
- Kuriakose, T.; Man, S.M.; Malireddi, R.K.; Karki, R.; Kesavardhana, S.; Place, D.E.; Neale, G.; Vogel, P.; Kanneganti, T.D. ZBP1/DAI is an innate sensor of influenza virus triggering the NLRP3 inflammasome and programmed cell death pathways. Sci. Immunol. 2016, 1. [Google Scholar] [CrossRef] [PubMed]
- Thapa, R.J.; Ingram, J.P.; Ragan, K.B.; Nogusa, S.; Boyd, D.F.; Benitez, A.A.; Sridharan, H.; Kosoff, R.; Shubina, M.; Landsteiner, V.J.; et al. DAI Senses Influenza A Virus Genomic RNA and Activates RIPK3-Dependent Cell Death. Cell Host Microbe 2016, 20, 674–681. [Google Scholar] [CrossRef]
- Upton, J.W.; Kaiser, W.J.; Mocarski, E.S. DAI/ZBP1/DLM-1 complexes with RIP3 to mediate virus-induced programmed necrosis that is targeted by murine cytomegalovirus vIRA. Cell Host Microbe 2012, 11, 290–297. [Google Scholar] [CrossRef] [PubMed]
- Cai, Z.; Jitkaew, S.; Zhao, J.; Chiang, H.C.; Choksi, S.; Liu, J.; Ward, Y.; Wu, L.G.; Liu, Z.G. Plasma membrane translocation of trimerized MLKL protein is required for TNF-induced necroptosis. Nat. Cell Biol. 2014, 16, 55–65. [Google Scholar] [CrossRef]
- Wang, H.; Sun, L.; Su, L.; Rizo, J.; Liu, L.; Wang, L.F.; Wang, F.S.; Wang, X. Mixed lineage kinase domain-like protein MLKL causes necrotic membrane disruption upon phosphorylation by RIP3. Mol. Cell 2014, 54, 133–146. [Google Scholar] [CrossRef]
- Zhao, J.; Jitkaew, S.; Cai, Z.; Choksi, S.; Li, Q.; Luo, J.; Liu, Z.G. Mixed lineage kinase domain-like is a key receptor interacting protein 3 downstream component of TNF-induced necrosis. Proc. Natl. Acad. Sci. USA 2012, 109, 5322–5327. [Google Scholar] [CrossRef] [PubMed]
- Arnez, K.H.; Kindlova, M.; Bokil, N.J.; Murphy, J.M.; Sweet, M.J.; Guncar, G. Analysis of the N-terminal region of human MLKL, as well as two distinct MLKL isoforms, reveals new insights into necroptotic cell death. Biosci. Rep. 2015, 36, e00291. [Google Scholar] [CrossRef] [PubMed]
- Callow, M.G.; Watanabe, C.; Wickliffe, K.E.; Bainer, R.; Kummerfield, S.; Weng, J.; Cuellar, T.; Janakiraman, V.; Chen, H.; Chih, B.; et al. CRISPR whole-genome screening identifies new necroptosis regulators and RIPK1 alternative splicing. Cell Death Dis. 2018, 9, 261. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Hu, W.; Feng, S.; Ma, J.; Wu, M. RIP3 beta and RIP3 gamma, two novel splice variants of receptor-interacting protein 3 (RIP3), downregulate RIP3-induced apoptosis. Biochem. Biophys. Res. Commun. 2005, 332, 181–187. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Zhao, W. NLRP3 Inflammasome-A Key Player in Antiviral Responses. Front. Immunol. 2020, 11, 211. [Google Scholar] [CrossRef] [PubMed]
- Zheng, D.; Liwinski, T.; Elinav, E. Inflammasome activation and regulation: Toward a better understanding of complex mechanisms. Cell Discov. 2020, 6, 36. [Google Scholar] [CrossRef]
- Platnich, J.M.; Muruve, D.A. NOD-like receptors and inflammasomes: A review of their canonical and non-canonical signaling pathways. Arch. Biochem. Biophys. 2019, 670, 4–14. [Google Scholar] [CrossRef] [PubMed]
- Hoss, F.; Mueller, J.L.; Rojas Ringeling, F.; Rodriguez-Alcazar, J.F.; Brinkschulte, R.; Seifert, G.; Stahl, R.; Broderick, L.; Putnam, C.D.; Kolodner, R.D.; et al. Alternative splicing regulates stochastic NLRP3 activity. Nat. Commun. 2019, 10, 3238. [Google Scholar] [CrossRef]
- Bryan, N.B.; Dorfleutner, A.; Kramer, S.J.; Yun, C.; Rojanasakul, Y.; Stehlik, C. Differential splicing of the apoptosis-associated speck like protein containing a caspase recruitment domain (ASC) regulates inflammasomes. J. Inflamm. 2010, 7, 23. [Google Scholar] [CrossRef] [PubMed]
- Suganuma, Y.; Tanaka, H.; Kawase, A.; Kishida, A.; Yamaguchi, M.; Yabuuchi, A.; Inoue, K.; Shiozawa, S.; Komai, K. Expression of a PYCARD/ASC variant lacking exon 2 in Japanese patients with palindromic rheumatism increases interleukin-1beta secretion. Asian Pac. J. Allergy Immunol. 2019. [Google Scholar] [CrossRef]
- Boudreault, S.; Roy, P.; Lemay, G.; Bisaillon, M. Viral modulation of cellular RNA alternative splicing: A new key player in virus-host interactions? Wiley Interdiscip. Rev. RNA 2019, 10, e1543. [Google Scholar] [CrossRef]
- Ashraf, U.; Benoit-Pilven, C.; Lacroix, V.; Navratil, V.; Naffakh, N. Advances in Analyzing Virus-Induced Alterations of Host Cell Splicing. Trends Microbiol. 2019, 27, 268–281. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liao, K.-C.; Garcia-Blanco, M.A. Role of Alternative Splicing in Regulating Host Response to Viral Infection. Cells 2021, 10, 1720. https://doi.org/10.3390/cells10071720
Liao K-C, Garcia-Blanco MA. Role of Alternative Splicing in Regulating Host Response to Viral Infection. Cells. 2021; 10(7):1720. https://doi.org/10.3390/cells10071720
Chicago/Turabian StyleLiao, Kuo-Chieh, and Mariano A. Garcia-Blanco. 2021. "Role of Alternative Splicing in Regulating Host Response to Viral Infection" Cells 10, no. 7: 1720. https://doi.org/10.3390/cells10071720
APA StyleLiao, K.-C., & Garcia-Blanco, M. A. (2021). Role of Alternative Splicing in Regulating Host Response to Viral Infection. Cells, 10(7), 1720. https://doi.org/10.3390/cells10071720





