The Immune Landscape of Osteosarcoma: Implications for Prognosis and Treatment Response
Abstract
1. Introduction
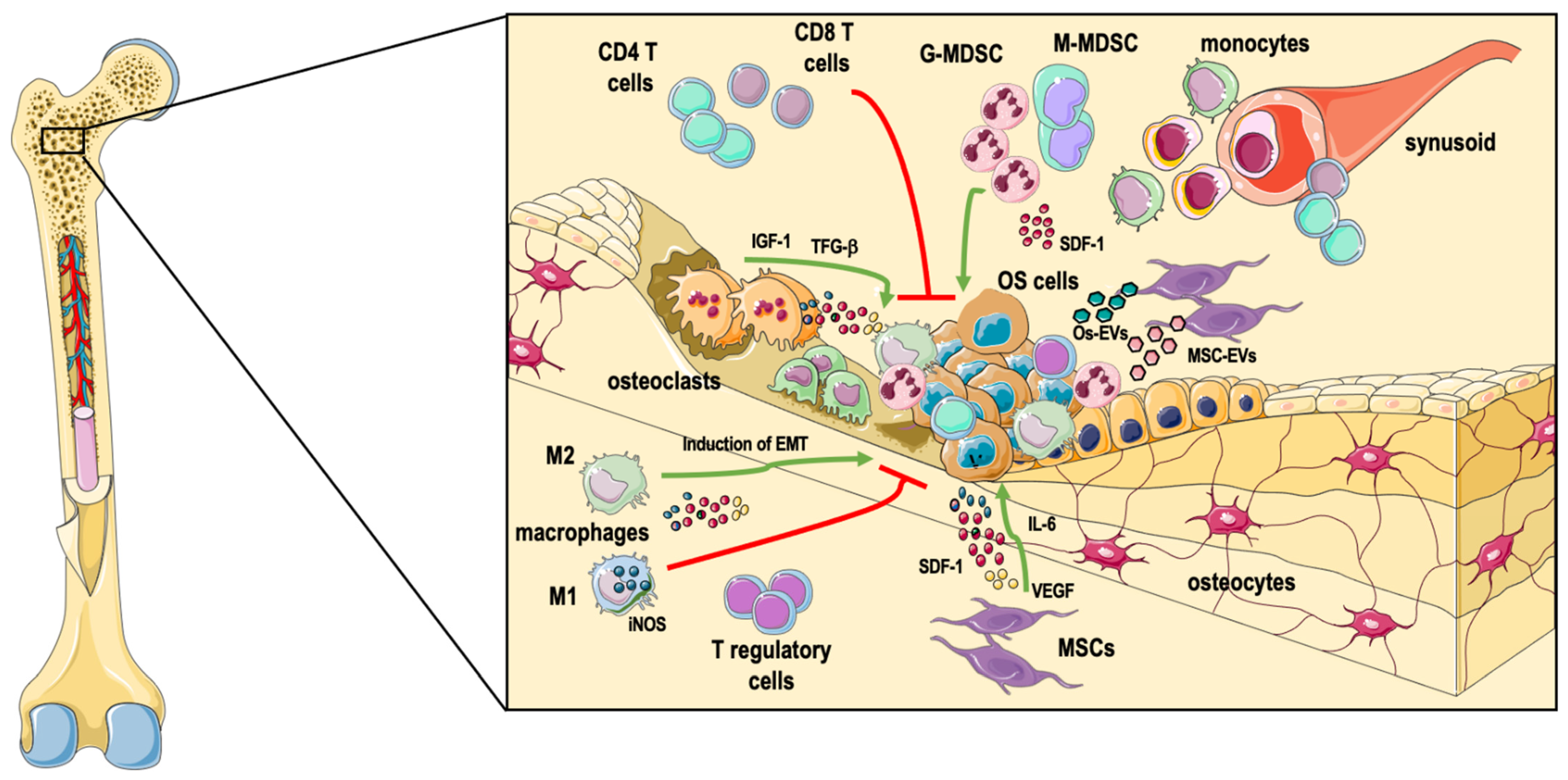
2. The Heterogenous Bone Marrow Microenvironment
2.1. Immune Cells
2.1.1. Macrophages
2.1.2. Other Myeloid Cells
2.1.3. T Lymphocytes and Other Immune Cell Types
2.2. Mesenchymal Stem Cells
2.3. Osteoclasts
3. Definition of Osteosarcoma Immune Landscape by Transcriptomic Analysis and Bioinformatics Tools
4. Recent Advances and Future Perspectives
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mirabello, L.; Troisi, R.J.; Savage, S.A. Osteosarcoma Incidence and Survival Rates from 1973 to 2004 Data from the Surveillance, Epidemiology, and End Results Program. Cancer 2009, 115, 1531–1543. [Google Scholar] [CrossRef]
- Ta, H.; Dass, C.; Choong, P.; Dunstan, D. Osteosarcoma treatment: State of the art. Cancer Metast. Rev. 2009, 28, 247–263. [Google Scholar] [CrossRef]
- Gaspar, N.; Occean, B.V.; Pacquement, H.; Bompas, E.; Bouvier, C.; Brisse, H.J.; Castex, M.P.; Cheurfa, N.; Corradini, N.; Delaye, J.; et al. Results of methotrexate-etoposide-ifosfamide based regimen (M-EI) in osteosarcoma patients included in the French OS2006/sarcome-09 study. Eur. J. Cancer 2018, 88, 57–66. [Google Scholar] [CrossRef]
- Mutsaers, A.J.; Walkley, C.R. Cells of origin in osteosarcoma: Mesenchymal stem cells or osteoblast committed cells? Bone 2014, 62, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Kuijjer, M.L.; Rydbeck, H.; Kresse, S.H.; Buddingh, E.P.; Roelofs, H.; Burger, H.; Myklebost, O.; Hogendoorn, P.C.W.; Meza-Zepeda, L.A.; Cleton-Jansen, A.M. Identification of osteosarcoma driver genes by integrative analysis of copy number and gene expression data. Cancer Res. 2012, 72. [Google Scholar] [CrossRef] [PubMed]
- Binnewies, M.; Roberts, E.W.; Kersten, K.; Chan, V.; Fearon, D.F.; Merad, M.; Coussens, L.M.; Gabrilovich, D.I.; Ostrand-Rosenberg, S.; Hedrick, C.C.; et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat. Med. 2018, 24, 541–550. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Coussens, L.M. Accessories to the Crime: Functions of Cells Recruited to the Tumor Microenvironment. Cancer Cell 2012, 21, 309–322. [Google Scholar] [CrossRef]
- Tsukasaki, M.; Takayanagi, H. Osteoimmunology: Evolving concepts in bone-immune interactions in health and disease. Nat. Rev. Immunol. 2019, 19, 626–642. [Google Scholar] [CrossRef]
- Heymann, M.F.; Lezot, F.; Heyman, D. The contribution of immune infiltrates and the local microenvironment in the pathogenesis of osteosarcoma. Cell. Immunol. 2019, 343, 103711. [Google Scholar] [CrossRef]
- Kim, H.J.; Cantor, H. CD4 T-cell Subsets and Tumor Immunity: The Helpful and the Not-so-Helpful. Cancer Immunol. Res. 2014, 2, 91–98. [Google Scholar] [CrossRef]
- Kennedy, R.; Celis, E. Multiple roles for CD4(+) T cells in anti-tumor immune responses. Immunol. Rev. 2008, 222, 129–144. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; McKay, D.; Pollard, J.W.; Lewis, C.E. Diverse Functions of Macrophages in Different Tumor Microenvironments. Cancer Res. 2018, 78, 5492–5503. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.X.; Huang, C.; Lin, Z.; Zhan, S.X.; Kong, L.N.; Fang, C.B.; Li, J. Macrophage polarization and function with emphasis on the evolving roles of coordinated regulation of cellular signaling pathways. Cell Signal. 2014, 26, 192–197. [Google Scholar] [CrossRef]
- Noy, R.; Pollard, J.W. Tumor-Associated Macrophages: From Mechanisms to Therapy. Immunity 2014, 41, 49–61. [Google Scholar] [CrossRef] [PubMed]
- Buddingh, E.P.; Kuijjer, M.L.; Duim, R.A.J.; Burger, H.; Agelopoulos, K.; Myklebost, O.; Serra, M.; Mertens, F.; Hogendoom, P.C.W.; Lankester, A.C.; et al. Tumor-Infiltrating Macrophages Are Associated with Metastasis Suppression in High-Grade Osteosarcoma: A Rationale for Treatment with Macrophage Activating Agents. Clin. Cancer Res. 2011, 17, 2110–2119. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Brouchet, A.; Illac, C.; Gilhodes, J.; Bouvier, C.; Aubert, S.; Guinebretiere, J.M.; Marie, B.; Larousserie, F.; Entz-Werle, N.; de Pinieux, G.; et al. CD163-positive tumor-associated macrophages and CD8-positive cytotoxic lymphocytes are powerful diagnostic markers for the therapeutic stratification of osteosarcoma patients: An immunohistochemical analysis of the biopsies fromthe French OS2006 phase 3 trial. Oncoimmunology 2017, 6, e1331193. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Guo, W.; Ren, T.T.; Huang, Y.; Wang, S.D.; Liu, K.S.; Zheng, B.X.; Yang, K.; Zhang, H.L.; Liang, X. Tumor-associated macrophages promote lung metastasis and induce epithelial-mesenchymal transition in osteosarcoma by activating the COX-2/STAT3 axis. Cancer Lett. 2019, 440, 116–125. [Google Scholar] [CrossRef] [PubMed]
- Han, Q.L.; Shi, H.G.; Liu, F. CD163(+) M2-type tumor-associated macrophage support the suppression of tumor-infiltrating T cells in osteosarcoma. Int. Immunopharmacol. 2016, 34, 101–106. [Google Scholar] [CrossRef]
- Dumars, C.; Ngyuen, J.M.; Gaultier, A.; Lanel, R.; Corradini, N.; Gouin, F.; Heymann, D.; Heymann, M.F. Dysregulation of macrophage polarization is associated with the metastatic process in osteosarcoma. Oncotarget 2016, 7, 78343–78354. [Google Scholar] [CrossRef]
- Xiao, Q.; Zhang, X.P.; Wu, Y.X.; Yang, Y. Inhibition of macrophage polarization prohibits growth of human osteosarcoma. Tumor Biol. 2014, 35, 7611–7616. [Google Scholar] [CrossRef]
- Shao, X.J.; Xiang, S.F.; Chen, Y.Q.; Zhang, N.; Cao, J.; Zhu, H.; Yang, B.; Zhou, Q.; Ying, M.D.; He, Q.J. Inhibition of M2-like macrophages by all-trans retinoic acid prevents cancer initiation and stemness in osteosarcoma cells. Acta Pharm. Sin. 2019, 40, 1343–1350. [Google Scholar] [CrossRef]
- Zhou, Y.; Yang, D.; Yang, Q.B.; Lv, X.B.; Huang, W.T.; Zhou, Z.H.; Wang, Y.L.; Zhang, Z.C.; Yuan, T.; Ding, X.M.; et al. Single-cell RNA landscape of intratumoral heterogeneity and immunosuppressive microenvironment in advanced osteosarcoma. Nat. Commun. 2020, 11, 6322. [Google Scholar] [CrossRef]
- Niu, J.F.; Yan, T.Q.; Guo, W.; Wang, W.; Zhao, Z.Q.; Ren, T.T.; Huang, Y.; Zhang, H.L.; Yu, Y.Y.; Liang, X. Identification of Potential Therapeutic Targets and Immune Cell Infiltration Characteristics in Osteosarcoma Using Bioinformatics Strategy. Front. Oncol. 2020, 10, 1628. [Google Scholar] [CrossRef]
- Cao, H.X.; Quan, S.; Zhang, L.; Chen, Y.Z.; Jiao, G.J. BMPR2 expression level is correlated with low immune infiltration and predicts metastasis and poor survival in osteosarcoma. Oncol. Lett. 2021, 21, 391. [Google Scholar] [CrossRef]
- Deng, C.Z.; Xu, Y.Y.; Fu, J.H.; Zhu, X.J.; Chen, H.M.; Xu, H.Y.; Wang, G.Y.; Song, Y.J.; Song, G.H.; Lu, J.H.; et al. Reprograming the tumor immunologic microenvironment using neoadjuvant chemotherapy in osteosarcoma. Cancer Sci. 2020, 111, 1899–1909. [Google Scholar] [CrossRef] [PubMed]
- Veglia, F.; Sanseviero, E.; Gabrilovich, D.I. Myeloid-derived suppressor cells in the era of increasing myeloid cell diversity. Nat. Rev. Immunol. 2021, 1–14. [Google Scholar] [CrossRef]
- Tcyganov, E.; Mastio, J.; Chen, E.; Gabrilovich, D.I. Plasticity of myeloid-derived suppressor cells in cancer. Curr. Opin. Immunol. 2018, 51, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Uehara, T.; Eikawa, S.; Nishida, M.; Kunisada, Y.; Yoshida, A.; Fujiwara, T.; Kunisada, T.; Ozaki, T.; Udono, H. Metformin induces CD11b(+)-cell-mediated growth inhibition of an osteosarcoma: Implications for metabolic reprogramming of myeloid cells and anti-tumor effects. Int. Immunol. 2019, 31, 187–198. [Google Scholar] [CrossRef] [PubMed]
- Jiang, K.; Li, J.; Zhang, J.T.; Wang, L.; Zhang, Q.F.; Ge, J.L.; Guo, Y.S.; Wang, B.; Huang, Y.; Yang, T.M.; et al. SDF-1/CXCR4 axis facilitates myeloid-derived suppressor cells accumulation in osteosarcoma microenvironment and blunts the response to anti-PD-1 therapy. Int. Immunopharmacol. 2019, 75, 105818. [Google Scholar] [CrossRef]
- Schreiber, R.D.; Old, L.J.; Smyth, M.J. Cancer Immunoediting: Integrating Immunity’s Roles in Cancer Suppression and Promotion. Science 2011, 331, 1565–1570. [Google Scholar] [CrossRef]
- O’Donnell, J.S.; Teng, M.W.L.; Smyth, M.J. Cancer immunoediting and resistance to T cell-based immunotherapy. Nat. Rev. Clin. Oncol. 2019, 16, 151–167. [Google Scholar] [CrossRef]
- Teng, M.W.L.; Galon, J.; Fridman, W.H.; Smyth, M.J. From mice to humans: Developments in cancer immunoediting. J. Clin. Investig. 2015, 125, 3338–3346. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Allison, J.P. The future of immune checkpoint therapy. Science 2015, 348, 56–61. [Google Scholar] [CrossRef]
- Topalian, S.L.; Hodi, F.S.; Brahmer, J.R.; Gettinger, S.N.; Smith, D.C.; McDermott, D.F.; Powderly, J.D.; Carvajal, R.D.; Sosman, J.A.; Atkins, M.B.; et al. Safety, Activity, and Immune Correlates of Anti-PD-1 Antibody in Cancer. N. Engl. J. Med. 2012, 366, 2443–2454. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.M.T.; Shenasa, E.; Nielsen, T.O. Sarcomas: Immune biomarker expression and checkpoint inhibitor trials. Cancer Treat. Rev. 2020, 91, 102115. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.K.; Cote, G.M.; Choy, E.; Yang, P.; Harmon, D.; Schwab, J.; Nielsen, G.P.; Chebib, I.; Ferrone, S.; Wang, X.H.; et al. Programmed Cell Death Ligand 1 Expression in Osteosarcoma. Cancer Immunol. Res. 2014, 2, 690–698. [Google Scholar] [CrossRef] [PubMed]
- Koirala, P.; Roth, M.E.; Gill, J.; Piperdi, S.; Chinai, J.M.; Geller, D.S.; Hoang, B.H.; Park, A.; Fremed, M.A.; Zang, X.X.; et al. Immune infiltration and PD-L1 expression in the tumor microenvironment are prognostic in osteosarcoma. Sci. Rep. 2016, 6, 30093. [Google Scholar] [CrossRef]
- Toda, Y.; Kohashi, K.; Yamada, Y.; Yoshimoto, M.; Ishihara, S.; Ito, Y.; Iwasaki, T.; Yamamoto, H.; Matsumoto, Y.; Nakashima, Y.; et al. PD-L1 and IDO1 expression and tumor-infiltrating lymphocytes in osteosarcoma patients: Comparative study of primary and metastatic lesions. J. Cancer Res. Clin. 2020, 146, 2607–2620. [Google Scholar] [CrossRef]
- Palmerini, E.; Agostinelli, C.; Picci, P.; Pileri, S.; Marafioti, T.; Lollini, P.L.; Scotlandi, K.; Longhi, A.; Benassi, M.S.; Ferrari, S. Tumoral immune-infiltrate (IF), PD-L1 expression and role of CD8/TIA-1 lymphocytes in localized osteosarcoma patients treated within protocol ISG-OS1. Oncotarget 2017, 8, 111836–111846. [Google Scholar] [CrossRef]
- Fritzsching, B.; Fellenberg, J.; Moskovszky, L.; Sapi, Z.; Krenacs, T.; Machado, I.; Poeschl, J.; Lehner, B.; Szendroi, M.; Bosch, A.L.; et al. CD8(+)/FOXP3(+)-ratio in osteosarcoma microenvironment separates survivors from non-survivors: A multicenter validated retrospective study. Oncoimmunology 2015, 4, e990800. [Google Scholar] [CrossRef]
- Yoshida, K.; Okamoto, M.; Sasaki, J.; Kuroda, C.; Ishida, H.; Ueda, K.; Ideta, H.; Kamanaka, T.; Sobajima, A.; Takizawa, T.; et al. Anti-PD-1 antibody decreases tumour-infiltrating regulatory T cells. BMC Cancer 2020, 20, 25. [Google Scholar] [CrossRef] [PubMed]
- Takenaka, W.; Takahashi, Y.; Tamari, K.; Minami, K.; Katsuki, S.; Seo, Y.; Isohashi, F.; Koizumi, M.; Ogawa, K. Radiation Dose Escalation Is Crucial in Anti-CTLA-4 Antibody Therapy to Enhance Local and Distant Antitumor Effect in Murine Osteosarcoma. Cancers 2020, 12, 1546. [Google Scholar] [CrossRef] [PubMed]
- Asadzadeh, Z.; Mohammadi, H.; Safarzadeh, E.; Hemmatzadeh, M.; Mahdian-Shakib, A.; Jadidi-Niaragh, F.; Azizi, G.; Baradaran, B. The paradox of Th17 cell functions in tumor immunity. Cell. Immunol. 2017, 322, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Honorati, M.C.; Cattini, L.; Facchini, A. Possible prognostic role of IL-17R in osteosarcoma. J. Cancer Res. Clin. 2007, 133, 1017–1021. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.N.; Wang, Z.; Li, B.H.; Wang, S.D.; Chen, T.; Ye, Z.M. Innate Immune Cells: A Potential and Promising Cell Population for Treating Osteosarcoma. Front. Immunol. 2019, 10, 1114. [Google Scholar] [CrossRef]
- Li, Z.X.; Peng, H.Q.; Xu, Q.; Ye, Z.M. Sensitization of human osteosarcoma cells to Vγ9Vδ2 T-cell-mediated cytotoxicity by zoledronate. J. Orthop. Res. 2012, 30, 824–830. [Google Scholar] [CrossRef]
- Muraro, M.; Mereuta, O.M.; Carraro, F.; Madon, E.; Fagioll, F. Osteosarcoma cell line growth inhibition by zoledronate-stimulated effector cells. Cell. Immunol. 2007, 249, 63–72. [Google Scholar] [CrossRef][Green Version]
- Wang, S.D.; Li, H.Y.; Ye, C.Y.; Lin, P.; Li, B.H.; Zhang, W.; Sun, L.L.; Wang, Z.; Xue, D.T.; Teng, W.S.Y.; et al. Valproic Acid Combined with Zoledronate Enhance gamma delta T Cell-Mediated Cytotoxicity against Osteosarcoma Cells via the Accumulation of Mevalonate Pathway Intermediates. Front. Immunol. 2018, 9, 377. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, Z.A.; Li, S.; Li, B.H.; Sun, L.L.; Li, H.Y.; Lin, P.; Wang, S.D.; Teng, W.S.Y.; Zhou, X.Z.; et al. Decitabine Enhances Vγ9Vδ2 T Cell-Mediated Cytotoxic Effects on Osteosarcoma Cells via the NKG2DL-NKG2D Axis. Front. Immunol. 2018, 9, 1239. [Google Scholar] [CrossRef]
- Fleming, C.; Morrissey, S.; Cai, Y.H.; Yan, J. γδ T Cells: Unexpected Regulators of Cancer Development and Progression. Trends Cancer 2017, 3, 561–570. [Google Scholar] [CrossRef]
- Morrison, S.J.; Scadden, D.T. The bone marrow niche for haematopoietic stem cells. Nature 2014, 505, 327–334. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Wang, G.Y.; Chen, R.L.; Hua, Y.Q.; Cai, Z.D. Mesenchymal stem cells in the osteosarcoma microenvironment: Their biological properties, influence on tumor growth, and therapeutic implications. Stem Cell Res. Ther. 2018, 9, 22. [Google Scholar] [CrossRef] [PubMed]
- Cortini, M.; Massa, A.; Avnet, S.; Bonuccelli, G.; Baldini, N. Tumor-Activated Mesenchymal Stromal Cells Promote Osteosarcoma Stemness and Migratory Potential via IL-6 Secretion. PLoS ONE 2016, 11, e0166500. [Google Scholar] [CrossRef]
- Zhang, P.; Dong, L.; Yan, K.; Long, H.; Yang, T.T.; Dong, M.Q.; Zhou, Y.; Fan, Q.Y.; Ma, B.A. CXCR4-mediated osteosarcoma growth and pulmonary metastasis is promoted by mesenchymal stem cells through VEGF. Oncol. Rep. 2013, 30, 1753–1761. [Google Scholar] [CrossRef] [PubMed]
- Pietrovito, L.; Leo, A.; Gori, V.; Lulli, M.; Parri, M.; Becherucci, V.; Piccini, L.; Bambi, F.; Taddei, M.L.; Chiarugi, P. Bone marrow-derived mesenchymal stem cells promote invasiveness and transendothelial migration of osteosarcoma cells via a mesenchymal to amoeboid transition. Mol. Oncol. 2018, 12, 659–676. [Google Scholar] [CrossRef]
- Bonuccelli, G.; Avnet, S.; Grisendi, G.; Salerno, M.; Granchi, D.; Dominici, M.; Kusuzaki, K.; Baldini, N. Role of mesenchymal stem cells in osteosarcoma and metabolic reprogramming of tumor cells. Oncotarget 2014, 5, 7575–7588. [Google Scholar] [CrossRef] [PubMed]
- Baglio, S.R.; Lagerweij, T.; Perez-Lanzon, M.; Ho, X.D.; Leveille, N.; Melo, S.A.; Cleton-Jansen, A.M.; Jordanova, E.S.; Roncuzzi, L.; Greco, M.; et al. Blocking Tumor-Educated MSC Paracrine Activity Halts Osteosarcoma Progression. Clin. Cancer Res. 2017, 23, 3721–3733. [Google Scholar] [CrossRef] [PubMed]
- Endo-Munoz, L.; Evdokiou, A.; Saunders, N.A. The role of osteoclasts and tumour-associated macrophages in osteosarcoma metastasis. Biochim. Biophys. Acta 2012, 1826, 434–442. [Google Scholar] [CrossRef]
- Avnet, S.; Longhi, A.; Salerno, M.; Halleen, J.M.; Perut, F.; Granchi, D.; Ferrari, S.; Bertoni, F.; Giuti, A.; Baldini, N. Increased osteoclast activity is associated with aggressiveness of osteosarcoma. Int. J. Oncol. 2008, 33, 1231–1238. [Google Scholar] [CrossRef]
- Akiyama, T.; Dass, C.R.; Choong, P.F.M. Novel therapeutic strategy for osteosarcoma targeting osteoclast differentiation, bone-resorbing activity, and apoptosis pathway. Mol. Cancer Ther. 2008, 7, 3461–3469. [Google Scholar] [CrossRef] [PubMed]
- Akiyama, T.; Dass, C.R.; Shinoda, Y.; Kawano, H.; Tanaka, S.; Choong, P.F.M. Systemic RANK-Fc protein therapy is efficacious against primary osteosarcoma growth in a murine model via activity against osteoclasts. J. Pharm. Pharmacol. 2010, 62, 470–476. [Google Scholar] [CrossRef]
- Chen, Y.; Di Grappa, M.A.; Molyneux, S.D.; McKee, T.D.; Waterhouse, P.; Penninger, J.M.; Khokha, R. RANKL blockade prevents and treats aggressive osteosarcomas. Sci. Transl. Med. 2015, 7, 317ra197. [Google Scholar] [CrossRef] [PubMed]
- Lu, K.H.; Lu, E.W.H.; Lin, C.W.; Yang, J.S.; Yang, S.F. New insights into molecular and cellular mechanisms of zoledronate in human osteosarcoma. Pharm. Ther. 2020, 214, 107611. [Google Scholar] [CrossRef] [PubMed]
- Endo-Munoz, L.; Cumming, A.; Rickwood, D.; Wilson, D.; Cueva, C.; Ng, C.; Strutton, G.; Cassady, A.I.; Evdokiou, A.; Sommerville, S.; et al. Loss of Osteoclasts Contributes to Development of Osteosarcoma Pulmonary Metastases. Cancer Res. 2010, 70, 7063–7072. [Google Scholar] [CrossRef] [PubMed]
- Newman, A.M.; Liu, C.L.; Green, M.R.; Gentles, A.J.; Feng, W.G.; Xu, Y.; Hoang, C.D.; Diehn, M.; Alizadeh, A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 2015, 12, 453–457. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Zheng, J.H.; Lin, Z.H.; Lv, H.Y.; Ye, Z.M.; Chen, Y.P.; Zhang, X.Y. Profiles of immune cell infiltration and immune-related genes in the tumor microenvironment of osteosarcoma. Aging 2020, 12, 3486–3501. [Google Scholar] [CrossRef]
- Zhang, T.; Nie, Y.L.; Xia, H.F.; Zhang, Y.B.; Cai, K.L.; Chen, X.D.; Li, H.L.; Wang, J.L. Identification of Immune-Related Prognostic Genes and LncRNAs Biomarkers Associated with Osteosarcoma Microenvironment. Front. Oncol. 2020, 10, 1109. [Google Scholar] [CrossRef]
- Hong, W.F.; Yuan, H.; Gu, Y.J.; Liu, M.Y.; Ji, Y.Y.; Huang, Z.F.; Yang, J.L.; Ma, L.H. Immune-related prognosis biomarkers associated with osteosarcoma microenvironment. Cancer Cell Int. 2020, 20, 83. [Google Scholar] [CrossRef]
- Song, Y.J.; Xu, Y.Y.; Zhu, X.J.; Fu, J.C.; Deng, C.Z.; Chen, H.M.; Xu, H.Y.; Song, G.H.; Lu, J.C.; Tang, Q.L.; et al. Immune Landscape of the Tumor Microenvironment Identifies Prognostic Gene Signature CD4/CD68/CSF1R in Osteosarcoma. Front. Oncol. 2020, 10, 1198. [Google Scholar] [CrossRef]
- Landuzzi, L.; Manara, M.C.; Lollini, P.L.; Scotlandi, K. Patient Derived Xenografts for Genome-Driven Therapy of Osteosarcoma. Cells 2021, 10, 416. [Google Scholar] [CrossRef]
- Sayles, L.C.; Breese, M.R.; Koehne, A.L.; Leung, S.G.; Lee, A.G.; Liu, H.Y.; Spillinger, A.; Shah, A.T.; Tanasa, B.; Straessler, K.; et al. Genome-Informed Targeted Therapy for Osteosarcoma. Cancer Discov. 2019, 9, 46–63. [Google Scholar] [CrossRef]
| Type of Study | Markers and TAM Phenotype | Findings | References |
|---|---|---|---|
| Human samples, GEP and IHC analysis | CD14/HLA-DRα (M1); CD14/CD163 (M2) | Higher CD14+ TAMs levels correlated with better OS and metastasis suppression with no clinical relevance between M1/M2 phenotype | [15] |
| Human samples, IHC analysis | CD163 (M2); | Better overall survival and prolonged metastasis progression-free survival. | [16] |
| Human samples, IHC analysis | CD68/iNOS/COX-2 (M1); CD163 (M2); | Higher CD68+/COX-2 levels in lung metastasis, unchanged CD163+ compared to paients with primary tumours while iNOS+ was relatively higher. | [17] |
| Pre-clinical study (THP-1 human cell line) | CD206/CD163 (M2); | BALB/c mice subcutaneously injected with THP-1 cell line promoted migration/invasion of OS cells through MO/M2 TAMs, showing higher levels of ZEB-1 and SNAIL toward an EMT phenotype. | [17] |
| Human samples, FACS analysis | CD14/CD163 (M2); | Higher CD163+ TAMs levels in primary tumour than PB and TILs correlated with lower levels of TIM-3+/PD-1+ T cells. | [18] |
| Human samples, IHC analysis | CD68/iNOS (M1); | Higher CD68/iNOS+ level in non metastatic patients’ group, with better OS | [19] |
| Pre-clinical study (U2OS human cell line) | F4/F80/CD163 (M2) | NOD/SCID mice orthotopically injected with OS cells show the recruitment of CD163+ M2 TAMs subtype and a higher tumour growth. | [20] |
| Pre-clinical study (K7M2 mouse cell line) | CD163/CD209/MRC1/CCR2/F4/80 (M2); | BALB/c mice orthotopically co-injected with OS with or w/out RAW264.7 cells treated with ATRA show reduced M2-polarization through suppressing colony/sarcosphere formation and tumour growth. | [21] |
| Human samples, single cell RNA seq analysis | CD163/MRC1/MS4A4/MAF (M2); FABP4+ (M3); | M1-, M2- and M3 TAMs are the 80% of the major macrophages cell subtypes in the myeloid compartment of OS lesions, while the FABP4+ M3 levels are directly correlated with the metastatic spread through the lung. | [22] |
| Human samples, CIBERSORT algorithm | Defined by CIBERSORT | M0 and M2 are the most relevant cell subtypes in patients’ metastatic samples. | [23] |
| Human samples, CIBERSORT and TIMER analysis | Defined by CIBERSORT | Lower levels of M2 TAMs directly associated with patients’ poor survival. | [24] |
| Human samples, GEP and TIMER analysis | Defined by CIBERSORT | Higher M1/M2 TAMs levels in the infiltrating microenvironment showed an improved overall survival of patients; better OS | [25] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cascini, C.; Chiodoni, C. The Immune Landscape of Osteosarcoma: Implications for Prognosis and Treatment Response. Cells 2021, 10, 1668. https://doi.org/10.3390/cells10071668
Cascini C, Chiodoni C. The Immune Landscape of Osteosarcoma: Implications for Prognosis and Treatment Response. Cells. 2021; 10(7):1668. https://doi.org/10.3390/cells10071668
Chicago/Turabian StyleCascini, Caterina, and Claudia Chiodoni. 2021. "The Immune Landscape of Osteosarcoma: Implications for Prognosis and Treatment Response" Cells 10, no. 7: 1668. https://doi.org/10.3390/cells10071668
APA StyleCascini, C., & Chiodoni, C. (2021). The Immune Landscape of Osteosarcoma: Implications for Prognosis and Treatment Response. Cells, 10(7), 1668. https://doi.org/10.3390/cells10071668






