Regulatory and Functional Involvement of Long Non-Coding RNAs in DNA Double-Strand Break Repair Mechanisms
Abstract
1. Introduction
2. Transcriptome and Long Non-Coding RNA
- (i)
- Genomic location of lncRNAs. This classification involves: (a) intergenic lncRNAs (lincRNAs)—loci encoding such transcripts are located in the intervening regions among protein coding genes; (b) intronic lncRNAs—these transcripts are derived from introns that separate the coding exons of a gene.
- (ii)
- Location context. In this respect, lncRNAs relate to sense- or antisense-derived transcripts: (a) the former are transcribed from the sense strand and may overlap partly or completely with the entire sequence of a protein coding gene; (b) the antisense-derived transcripts originate from the antisense strand and can emerge from three routes: (1) antisense strand-derived transcripts may overlap with an exon of a sense gene, (2) transcripts may overlap with the intron of a sense gene, and (3) transcripts may cover the entire protein-coding sequence.
- (iii)
- Effect exerted on DNA sequences. Cis- and trans-lncRNAs: the former regulate the expression of genes in close genomic proximity, while the latter regulate the expression of distant genes.
- (iv)
- Mechanism of function. According to the function that they exert, lncRNAs are categorized into the following groups: (a) Transcriptional regulation. This group involves lncRNAs that are implicated in chromatin remodeling and transcriptional interference, including enhancer lncRNAs (eRNAs) that are transcribed from enhancers; (b) Post-transcriptional regulation. This group comprises lncRNAs that participate in splicing regulation and post-transcriptional regulation (mRNA stability); (c) Translational control, by facilitating or repressing this process; (d) Other functions, such as protein localization, telomere replication and RNA interference.
- (v)
- Targeting mechanism. This is a heterogeneous group of lncRNAs due to the multiple mechanisms of action and interactionsubstrates that they present or recognize, respectively. There are several subdivisions in this category: (a) signal dependency as lncRNAs show cell type-specific expression and response to diverse stimuli; (b) decoy role, since certain lncRNAs can bind and titrate away an RNA or protein target, without any further function(s); (c) guiding ability as they bind proteins and then direct the localization of the ribonucleoprotein complex to specific targets; (d) scaffolding capacity, since such lncRNAs serve as central platforms for the concurrent binding of multiple proteins to form ribonucleoprotein complexes. LncRNAs can also be grouped together based on the nature of the interactions that they establish with their targets: RNA–RNA pairing, RNA–DNA hybrids, RNA structure-mediated interactions and protein linkers.
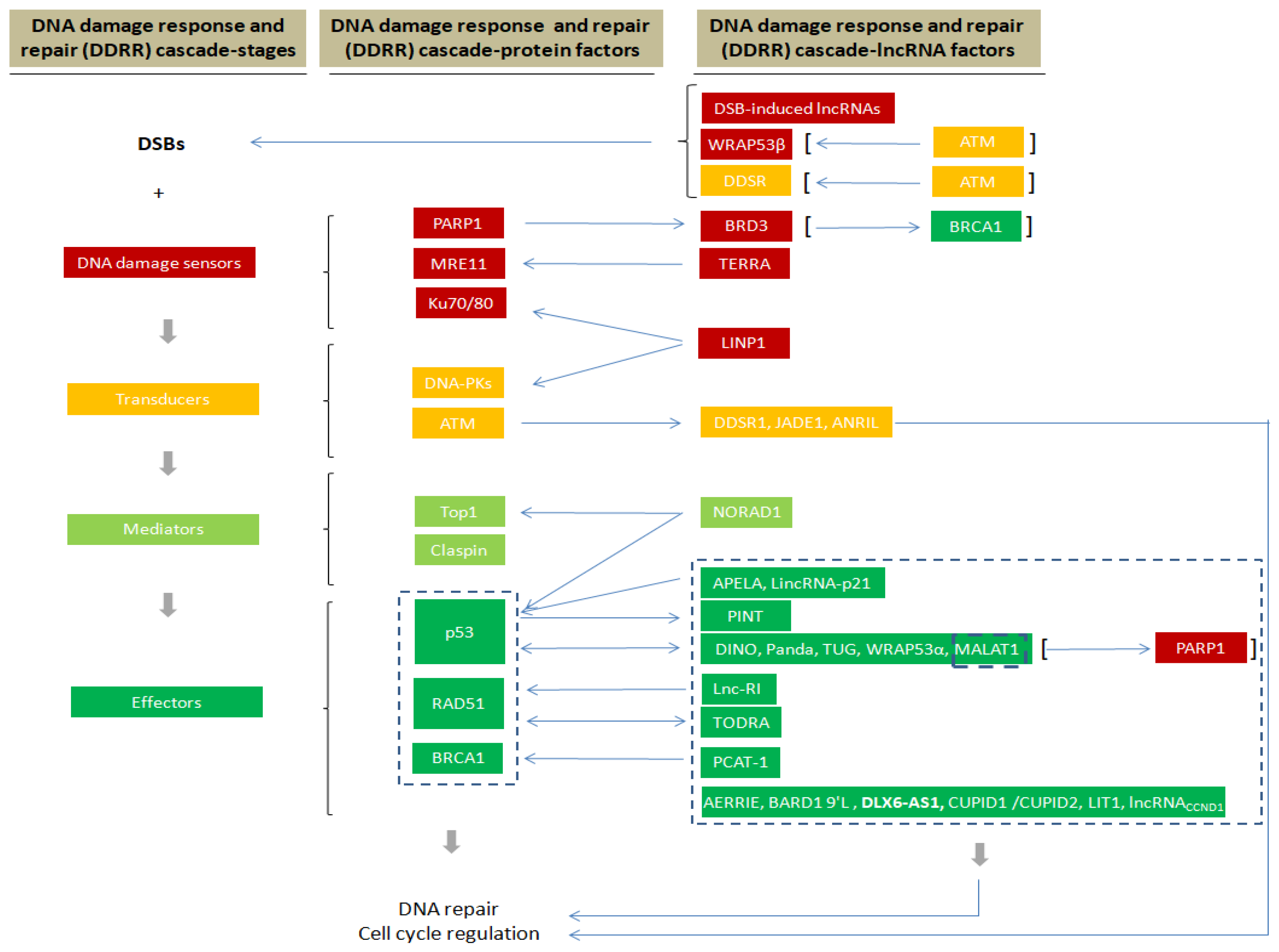
3. The DNA Damage Response and Repair (DDRR) Network
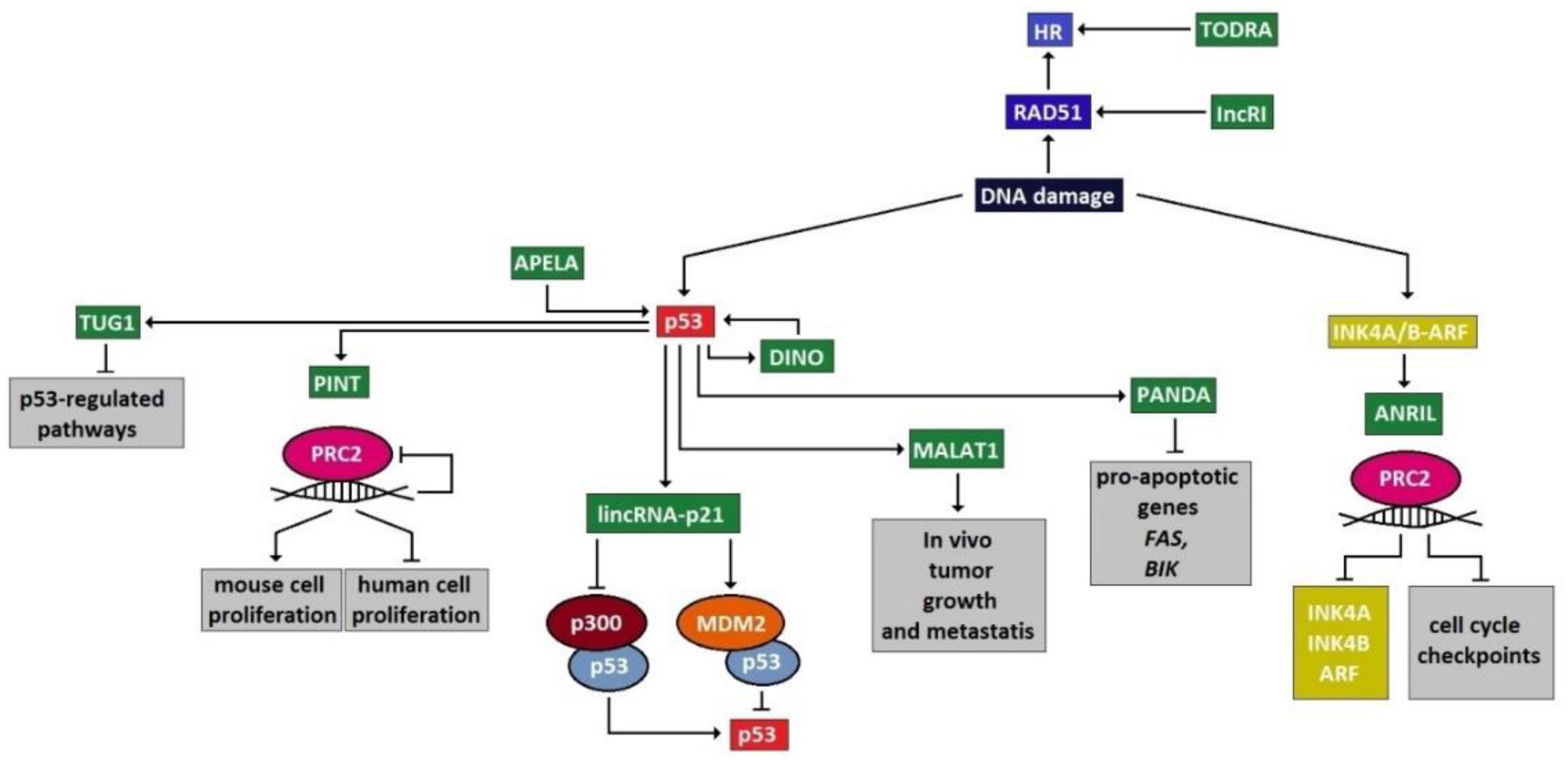
4. LncRNA and DSB Repair
4.1. LncRNAs Involved in DSB Sensing
4.2. LncRNAs Acting at Transducer/Mediator Level
4.3. LncRNAs Acting at the Effector Level
5. Significance of lncRNAs in DDRR
6. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Gorgoulis, V.G.; Pefani, D.E.; Pateras, I.S.; Trougakos, I.P. Integrating the DNA damage and protein stress responses during cancer development and treatment. J. Pathol. 2018, 246, 12–40. [Google Scholar] [CrossRef]
- Pateras, I.S.; Havaki, S.; Nikitopoulou, X.; Vougas, K.; Townsend, P.A.; Panayiotidis, M.I.; Georgakilas, A.G.; Gorgoulis, V.G. The DNA damage response and immune signaling alliance: Is it good or bad? Nature decides when and where. Pharmacol. Ther. 2015, 154, 36–56. [Google Scholar] [CrossRef]
- Michelini, F.; Jalihal, A.P.; Francia, S.; Meers, C.; Neeb, Z.T.; Rossiello, F.; Gioia, U.; Aguado, J.; Jones-Weinert, C.; Luke, B.; et al. From “Cellular” RNA to “Smart” RNA: Multiple Roles of RNA in Genome Stability and Beyond. Chem. Rev. 2018, 118, 4365–4403. [Google Scholar] [CrossRef] [PubMed]
- Halazonetis, T.D.; Gorgoulis, V.G.; Bartek, J. An oncogene-induced DNA damage model for cancer development. Science 2008, 319, 1352–1355. [Google Scholar] [CrossRef] [PubMed]
- Carninci, P.; Kasukawa, T.; Katayama, S.; Gough, J.; Frith, M.C.; Maeda, N.; Oyama, R.; Ravasi, T.; Lenhard, B.; Wells, C.; et al. The transcriptional landscape of the mammalian genome. Science 2005, 309, 1559–1563. [Google Scholar] [CrossRef] [PubMed]
- Willingham, A.T.; Orth, A.P.; Batalov, S.; Peters, E.C.; Wen, B.G.; Aza-Blanc, P.; Hogenesch, J.B.; Schultz, P.G. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science 2005, 309, 1570–1573. [Google Scholar] [CrossRef]
- Bertone, P.; Stolc, V.; Royce, T.E.; Rozowsky, J.S.; Urban, A.E.; Zhu, X.; Rinn, J.L.; Tongprasit, W.; Samanta, M.; Weissman, S.; et al. Global identification of human transcribed sequences with genome tiling arrays. Science 2004, 306, 2242–2246. [Google Scholar] [CrossRef]
- Ransohoff, J.D.; Wei, Y.; Khavari, P.A. The functions and unique features of long intergenic non-coding RNA. Nat. Rev. Mol. Cell Biol. 2018, 19, 143–157. [Google Scholar] [CrossRef]
- Johnson, J.M.; Edwards, S.; Shoemaker, D.; Schadt, E.E. Dark matter in the genome: Evidence of widespread transcription detected by microarray tiling experiments. Trends Genet. 2005, 21, 93–102. [Google Scholar] [CrossRef]
- Ma, L.; Bajic, V.B.; Zhang, Z. On the classification of long non-coding RNAs. RNA Biol. 2013, 10, 925–933. [Google Scholar] [CrossRef]
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef]
- Ma, L.; Cao, J.; Liu, L.; Du, Q.; Li, Z.; Zou, D.; Bajic, V.B.; Zhang, Z. LncBook: A curated knowledgebase of human long non-coding RNAs. Nucleic Acids Res. 2019, 47, 2699. [Google Scholar] [CrossRef]
- Centonze, D.; Rossi, S.; Napoli, I.; Mercaldo, V.; Lacoux, C.; Ferrari, F.; Ciotti, M.T.; De Chiara, V.; Prosperetti, C.; Maccarrone, M.; et al. The brain cytoplasmic RNA BC1 regulates dopamine D2 receptor-mediated transmission in the striatum. J. Neurosci. 2007, 27, 8885–8892. [Google Scholar] [CrossRef]
- Donley, N.; Smith, L.; Thayer, M.J. ASAR15, A cis-acting locus that controls chromosome-wide replication timing and stability of human chromosome 15. PLoS Genet. 2015, 11, e1004923. [Google Scholar] [CrossRef]
- Donley, N.; Stoffregen, E.P.; Smith, L.; Montagna, C.; Thayer, M.J. Asynchronous replication, mono-allelic expression, and long range Cis-effects of ASAR6. PLoS Genet. 2013, 9, e1003423. [Google Scholar] [CrossRef] [PubMed]
- Heskett, M.B.; Smith, L.G.; Spellman, P.; Thayer, M.J. Reciprocal monoallelic expression of ASAR lncRNA genes controls replication timing of human chromosome 6. RNA 2020, 26, 724–738. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, J.C.R.; Acuna, S.M.; Aoki, J.I.; Floeter-Winter, L.M.; Muxel, S.M. Long Non-Coding RNAs in the Regulation of Gene Expression: Physiology and Disease. Noncoding RNA 2019, 5, 17. [Google Scholar] [CrossRef] [PubMed]
- Yunusov, D.; Anderson, L.; DaSilva, L.F.; Wysocka, J.; Ezashi, T.; Roberts, R.M.; Verjovski-Almeida, S. HIPSTR and thousands of lncRNAs are heterogeneously expressed in human embryos, primordial germ cells and stable cell lines. Sci. Rep. 2016, 6, 32753. [Google Scholar] [CrossRef] [PubMed]
- Cabili, M.N.; Trapnell, C.; Goff, L.; Koziol, M.; Tazon-Vega, B.; Regev, A.; Rinn, J.L. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011, 25, 1915–1927. [Google Scholar] [CrossRef]
- Yan, L.; Yang, M.; Guo, H.; Yang, L.; Wu, J.; Li, R.; Liu, P.; Lian, Y.; Zheng, X.; Yan, J.; et al. Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat. Struct. Mol. Biol. 2013, 20, 1131–1139. [Google Scholar] [CrossRef]
- Liu, S.J.; Nowakowski, T.J.; Pollen, A.A.; Lui, J.H.; Horlbeck, M.A.; Attenello, F.J.; He, D.; Weissman, J.S.; Kriegstein, A.R.; Diaz, A.A.; et al. Single-cell analysis of long non-coding RNAs in the developing human neocortex. Genome Biol. 2016, 17, 67. [Google Scholar] [CrossRef]
- Sun, Q.; Hao, Q.; Prasanth, K.V. Nuclear Long Noncoding RNAs: Key Regulators of Gene Expression. Trends Genet. 2018, 34, 142–157. [Google Scholar] [CrossRef]
- Pagano, J.M.; Farley, B.M.; McCoig, L.M.; Ryder, S.P. Molecular basis of RNA recognition by the embryonic polarity determinant MEX-5. J. Biol. Chem. 2007, 282, 8883–8894. [Google Scholar] [CrossRef]
- Negrini, S.; Gorgoulis, V.G.; Halazonetis, T.D. Genomic instability—An evolving hallmark of cancer. Nat. Rev. Mol. Cell Biol. 2010, 11, 220–228. [Google Scholar] [CrossRef]
- Galanos, P.; Pappas, G.; Polyzos, A.; Kotsinas, A.; Svolaki, I.; Giakoumakis, N.N.; Glytsou, C.; Pateras, I.S.; Swain, U.; Souliotis, V.L.; et al. Mutational signatures reveal the role of RAD52 in p53-independent p21-driven genomic instability. Genome Biol. 2018, 19, 37. [Google Scholar] [CrossRef] [PubMed]
- Lagopati, N.; Belogiannis, K.; Angelopoulou, A.; Papaspyropoulos, A.; Gorgoulis, V. Non-Canonical Functions of the ARF Tumor Suppressor in Development and Tumorigenesis. Biomolecules 2021, 11, 86. [Google Scholar] [CrossRef] [PubMed]
- Shibata, A.; Moiani, D.; Arvai, A.S.; Perry, J.; Harding, S.M.; Genois, M.M.; Maity, R.; van Rossum-Fikkert, S.; Kertokalio, A.; Romoli, F.; et al. DNA double-strand break repair pathway choice is directed by distinct MRE11 nuclease activities. Mol. Cell 2014, 53, 7–18. [Google Scholar] [CrossRef] [PubMed]
- Schipler, A.; Iliakis, G. DNA double-strand-break complexity levels and their possible contributions to the probability for error-prone processing and repair pathway choice. Nucleic Acids Res. 2013, 41, 7589–7605. [Google Scholar] [CrossRef] [PubMed]
- Branzei, D.; Foiani, M. Regulation of DNA repair throughout the cell cycle. Nat. Rev. Mol. Cell Biol. 2008, 9, 297–308. [Google Scholar] [CrossRef] [PubMed]
- Chapman, J.R.; Taylor, M.R.; Boulton, S.J. Playing the end game: DNA double-strand break repair pathway choice. Mol. Cell 2012, 47, 497–510. [Google Scholar] [CrossRef] [PubMed]
- Kakarougkas, A.; Jeggo, P.A. DNA DSB repair pathway choice: An orchestrated handover mechanism. Br. J. Radiol. 2014, 87, 20130685. [Google Scholar] [CrossRef]
- Domingo-Prim, J.; Bonath, F.; Visa, N. RNA at DNA Double-Strand Breaks: The Challenge of Dealing with DNA: RNA Hybrids. Bioessays 2020, 42, e1900225. [Google Scholar] [CrossRef]
- Henriksson, S.; Rassoolzadeh, H.; Hedstrom, E.; Coucoravas, C.; Julner, A.; Goldstein, M.; Imreh, G.; Zhivotovsky, B.; Kastan, M.B.; Helleday, T.; et al. The scaffold protein WRAP53beta orchestrates the ubiquitin response critical for DNA double-strand break repair. Genes Dev. 2014, 28, 2726–2738. [Google Scholar] [CrossRef] [PubMed]
- Wang, E.T.; Sandberg, R.; Luo, S.; Khrebtukova, I.; Zhang, L.; Mayr, C.; Kingsmore, S.F.; Schroth, G.P.; Burge, C.B. Alternative isoform regulation in human tissue transcriptomes. Nature 2008, 456, 470–476. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Wang, X.; Arai, S.; Kurokawa, R. Promoter-associated noncoding RNA from the CCND1 promoter. Methods Mol. Biol. 2012, 809, 609–622. [Google Scholar] [CrossRef] [PubMed]
- Ji, P.; Diederichs, S.; Wang, W.; Boing, S.; Metzger, R.; Schneider, P.M.; Tidow, N.; Brandt, B.; Buerger, H.; Bulk, E.; et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 2003, 22, 8031–8041. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Sun, T.; Ji, L.; Deng, W.; Roth, J.; Minna, J.; Arlinghaus, R. Oncogenic activation of c-Abl in non-small cell lung cancer cells lacking FUS1 expression: Inhibition of c-Abl by the tumor suppressor gene product Fus1. Oncogene 2007, 26, 6989–6996. [Google Scholar] [CrossRef]
- Gutschner, T.; Hammerle, M.; Diederichs, S. MALAT1—A paradigm for long noncoding RNA function in cancer. J. Mol. Med. 2013, 91, 791–801. [Google Scholar] [CrossRef]
- Dianatpour, A.; Ghafouri-Fard, S. Long Non Coding RNA Expression Intersecting Cancer and Spermatogenesis: A Systematic Review. Asian Pac. J. Cancer Prev. 2017, 18, 2601–2610. [Google Scholar] [CrossRef]
- Hu, Z.; Mi, S.; Zhao, T.; Peng, C.; Peng, Y.; Chen, L.; Zhu, W.; Yao, Y.; Song, Q.; Li, X.; et al. BGL3 lncRNA mediates retention of the BRCA1/BARD1 complex at DNA damage sites. EMBO J. 2020, 39, e104133. [Google Scholar] [CrossRef] [PubMed]
- Porro, A.; Feuerhahn, S.; Lingner, J. TERRA-reinforced association of LSD1 with MRE11 promotes processing of uncapped telomeres. Cell Rep. 2014, 6, 765–776. [Google Scholar] [CrossRef] [PubMed]
- Cajigas, I.; Leib, D.E.; Cochrane, J.; Luo, H.; Swyter, K.R.; Chen, S.; Clark, B.S.; Thompson, J.; Yates, J.R., 3rd; Kingston, R.E.; et al. Evf2 lncRNA/BRG1/DLX1 interactions reveal RNA-dependent inhibition of chromatin remodeling. Development 2015, 142, 2641–2652. [Google Scholar] [CrossRef] [PubMed]
- Shen, F.; Luo, Z.; Liu, H.; Wang, R.; Zhang, S.; Gan, J.; Sheng, J. Structural insights into RNA duplexes with multiple 2-5-linkages. Nucleic Acids Res. 2017, 45, 3537–3546. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y.; Hsieh, C.H.; Alonso, L.C. ANRIL: A lncRNA at the CDKN2A/B Locus with Roles in Cancer and Metabolic Disease. Front. Endocrinol. 2018, 9, 405. [Google Scholar] [CrossRef]
- Lukas, J.; Altmeyer, M. A lncRNA to repair DNA. EMBO Rep. 2015, 16, 1413–1414. [Google Scholar] [CrossRef]
- Wan, G.; Hu, X.; Liu, Y.; Han, C.; Sood, A.K.; Calin, G.A.; Zhang, X.; Lu, X. A novel non-coding RNA lncRNA-JADE connects DNA damage signalling to histone H4 acetylation. EMBO J. 2013, 32, 2833–2847. [Google Scholar] [CrossRef]
- Munschauer, M.; Nguyen, C.T.; Sirokman, K.; Hartigan, C.R.; Hogstrom, L.; Engreitz, J.M.; Ulirsch, J.C.; Fulco, C.P.; Subramanian, V.; Chen, J.; et al. The NORAD lncRNA assembles a topoisomerase complex critical for genome stability. Nature 2018, 561, 132–136. [Google Scholar] [CrossRef]
- Marin-Bejar, O.; Mas, A.M.; Gonzalez, J.; Martinez, D.; Athie, A.; Morales, X.; Galduroz, M.; Raimondi, I.; Grossi, E.; Guo, S.; et al. The human lncRNA LINC-PINT inhibits tumor cell invasion through a highly conserved sequence element. Genome Biol. 2017, 18, 202. [Google Scholar] [CrossRef]
- Marin-Bejar, O.; Marchese, F.P.; Athie, A.; Sanchez, Y.; Gonzalez, J.; Segura, V.; Huang, L.; Moreno, I.; Navarro, A.; Monzo, M.; et al. Pint lincRNA connects the p53 pathway with epigenetic silencing by the Polycomb repressive complex 2. Genome Biol. 2013, 14, R104. [Google Scholar] [CrossRef]
- Khalil, A.M.; Guttman, M.; Huarte, M.; Garber, M.; Raj, A.; Rivea Morales, D.; Thomas, K.; Presser, A.; Bernstein, B.E.; van Oudenaarden, A.; et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. USA 2009, 106, 11667–11672. [Google Scholar] [CrossRef]
- Yang, L.; Lin, C.; Liu, W.; Zhang, J.; Ohgi, K.A.; Grinstein, J.D.; Dorrestein, P.C.; Rosenfeld, M.G. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell 2011, 147, 773–788. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Liu, Y.; Zhuang, H.; Yang, B.; Hei, K.; Xiao, M.; Hou, C.; Gao, H.; Zhang, X.; Jia, C.; et al. Quantitative proteomics reveals that long non-coding RNA MALAT1 interacts with DBC1 to regulate p53 acetylation. Nucleic Acids Res. 2017, 45, 9947–9959. [Google Scholar] [CrossRef] [PubMed]
- Hung, T.; Wang, Y.; Lin, M.F.; Koegel, A.K.; Kotake, Y.; Grant, G.D.; Horlings, H.M.; Shah, N.; Umbricht, C.; Wang, P.; et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat. Genet. 2011, 43, 621–629. [Google Scholar] [CrossRef] [PubMed]
- Huarte, M.; Guttman, M.; Feldser, D.; Garber, M.; Koziol, M.J.; Kenzelmann-Broz, D.; Khalil, A.M.; Zuk, O.; Amit, I.; Rabani, M.; et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 2010, 142, 409–419. [Google Scholar] [CrossRef] [PubMed]
- Mahmoudi, S.; Henriksson, S.; Corcoran, M.; Mendez-Vidal, C.; Wiman, K.G.; Farnebo, M. Wrap53, a Natural p53 Antisense Transcript Required for p53 Induction upon DNA Damage. Mol. Cell 2016, 64, 1009. [Google Scholar] [CrossRef]
- Schmitt, A.M.; Garcia, J.T.; Hung, T.; Flynn, R.A.; Shen, Y.; Qu, K.; Payumo, A.Y.; Peres-da-Silva, A.; Broz, D.K.; Baum, R.; et al. An inducible long noncoding RNA amplifies DNA damage signaling. Nat. Genet. 2016, 48, 1370–1376. [Google Scholar] [CrossRef]
- Prensner, J.R.; Iyer, M.K.; Balbin, O.A.; Dhanasekaran, S.M.; Cao, Q.; Brenner, J.C.; Laxman, B.; Asangani, I.A.; Grasso, C.S.; Kominsky, H.D.; et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat. Biotechnol. 2011, 29, 742–749. [Google Scholar] [CrossRef]
- Prensner, J.R.; Chen, W.; Iyer, M.K.; Cao, Q.; Ma, T.; Han, S.; Sahu, A.; Malik, R.; Wilder-Romans, K.; Navone, N.; et al. PCAT-1, a long noncoding RNA, regulates BRCA2 and controls homologous recombination in cancer. Cancer Res. 2014, 74, 1651–1660. [Google Scholar] [CrossRef]
- Gazy, I.; Zeevi, D.A.; Renbaum, P.; Zeligson, S.; Eini, L.; Bashari, D.; Smith, Y.; Lahad, A.; Goldberg, M.; Ginsberg, D.; et al. TODRA, a lncRNA at the RAD51 Locus, Is Oppositely Regulated to RAD51, and Enhances RAD51-Dependent DSB (Double Strand Break) Repair. PLoS ONE 2015, 10, e0134120. [Google Scholar] [CrossRef]
- Liu, R.; Zhang, Q.; Shen, L.; Chen, S.; He, J.; Wang, D.; Wang, Q.; Qi, Z.; Zhou, M.; Wang, Z. Long noncoding RNA lnc-RI regulates DNA damage repair and radiation sensitivity of CRC cells through NHEJ pathway. Cell Biol. Toxicol. 2020, 36, 493–507. [Google Scholar] [CrossRef]
- Ozden, O.; Bishehsari, F.; Bauer, J.; Park, S.H.; Jana, A.; Baik, S.H.; Sporn, J.C.; Staudacher, J.J.; Yazici, C.; Krett, N.; et al. Expression of an Oncogenic BARD1 Splice Variant Impairs Homologous Recombination and Predicts Response to PARP-1 Inhibitor Therapy in Colon Cancer. Sci. Rep. 2016, 6, 26273. [Google Scholar] [CrossRef] [PubMed]
- Pilyugin, M.; Irminger-Finger, I. Long non-coding RNA and microRNAs might act in regulating the expression of BARD1 mRNAs. Int. J. BioChem. Cell Biol. 2014, 54, 356–367. [Google Scholar] [CrossRef] [PubMed]
- Betts, J.A.; Moradi Marjaneh, M.; Al-Ejeh, F.; Lim, Y.C.; Shi, W.; Sivakumaran, H.; Tropee, R.; Patch, A.M.; Clark, M.B.; Bartonicek, N.; et al. Long Noncoding RNAs CUPID1 and CUPID2 Mediate Breast Cancer Risk at 11q13 by Modulating the Response to DNA Damage. Am. J. Hum. Genet. 2017, 101, 255–266. [Google Scholar] [CrossRef] [PubMed]
- Kwon, S.J.; Park, J.H.; Park, E.J.; Lee, S.A.; Lee, H.S.; Kang, S.W.; Kwon, J. ATM-mediated phosphorylation of the chromatin remodeling enzyme BRG1 modulates DNA double-strand break repair. Oncogene 2015, 34, 303–313. [Google Scholar] [CrossRef]
- Pham, T.P.; Bink, D.I.; Stanicek, L.; van Bergen, A.; van Leeuwen, E.; Tran, Y.; Matic, L.; Hedin, U.; Wittig, I.; Dimmeler, S.; et al. Long Non-coding RNA Aerrie Controls DNA Damage Repair via YBX1 to Maintain Endothelial Cell Function. Front. Cell Dev. Biol. 2020, 8, 619079. [Google Scholar] [CrossRef] [PubMed]
- Pateras, I.S.; Apostolopoulou, K.; Niforou, K.; Kotsinas, A.; Gorgoulis, V.G. p57KIP2: “Kip”ing the cell under control. Mol. Cancer Res. 2009, 7, 1902–1919. [Google Scholar] [CrossRef]
- Pateras, I.S.; Apostolopoulou, K.; Koutsami, M.; Evangelou, K.; Tsantoulis, P.; Liloglou, T.; Nikolaidis, G.; Sigala, F.; Kittas, C.; Field, J.K.; et al. Downregulation of the KIP family members p27(KIP1) and p57(KIP2) by SKP2 and the role of methylation in p57(KIP2) inactivation in nonsmall cell lung cancer. Int. J. Cancer 2006, 119, 2546–2556. [Google Scholar] [CrossRef]
- Flynn, R.A.; Chang, H.Y. Long noncoding RNAs in cell-fate programming and reprogramming. Cell Stem Cell 2014, 14, 752–761. [Google Scholar] [CrossRef]
- Liu, S.J.; Malatesta, M.; Lien, B.V.; Saha, P.; Thombare, S.S.; Hong, S.J.; Pedraza, L.; Koontz, M.; Seo, K.; Horlbeck, M.A.; et al. CRISPRi-based radiation modifier screen identifies long non-coding RNA therapeutic targets in glioma. Genome Biol. 2020, 21, 83. [Google Scholar] [CrossRef]
- Papaspyropoulos, A.; Tsolaki, M.; Foroglou, N.; Pantazaki, A.A. Modeling and Targeting Alzheimer’s Disease with Organoids. Front. Pharmacol. 2020, 11, 396. [Google Scholar] [CrossRef]
- Sachs, N.; Papaspyropoulos, A.; Zomer-van Ommen, D.D.; Heo, I.; Bottinger, L.; Klay, D.; Weeber, F.; Huelsz-Prince, G.; Iakobachvili, N.; Amatngalim, G.D.; et al. Long-term expanding human airway organoids for disease modeling. EMBO J. 2019, 38. [Google Scholar] [CrossRef] [PubMed]
- Wiener, D.J.; Basak, O.; Asra, P.; Boonekamp, K.E.; Kretzschmar, K.; Papaspyropoulos, A.; Clevers, H. Establishment and characterization of a canine keratinocyte organoid culture system. Vet. Dermatol. 2018, 29, 375-e126. [Google Scholar] [CrossRef] [PubMed]
- Arun, G.; Diermeier, S.D.; Spector, D.L. Therapeutic Targeting of Long Non-Coding RNAs in Cancer. Trends Mol. Med. 2018, 24, 257–277. [Google Scholar] [CrossRef] [PubMed]
- Robertson, M.P.; Joyce, G.F. The origins of the RNA world. Cold Spring Harb. Perspect. Biol. 2012, 4. [Google Scholar] [CrossRef]
- Galanos, P.; Vougas, K.; Walter, D.; Polyzos, A.; Maya-Mendoza, A.; Haagensen, E.J.; Kokkalis, A.; Roumelioti, F.M.; Gagos, S.; Tzetis, M.; et al. Chronic p53-independent p21 expression causes genomic instability by deregulating replication licensing. Nat. Cell Biol. 2016, 18, 777–789. [Google Scholar] [CrossRef] [PubMed]
- Jackson, S.P.; Bartek, J. The DNA-damage response in human biology and disease. Nature 2009, 461, 1071–1078. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Papaspyropoulos, A.; Lagopati, N.; Mourkioti, I.; Angelopoulou, A.; Kyriazis, S.; Liontos, M.; Gorgoulis, V.; Kotsinas, A. Regulatory and Functional Involvement of Long Non-Coding RNAs in DNA Double-Strand Break Repair Mechanisms. Cells 2021, 10, 1506. https://doi.org/10.3390/cells10061506
Papaspyropoulos A, Lagopati N, Mourkioti I, Angelopoulou A, Kyriazis S, Liontos M, Gorgoulis V, Kotsinas A. Regulatory and Functional Involvement of Long Non-Coding RNAs in DNA Double-Strand Break Repair Mechanisms. Cells. 2021; 10(6):1506. https://doi.org/10.3390/cells10061506
Chicago/Turabian StylePapaspyropoulos, Angelos, Nefeli Lagopati, Ioanna Mourkioti, Andriani Angelopoulou, Spyridon Kyriazis, Michalis Liontos, Vassilis Gorgoulis, and Athanassios Kotsinas. 2021. "Regulatory and Functional Involvement of Long Non-Coding RNAs in DNA Double-Strand Break Repair Mechanisms" Cells 10, no. 6: 1506. https://doi.org/10.3390/cells10061506
APA StylePapaspyropoulos, A., Lagopati, N., Mourkioti, I., Angelopoulou, A., Kyriazis, S., Liontos, M., Gorgoulis, V., & Kotsinas, A. (2021). Regulatory and Functional Involvement of Long Non-Coding RNAs in DNA Double-Strand Break Repair Mechanisms. Cells, 10(6), 1506. https://doi.org/10.3390/cells10061506






