Ubiquitin-Conjugating Enzymes in Cancer
Abstract
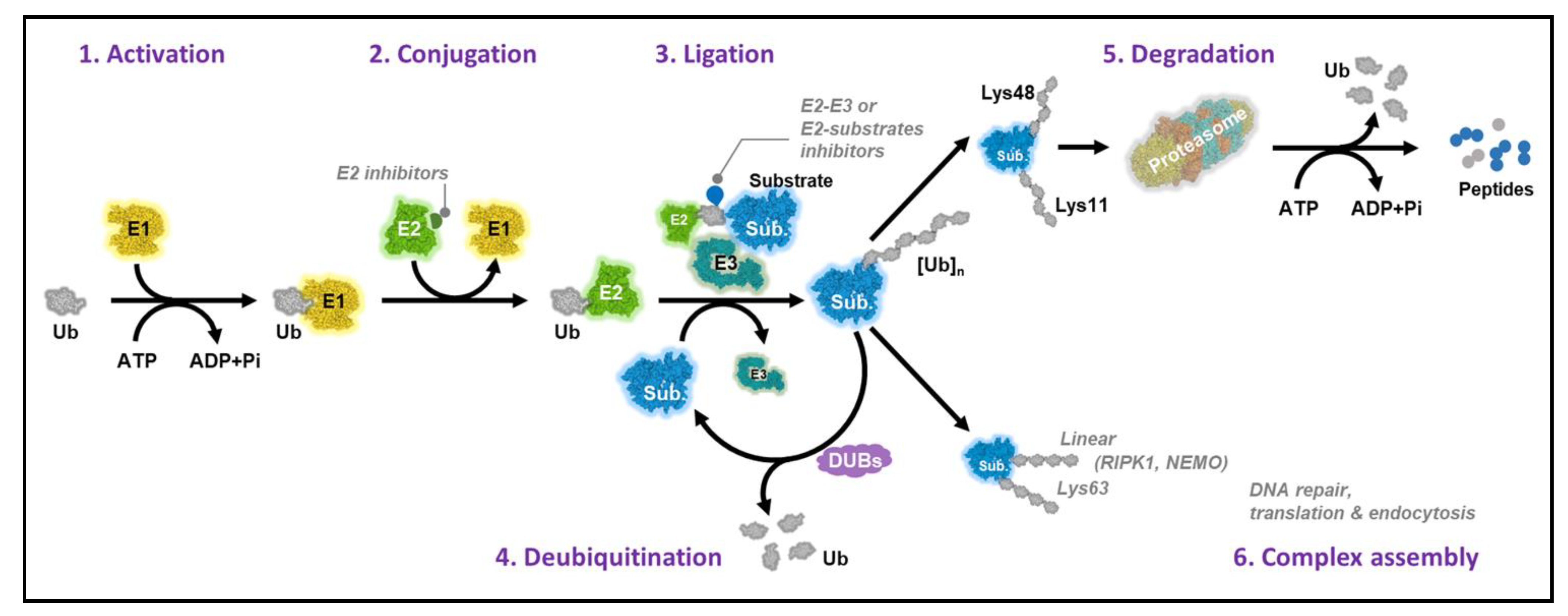
1. Introduction
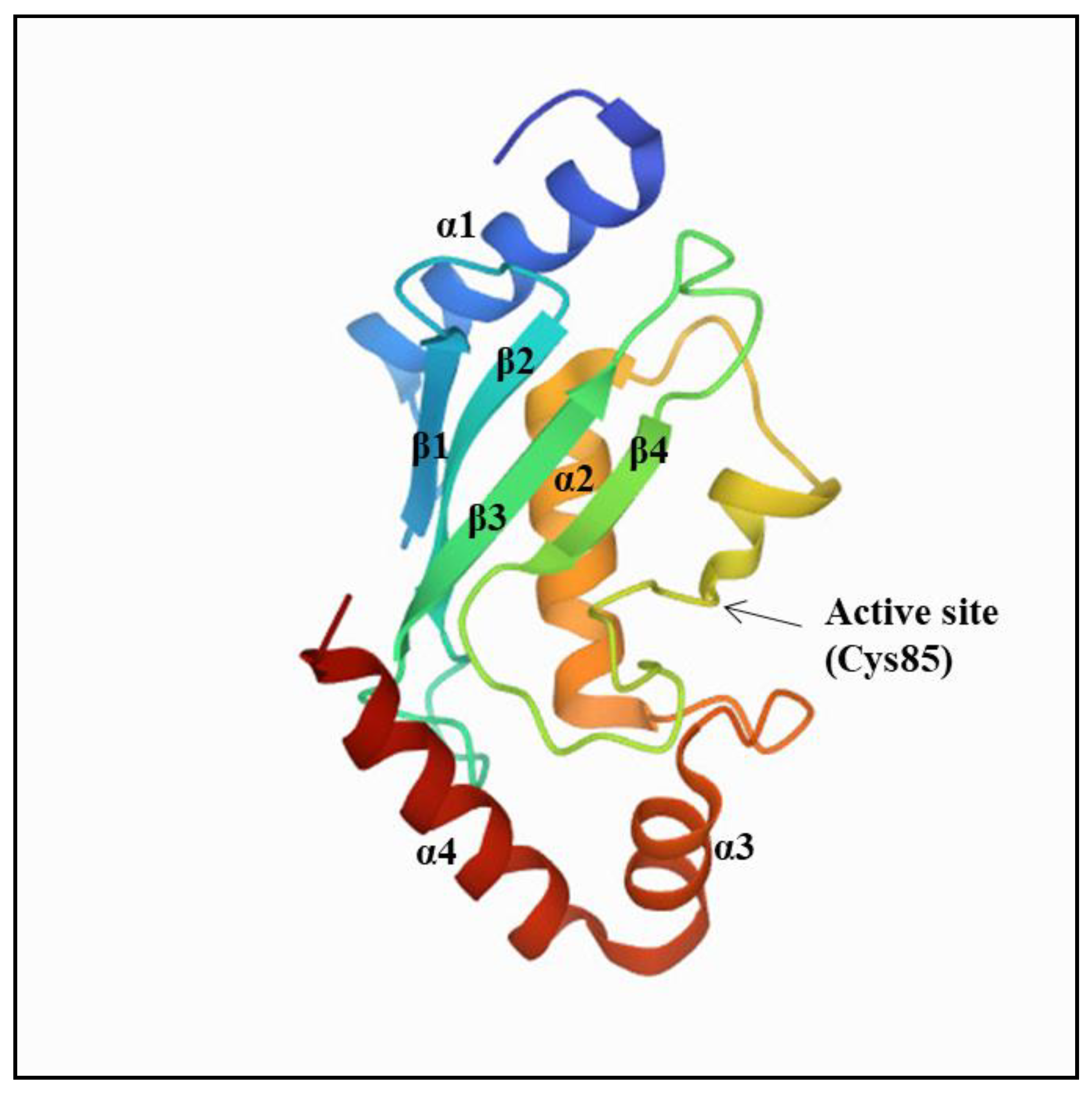
2. Ubiquitin-Conjugating Enzymes
2.1. UBE2A (RAD6A) and UBE2B (RAD6B)
2.2. UBE2C (UbcH10)
2.3. UBE2D
2.4. UBE2F
2.5. UBE2I (UBC9)
2.6. UBE2J2
2.7. UBE2L3 (UBCH7)
2.8. UBE2N
2.9. UBE2O
2.10. UBE2R1 (CDC34)
2.11. UBE2S
2.12. UBE2T
2.13. UBE2Z (USE1)
3. Conclusions and Perspective
4. Targeting Specific E2s with Small-Molecule Inhibitors
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tan, C.S.; Gilligan, D.; Pacey, S. Treatment approaches for EGFR-inhibitor-resistant patients with non-small-cell lung cancer. Lancet Oncol. 2015, 16, e447–e459. [Google Scholar] [CrossRef]
- Minguet, J.; Smith, K.H.; Bramlage, P. Targeted therapies for treatment of non-small cell lung cancer—Recent advances and future perspectives. Int. J. Cancer 2016, 138, 2549–2561. [Google Scholar] [CrossRef] [PubMed]
- Lynch, T.J.; Wright, C.D.; Choi, N.C.; Aquino, S.L.; Mark, E.J. Case records of the Massachusetts General Hospital. Weekly clinicopathological exercises. Case 26-2004. A 56-year-old woman with cough and a lung nodule. N. Engl. J. Med. 2004, 351, 809–817. [Google Scholar] [CrossRef] [PubMed]
- Soda, M.; Choi, Y.L.; Enomoto, M.; Takada, S.; Yamashita, Y.; Ishikawa, S.; Fujiwara, S.; Watanabe, H.; Kurashina, K.; Hatanaka, H.; et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 2007, 448, 561–566. [Google Scholar] [CrossRef] [PubMed]
- Meador, C.B.; Micheel, C.M.; Levy, M.A.; Lovly, C.M.; Horn, L.; Warner, J.L.; Johnson, D.B.; Zhao, Z.; Anderson, I.A.; Sosman, J.A.; et al. Beyond histology: Translating tumor genotypes into clinically effective targeted therapies. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2014, 20, 2264–2275. [Google Scholar] [CrossRef] [PubMed]
- Chondrogianni, N.; Petropoulos, I.; Grimm, S.; Georgila, K.; Catalgol, B.; Friguet, B.; Grune, T.; Gonos, E.S. Protein damage, repair and proteolysis. Mol. Asp. Med. 2014, 35, 1–71. [Google Scholar] [CrossRef]
- MacGurn, J.A.; Hsu, P.C.; Emr, S.D. Ubiquitin and membrane protein turnover: From cradle to grave. Annu. Rev. Biochem. 2012, 81, 231–259. [Google Scholar] [CrossRef] [PubMed]
- Ciechanover, A. Intracellular protein degradation: From a vague idea through the lysosome and the ubiquitin-proteasome system and onto human diseases and drug targeting. Bioorg. Med. Chem. 2013, 21, 3400–3410. [Google Scholar] [CrossRef]
- Grabbe, C.; Husnjak, K.; Dikic, I. The spatial and temporal organization of ubiquitin networks. Nat. Rev. Mol. Cell Biol. 2011, 12, 295–307. [Google Scholar] [CrossRef] [PubMed]
- Stewart, M.D.; Ritterhoff, T.; Klevit, R.E.; Brzovic, P.S. E2 enzymes: More than just middle men. Cell Res. 2016, 26, 423–440. [Google Scholar] [CrossRef]
- Groettrup, M.; Pelzer, C.; Schmidtke, G.; Hofmann, K. Activating the ubiquitin family: UBA6 challenges the field. Trends Biochem. Sci. 2008, 33, 230–237. [Google Scholar] [CrossRef] [PubMed]
- Hormaechea-Agulla, D.; Kim, Y.; Song, M.S.; Song, S.J. New Insights into the Role of E2s in the Pathogenesis of Diseases: Lessons Learned from UBE2O. Mol. Cells 2018, 41, 168–178. [Google Scholar] [CrossRef] [PubMed]
- Deshaies, R.J.; Joazeiro, C.A. RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 2009, 78, 399–434. [Google Scholar] [CrossRef]
- Ye, Y.; Rape, M. Building ubiquitin chains: E2 enzymes at work. Nat. Rev. Mol. Cell Biol. 2009, 10, 755–764. [Google Scholar] [CrossRef]
- Streich, F.C., Jr.; Lima, C.D. Structural and functional insights to ubiquitin-like protein conjugation. Annu. Rev. Biophys. 2014, 43, 357–379. [Google Scholar] [CrossRef] [PubMed]
- Hammond-Martel, I.; Yu, H.; Affar el, B. Roles of ubiquitin signaling in transcription regulation. Cell. Signal. 2012, 24, 410–421. [Google Scholar] [CrossRef]
- Hofmann, K. Ubiquitin-binding domains and their role in the DNA damage response. DNA Repair 2009, 8, 544–556. [Google Scholar] [CrossRef] [PubMed]
- Ravid, T.; Hochstrasser, M. Diversity of degradation signals in the ubiquitin-proteasome system. Nat. Rev. Mol. Cell Biol. 2008, 9, 679–690. [Google Scholar] [CrossRef]
- Dikic, I.; Wakatsuki, S.; Walters, K.J. Ubiquitin-binding domains—From structures to functions. Nat. Rev. Mol. Cell Biol. 2009, 10, 659–671. [Google Scholar] [CrossRef]
- van Wijk, S.J.; Timmers, H.T. The family of ubiquitin-conjugating enzymes (E2s): Deciding between life and death of proteins. FASEB J. 2010, 24, 981–993. [Google Scholar] [CrossRef]
- Huang, D.T.; Hunt, H.W.; Zhuang, M.; Ohi, M.D.; Holton, J.M.; Schulman, B.A. Basis for a ubiquitin-like protein thioester switch toggling E1-E2 affinity. Nature 2007, 445, 394–398. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.; Schindelin, H. Structural insights into E1-catalyzed ubiquitin activation and transfer to conjugating enzymes. Cell 2008, 134, 268–278. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Li, X.; Gygi, S.P.; Harper, J.W. Dual E1 activation systems for ubiquitin differentially regulate E2 enzyme charging. Nature 2007, 447, 1135–1138. [Google Scholar] [CrossRef]
- Somasagara, R.R.; Tripathi, K.; Spencer, S.M.; Clark, D.W.; Barnett, R.; Bachaboina, L.; Scalici, J.; Rocconi, R.P.; Piazza, G.A.; Palle, K. Rad6 upregulation promotes stem cell-like characteristics and platinum resistance in ovarian cancer. Biochem. Biophys. Res. Commun. 2016, 469, 449–455. [Google Scholar] [CrossRef]
- Lyakhovich, A.; Shekhar, M.P. RAD6B overexpression confers chemoresistance: RAD6 expression during cell cycle and its redistribution to chromatin during DNA damage-induced response. Oncogene 2004, 23, 3097–3106. [Google Scholar] [CrossRef][Green Version]
- Shekhar, M.P.; Lyakhovich, A.; Visscher, D.W.; Heng, H.; Kondrat, N. Rad6 overexpression induces multinucleation, centrosome amplification, abnormal mitosis, aneuploidy, and transformation. Cancer Res. 2002, 62, 2115–2124. [Google Scholar] [PubMed]
- Lyakhovich, A.; Shekhar, M.P. Supramolecular complex formation between Rad6 and proteins of the p53 pathway during DNA damage-induced response. Mol. Cell. Biol. 2003, 23, 2463–2475. [Google Scholar] [CrossRef]
- Rosner, K.; Mehregan, D.R.; Kirou, E.; Abrams, J.; Kim, S.; Campbell, M.; Frieder, J.; Lawrence, K.; Haynes, B.; Shekhar, M.P. Melanoma Development and Progression Are Associated with Rad6 Upregulation and β -Catenin Relocation to the Cell Membrane. J. Ski. Cancer 2014, 2014, 439205. [Google Scholar] [CrossRef]
- Qin, T.; Huang, G.; Chi, L.; Sui, S.; Song, C.; Li, N.; Sun, S.; Li, N.; Zhang, M.; Zhao, Z.; et al. Exceptionally high UBE2C expression is a unique phenomenon in basal-like type breast cancer and is regulated by BRCA1. Biomed. Pharmacother. 2017, 95, 649–655. [Google Scholar] [CrossRef]
- Dastsooz, H.; Cereda, M.; Donna, D.; Oliviero, S. A Comprehensive Bioinformatics Analysis of UBE2C in Cancers. Int. J. Mol. Sci. 2019, 20, 2228. [Google Scholar] [CrossRef]
- Psyrri, A.; Kalogeras, K.T.; Kronenwett, R.; Wirtz, R.M.; Batistatou, A.; Bournakis, E.; Timotheadou, E.; Gogas, H.; Aravantinos, G.; Christodoulou, C.; et al. Prognostic significance of UBE2C mRNA expression in high-risk early breast cancer. A Hellenic Cooperative Oncology Group (HeCOG) Study. Ann. Oncol. 2012, 23, 1422–1427. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, A.; Ishibashi, Y.; Urashima, M.; Omura, N.; Nakada, K.; Nishikawa, K.; Shida, A.; Takada, K.; Kashiwagi, H.; Yanaga, K. High UBCH10 protein expression as a marker of poor prognosis in esophageal squamous cell carcinoma. Anticancer Res. 2014, 34, 955–961. [Google Scholar] [PubMed]
- Han, S.S.; Liu, Q.G.; Yao, Y.M.; Sun, H.; Zan, X.F.; Song, T.; Yang, X.; Zheng, X. [UbcH10 expression in hepatocellular carcinoma and its clinicopathological significance]. Nan Fang Yi Ke Da Xue Xue Bao J. South Med. Univ. 2011, 31, 280–284. [Google Scholar]
- Kadara, H.; Lacroix, L.; Behrens, C.; Solis, L.; Gu, X.; Lee, J.J.; Tahara, E.; Lotan, D.; Hong, W.K.; Wistuba, I.I.; et al. Identification of gene signatures and molecular markers for human lung cancer prognosis using an in vitro lung carcinogenesis system. Cancer Prev. Res. 2009, 2, 702–711. [Google Scholar] [CrossRef]
- Pallante, P.; Berlingieri, M.T.; Troncone, G.; Kruhoffer, M.; Orntoft, T.F.; Viglietto, G.; Caleo, A.; Migliaccio, I.; Decaussin-Petrucci, M.; Santoro, M.; et al. UbcH10 overexpression may represent a marker of anaplastic thyroid carcinomas. Br. J. Cancer 2005, 93, 464–471. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.K.; Wu, W.G.; Chen, L.; Dong, P.; Gu, J.; Mu, J.S.; Yang, J.H.; Liu, Y.B. Expression of UbcH10 in pancreatic ductal adenocarcinoma and its correlation with prognosis. Tumour Biol. J. Int. Soc. Oncodev. Biol. Med. 2013, 34, 1473–1477. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.; Jiang, X.; Zeng, C.; Zheng, S.; Luo, B.; Zeng, Y.; Ding, R.; Jiang, H.; He, Q.; Guo, J.; et al. High expression of ubiquitin-conjugating enzyme 2C (UBE2C) correlates with nasopharyngeal carcinoma progression. BMC Cancer 2013, 13, 192. [Google Scholar] [CrossRef]
- Hou, L.; Li, Y.; Wang, Y.; Xu, D.; Cui, H.; Xu, X.; Cong, Y.; Yu, C. UBE2D1 RNA Expression Was an Independent Unfavorable Prognostic Indicator in Lung Adenocarcinoma, but Not in Lung Squamous Cell Carcinoma. Dis. Markers 2018, 2018, 4108919. [Google Scholar] [CrossRef]
- Zhou, C.; Bi, F.; Yuan, J.; Yang, F.; Sun, S. Gain of UBE2D1 facilitates hepatocellular carcinoma progression and is associated with DNA damage caused by continuous IL-6. J. Exp. Clin. Cancer Res. 2018, 37, 290. [Google Scholar] [CrossRef]
- Shukla, S.; Allam, U.S.; Ahsan, A.; Chen, G.; Krishnamurthy, P.M.; Marsh, K.; Rumschlag, M.; Shankar, S.; Whitehead, C.; Schipper, M.; et al. KRAS protein stability is regulated through SMURF2: UBCH5 complex-mediated β-TrCP1 degradation. Neoplasia 2014, 16, 115–128. [Google Scholar] [CrossRef]
- Zhou, W.; Xu, J.; Li, H.; Xu, M.; Chen, Z.J.; Wei, W.; Pan, Z.; Sun, Y. Neddylation E2 UBE2F Promotes the Survival of Lung Cancer Cells by Activating CRL5 to Degrade NOXA via the K11 Linkage. Clin. Cancer Res. 2017, 23, 1104–1116. [Google Scholar] [CrossRef]
- Zhou, L.; Zhu, J.; Chen, W.; Jiang, Y.; Hu, T.; Wang, Y.; Ye, X.; Zhan, M.; Ji, C.; Xu, Z.; et al. Induction of NEDD8-conjugating enzyme E2 UBE2F by platinum protects lung cancer cells from apoptosis and confers to platinum-insensitivity. Cell Death Dis. 2020, 11, 975. [Google Scholar] [CrossRef]
- Lin, D.; Tatham, M.H.; Yu, B.; Kim, S.; Hay, R.T.; Chen, Y. Identification of a substrate recognition site on Ubc9. J. Biol. Chem. 2002, 277, 21740–21748. [Google Scholar] [CrossRef] [PubMed]
- Mo, Y.Y.; Yu, Y.; Theodosiou, E.; Ee, P.L.; Beck, W.T. A role for Ubc9 in tumorigenesis. Oncogene 2005, 24, 2677–2683. [Google Scholar] [CrossRef]
- Li, H.; Niu, H.; Peng, Y.; Wang, J.; He, P. Ubc9 promotes invasion and metastasis of lung cancer cells. Oncol. Rep. 2013, 29, 1588–1594. [Google Scholar] [CrossRef]
- Chen, S.; Tan, Y.; Deng, H.; Shen, Z.; Liu, Y.; Wu, P.; Tan, C.; Jiang, Y. UBE2J2 promotes hepatocellular carcinoma cell epithelial-mesenchymal transition and invasion in vitro. Oncotarget 2017, 8, 71736–71749. [Google Scholar] [CrossRef]
- Wang, X.; Herr, R.A.; Rabelink, M.; Hoeben, R.C.; Wiertz, E.J.; Hansen, T.H. Ube2j2 ubiquitinates hydroxylated amino acids on ER-associated degradation substrates. J. Cell Biol. 2009, 187, 655–668. [Google Scholar] [CrossRef]
- Ma, X.; Zhao, J.; Yang, F.; Liu, H.; Qi, W. Ubiquitin conjugating enzyme E2 L3 promoted tumor growth of NSCLC through accelerating p27kip1 ubiquitination and degradation. Oncotarget 2017, 8, 84193–84203. [Google Scholar] [CrossRef]
- Pulvino, M.; Liang, Y.; Oleksyn, D.; DeRan, M.; Van Pelt, E.; Shapiro, J.; Sanz, I.; Chen, L.; Zhao, J. Inhibition of proliferation and survival of diffuse large B-cell lymphoma cells by a small-molecule inhibitor of the ubiquitin-conjugating enzyme Ubc13-Uev1A. Blood 2012, 120, 1668–1677. [Google Scholar] [CrossRef]
- Cheng, J.; Fan, Y.H.; Xu, X.; Zhang, H.; Dou, J.; Tang, Y.; Zhong, X.; Rojas, Y.; Yu, Y.; Zhao, Y.; et al. A small-molecule inhibitor of UBE2N induces neuroblastoma cell death via activation of p53 and JNK pathways. Cell Death Dis. 2014, 5, e1079. [Google Scholar] [CrossRef]
- Gombodorj, N.; Yokobori, T.; Yoshiyama, S.; Kawabata-Iwakawa, R.; Rokudai, S.; Horikoshi, I.; Nishiyama, M.; Nakano, T. Inhibition of Ubiquitin-conjugating Enzyme E2 May Activate the Degradation of Hypoxia-inducible Factors and, thus, Overcome Cellular Resistance to Radiation in Colorectal Cancer. Anticancer Res. 2017, 37, 2425–2436. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Zhang, W.; Font-Burgada, J.; Palmer, T.; Hamil, A.S.; Biswas, S.K.; Poidinger, M.; Borcherding, N.; Xie, Q.; Ellies, L.G.; et al. Ubiquitin-conjugating enzyme Ubc13 controls breast cancer metastasis through a TAK1-p38 MAP kinase cascade. Proc. Natl. Acad. Sci. USA 2014, 111, 13870–13875. [Google Scholar] [CrossRef]
- Dikshit, A.; Jin, Y.J.; Degan, S.; Hwang, J.; Foster, M.W.; Li, C.Y.; Zhang, J.Y. UBE2N Promotes Melanoma Growth via MEK/FRA1/SOX10 Signaling. Cancer Res. 2018, 78, 6462–6472. [Google Scholar] [CrossRef] [PubMed]
- Song, T.T.; Xu, F.; Wang, W. Inhibiting ubiquitin conjugating enzyme E2 N by microRNA-590-3p reduced cell growth of cervical carcinoma. Kaohsiung J. Med. Sci. 2020, 36, 501–507. [Google Scholar] [CrossRef]
- Vila, I.K.; Yao, Y.; Kim, G.; Xia, W.; Kim, H.; Kim, S.J.; Park, M.K.; Hwang, J.P.; González-Billalabeitia, E.; Hung, M.C.; et al. A UBE2O-AMPKα2 Axis that Promotes Tumor Initiation and Progression Offers Opportunities for Therapy. Cancer Cell 2017, 31, 208–224. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Ma, F.; Liu, C.; Zhu, K.; Li, W.; Xu, Y.; Li, G.; Niu, Z.; Liu, J.; Chen, D.; et al. UBE2O promotes the proliferation, EMT and stemness properties of breast cancer cells through the UBE2O/AMPKα2/mTORC1-MYC positive feedback loop. Cell Death Dis. 2020, 11, 10. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, S.; Liu, C.; Li, G.; Lu, S.; Wang, Y.; Zhang, X.; Huang, D.; Qiu, Y.; Liu, Y. UBE2O Promotes Progression and Epithelial-Mesenchymal Transition in Head and Neck Squamous Cell Carcinoma. OncoTargets Ther. 2020, 13, 6191–6202. [Google Scholar] [CrossRef]
- Tanaka, K.; Kondoh, N.; Shuda, M.; Matsubara, O.; Imazeki, N.; Ryo, A.; Wakatsuki, T.; Hada, A.; Goseki, N.; Igari, T.; et al. Enhanced expression of mRNAs of antisecretory factor-1, gp96, DAD1 and CDC34 in human hepatocellular carcinomas. Biochim. Biophys. Acta 2001, 1536, 1–12. [Google Scholar] [CrossRef]
- Price, G.R.; Armes, J.E.; Ramus, S.J.; Provenzano, E.; Kumar, B.; Cowie, T.F.; Ciciulla, J.; Hutchins, A.M.; Thomas, M.; Venter, D.J. Phenotype-directed analysis of genotype in early-onset, familial breast cancers. Pathology 2006, 38, 520–527. [Google Scholar] [CrossRef]
- Zhao, X.C.; Wang, G.Z.; Wen, Z.S.; Zhou, Y.C.; Hu, Q.; Zhang, B.; Qu, L.W.; Gao, S.H.; Liu, J.; Ma, L.; et al. Systematic identification of CDC34 that functions to stabilize EGFR and promote lung carcinogenesis. EBioMedicine 2020, 53, 102689. [Google Scholar] [CrossRef]
- Li, Z.; Wang, Y.; Li, Y.; Yin, W.; Mo, L.; Qian, X.; Zhang, Y.; Wang, G.; Bu, F.; Zhang, Z.; et al. Ube2s stabilizes β-Catenin through K11-linked polyubiquitination to promote mesendoderm specification and colorectal cancer development. Cell Death Dis. 2018, 9, 456. [Google Scholar] [CrossRef]
- Ayesha, A.K.; Hyodo, T.; Asano, E.; Sato, N.; Mansour, M.A.; Ito, S.; Hamaguchi, M.; Senga, T. UBE2S is associated with malignant characteristics of breast cancer cells. Tumour Biol. J. Int. Soc. Oncodev. Biol. Med. 2016, 37, 763–772. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.; Lei, T.; Zheng, J.; Chen, S.; Du, L.; Xie, H. UBE2S mediates tumor progression via SOX6/β-Catenin signaling in endometrial cancer. Int. J. Biochem. Cell Biol. 2019, 109, 17–22. [Google Scholar] [CrossRef]
- Liu, Z.; Xu, L. UBE2S promotes the proliferation and survival of human lung adenocarcinoma cells. BMB Rep. 2018, 51, 642–647. [Google Scholar] [CrossRef]
- Pan, Y.H.; Yang, M.; Liu, L.P.; Wu, D.C.; Li, M.Y.; Su, S.G. UBE2S enhances the ubiquitination of p53 and exerts oncogenic activities in hepatocellular carcinoma. Biochem. Biophys. Res. Commun. 2018, 503, 895–902. [Google Scholar] [CrossRef] [PubMed]
- Ueki, T.; Park, J.H.; Nishidate, T.; Kijima, K.; Hirata, K.; Nakamura, Y.; Katagiri, T. Ubiquitination and downregulation of BRCA1 by ubiquitin-conjugating enzyme E2T overexpression in human breast cancer cells. Cancer Res. 2009, 69, 8752–8760. [Google Scholar] [CrossRef]
- Hu, W.; Xiao, L.; Cao, C.; Hua, S.; Wu, D. UBE2T promotes nasopharyngeal carcinoma cell proliferation, invasion, and metastasis by activating the AKT/GSK3β/β-catenin pathway. Oncotarget 2016, 7, 15161–15172. [Google Scholar] [CrossRef] [PubMed]
- Gong, Y.Q.; Peng, D.; Ning, X.H.; Yang, X.Y.; Li, X.S.; Zhou, L.Q.; Guo, Y.L. UBE2T silencing suppresses proliferation and induces cell cycle arrest and apoptosis in bladder cancer cells. Oncol. Lett. 2016, 12, 4485–4492. [Google Scholar] [CrossRef]
- Liu, L.P.; Yang, M.; Peng, Q.Z.; Li, M.Y.; Zhang, Y.S.; Guo, Y.H.; Chen, Y.; Bao, S.Y. UBE2T promotes hepatocellular carcinoma cell growth via ubiquitination of p53. Biochem. Biophys. Res. Commun. 2017, 493, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Luo, C.; Yao, Y.; Yu, Z.; Zhou, H.; Guo, L.; Zhang, J.; Cao, H.; Zhang, G.; Li, Y.; Jiao, Z. UBE2T knockdown inhibits gastric cancer progression. Oncotarget 2017, 8, 32639–32654. [Google Scholar] [CrossRef]
- Lian, J.H.; Wang, W.H.; Wang, J.Q.; Zhang, Y.H.; Li, Y. MicroRNA-122 promotes proliferation, invasion and migration of renal cell carcinoma cells through the PI3K/Akt signaling pathway. Asian Pac. J. Cancer Prev. APJCP 2013, 14, 5017–5021. [Google Scholar] [CrossRef]
- Wu, Z.H.; Zhang, Y.J.; Sun, H.Y. High ubiquitin conjugating enzyme E2 T mRNA expression and its prognostic significance in lung adenocarcinoma: A study based on the TCGA database. Medicine 2020, 99, e18543. [Google Scholar] [CrossRef]
- Kim, S.J.; Hyeong Lee, T.; Hee Nam, S.; Kim, J.H.; Oh, S.; Sook Cho, Y.; Sup Lee, M.; Choi, S.; Lee, P.C. Association of Uba6-Specific-E2 (USE1) with Lung Tumorigenesis. J. Natl. Cancer Inst. 2017, 109, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.H.; Kaustov, L.; Coyaud, E.; Srikumar, T.; Wan, J.; Arrowsmith, C.; Raught, B. KCMF1 (potassium channel modulatory factor 1) Links RAD6 to UBR4 (ubiquitin N-recognin domain-containing E3 ligase 4) and lysosome-mediated degradation. Mol. Cell. Proteom. 2015, 14, 674–685. [Google Scholar] [CrossRef]
- Koken, M.H.; Hoogerbrugge, J.W.; Jasper-Dekker, I.; de Wit, J.; Willemsen, R.; Roest, H.P.; Grootegoed, J.A.; Hoeijmakers, J.H. Expression of the ubiquitin-conjugating DNA repair enzymes HHR6A and B suggests a role in spermatogenesis and chromatin modification. Dev. Biol. 1996, 173, 119–132. [Google Scholar] [CrossRef] [PubMed]
- Notenboom, V.; Hibbert, R.G.; van Rossum-Fikkert, S.E.; Olsen, J.V.; Mann, M.; Sixma, T.K. Functional characterization of Rad18 domains for Rad6, ubiquitin, DNA binding and PCNA modification. Nucleic Acids Res. 2007, 35, 5819–5830. [Google Scholar] [CrossRef] [PubMed]
- Hedglin, M.; Benkovic, S.J. Regulation of Rad6/Rad18 Activity During DNA Damage Tolerance. Annu. Rev. Biophys. 2015, 44, 207–228. [Google Scholar] [CrossRef]
- Lee, P.C.; Sowa, M.E.; Gygi, S.P.; Harper, J.W. Alternative ubiquitin activation/conjugation cascades interact with N-end rule ubiquitin ligases to control degradation of RGS proteins. Mol. Cell 2011, 43, 392–405. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; Yue, W.; Li, L.; Li, S.; Gao, C.; Si, L.; Tian, H. Regulator of G-protein signaling 4: A novel tumor suppressor with prognostic significance in non-small cell lung cancer. Biochem. Biophys. Res. Commun. 2016, 469, 384–391. [Google Scholar] [CrossRef]
- Huang, G.; Song, H.; Wang, R.; Han, X.; Chen, L. The relationship between RGS5 expression and cancer differentiation and metastasis in non-small cell lung cancer. J. Surg. Oncol. 2012, 105, 420–424. [Google Scholar] [CrossRef] [PubMed]
- Park, H.J.; Kim, S.H.; Moon, D.O. Growth inhibition of human breast carcinoma cells by overexpression of regulator of G-protein signaling 4. Oncol. Lett. 2017, 13, 4357–4363. [Google Scholar] [CrossRef]
- van Ree, J.H.; Jeganathan, K.B.; Malureanu, L.; van Deursen, J.M. Overexpression of the E2 ubiquitin-conjugating enzyme UbcH10 causes chromosome missegregation and tumor formation. J. Cell Biol. 2010, 188, 83–100. [Google Scholar] [CrossRef]
- Rawat, A.; Gopal, G.; Selvaluxmy, G.; Rajkumar, T. Inhibition of ubiquitin conjugating enzyme UBE2C reduces proliferation and sensitizes breast cancer cells to radiation, doxorubicin, tamoxifen and letrozole. Cell. Oncol. 2013, 36, 459–467. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Pan, Y.H.; Shan, M.; Xu, M.; Bao, J.L.; Zhao, L.M. Knockdown of UbcH10 enhances the chemosensitivity of dual drug resistant breast cancer cells to epirubicin and docetaxel. Int. J. Mol. Sci. 2015, 16, 4698–4712. [Google Scholar] [CrossRef]
- Cacciola, N.A.; Calabrese, C.; Malapelle, U.; Pellino, G.; De Stefano, A.; Sepe, R.; Sgariglia, R.; Quintavalle, C.; Federico, A.; Bianco, A.; et al. UbcH10 expression can predict prognosis and sensitivity to the antineoplastic treatment for colorectal cancer patients. Mol. Carcinog. 2016, 55, 793–807. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Lu, J.; Fang, Q.; Lu, Y.; Xie, C.; Wu, H.; Yin, Z. UBE2C functions as a potential oncogene by enhancing cell proliferation, migration, invasion, and drug resistance in hepatocellular carcinoma cells. Biosci. Rep. 2019, 39. [Google Scholar] [CrossRef]
- Grou, C.P.; Carvalho, A.F.; Pinto, M.P.; Wiese, S.; Piechura, H.; Meyer, H.E.; Warscheid, B.; Sá-Miranda, C.; Azevedo, J.E. Members of the E2D (UbcH5) family mediate the ubiquitination of the conserved cysteine of Pex5p, the peroxisomal import receptor. J. Biol. Chem. 2008, 283, 14190–14197. [Google Scholar] [CrossRef] [PubMed]
- Brzovic, P.S.; Lissounov, A.; Christensen, D.E.; Hoyt, D.W.; Klevit, R.E. A UbcH5/ubiquitin noncovalent complex is required for processive BRCA1-directed ubiquitination. Mol. Cell 2006, 21, 873–880. [Google Scholar] [CrossRef]
- Ranaweera, R.S.; Yang, X. Auto-ubiquitination of Mdm2 enhances its substrate ubiquitin ligase activity. J. Biol. Chem. 2013, 288, 18939–18946. [Google Scholar] [CrossRef]
- Saville, M.K.; Sparks, A.; Xirodimas, D.P.; Wardrop, J.; Stevenson, L.F.; Bourdon, J.C.; Woods, Y.L.; Lane, D.P. Regulation of p53 by the ubiquitin-conjugating enzymes UbcH5B/C in vivo. J. Biol. Chem. 2004, 279, 42169–42181. [Google Scholar] [CrossRef]
- Tokumoto, M.; Fujiwara, Y.; Shimada, A.; Hasegawa, T.; Seko, Y.; Nagase, H.; Satoh, M. Cadmium toxicity is caused by accumulation of p53 through the down-regulation of Ube2d family genes in vitro and in vivo. J. Toxicol. Sci. 2011, 36, 191–200. [Google Scholar] [CrossRef]
- Sun, X.X.; Challagundla, K.B.; Dai, M.S. Positive regulation of p53 stability and activity by the deubiquitinating enzyme Otubain 1. EMBO J. 2012, 31, 576–592. [Google Scholar] [CrossRef]
- Varfolomeev, E.; Goncharov, T.; Fedorova, A.V.; Dynek, J.N.; Zobel, K.; Deshayes, K.; Fairbrother, W.J.; Vucic, D. c-IAP1 and c-IAP2 are critical mediators of tumor necrosis factor alpha (TNFalpha)-induced NF-kappaB activation. J. Biol. Chem. 2008, 283, 24295–24299. [Google Scholar] [CrossRef]
- David, Y.; Ziv, T.; Admon, A.; Navon, A. The E2 ubiquitin-conjugating enzymes direct polyubiquitination to preferred lysines. J. Biol. Chem. 2010, 285, 8595–8604. [Google Scholar] [CrossRef] [PubMed]
- Komander, D.; Rape, M. The ubiquitin code. Annu. Rev. Biochem. 2012, 81, 203–229. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Shen, S.; Zhang, Z.; Zhang, W.; Xiao, W. Ubiquitin-conjugating enzyme complex Uev1A-Ubc13 promotes breast cancer metastasis through nuclear factor-кB mediated matrix metalloproteinase-1 gene regulation. Breast Cancer Res. 2014, 16, R75. [Google Scholar] [CrossRef]
- Deng, L.; Wang, C.; Spencer, E.; Yang, L.; Braun, A.; You, J.; Slaughter, C.; Pickart, C.; Chen, Z.J. Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell 2000, 103, 351–361. [Google Scholar] [CrossRef]
- Andersen, P.L.; Zhou, H.; Pastushok, L.; Moraes, T.; McKenna, S.; Ziola, B.; Ellison, M.J.; Dixit, V.M.; Xiao, W. Distinct regulation of Ubc13 functions by the two ubiquitin-conjugating enzyme variants Mms2 and Uev1A. J. Cell Biol. 2005, 170, 745–755. [Google Scholar] [CrossRef]
- Mashtalir, N.; Daou, S.; Barbour, H.; Sen, N.N.; Gagnon, J.; Hammond-Martel, I.; Dar, H.H.; Therrien, M.; Affar el, B. Autodeubiquitination protects the tumor suppressor BAP1 from cytoplasmic sequestration mediated by the atypical ubiquitin ligase UBE2O. Mol. Cell 2014, 54, 392–406. [Google Scholar] [CrossRef]
- Goebl, M.G.; Yochem, J.; Jentsch, S.; McGrath, J.P.; Varshavsky, A.; Byers, B. The yeast cell cycle gene CDC34 encodes a ubiquitin-conjugating enzyme. Science 1988, 241, 1331–1335. [Google Scholar] [CrossRef] [PubMed]
- Plon, S.E.; Leppig, K.A.; Do, H.N.; Groudine, M. Cloning of the human homolog of the CDC34 cell cycle gene by complementation in yeast. Proc. Natl. Acad. Sci. USA 1993, 90, 10484–10488. [Google Scholar] [CrossRef] [PubMed]
- Gazdoiu, S.; Yamoah, K.; Wu, K.; Escalante, C.R.; Tappin, I.; Bermudez, V.; Aggarwal, A.K.; Hurwitz, J.; Pan, Z.Q. Proximity-induced activation of human Cdc34 through heterologous dimerization. Proc. Natl. Acad. Sci. USA 2005, 102, 15053–15058. [Google Scholar] [CrossRef]
- Kleiger, G.; Saha, A.; Lewis, S.; Kuhlman, B.; Deshaies, R.J. Rapid E2-E3 assembly and disassembly enable processive ubiquitylation of cullin-RING ubiquitin ligase substrates. Cell 2009, 139, 957–968. [Google Scholar] [CrossRef]
- Chong, R.A.; Wu, K.; Spratt, D.E.; Yang, Y.; Lee, C.; Nayak, J.; Xu, M.; Elkholi, R.; Tappin, I.; Li, J.; et al. Pivotal role for the ubiquitin Y59-E51 loop in lysine 48 polyubiquitination. Proc. Natl. Acad. Sci. USA 2014, 111, 8434–8439. [Google Scholar] [CrossRef]
- Pagano, M.; Tam, S.W.; Theodoras, A.M.; Beer-Romero, P.; Del Sal, G.; Chau, V.; Yew, P.R.; Draetta, G.F.; Rolfe, M. Role of the ubiquitin-proteasome pathway in regulating abundance of the cyclin-dependent kinase inhibitor p27. Science 1995, 269, 682–685. [Google Scholar] [CrossRef] [PubMed]
- Tam, S.W.; Theodoras, A.M.; Pagano, M. Kip1 degradation via the ubiquitin-proteasome pathway. Leukemia 1997, 11 (Suppl. 3), 363–366. [Google Scholar]
- Verma, R.; Feldman, R.M.; Deshaies, R.J. SIC1 is ubiquitinated in vitro by a pathway that requires CDC4, CDC34, and cyclin/CDK activities. Mol. Biol. Cell 1997, 8, 1427–1437. [Google Scholar] [CrossRef][Green Version]
- Henchoz, S.; Chi, Y.; Catarin, B.; Herskowitz, I.; Deshaies, R.J.; Peter, M. Phosphorylation- and ubiquitin-dependent degradation of the cyclin-dependent kinase inhibitor Far1p in budding yeast. Genes Dev. 1997, 11, 3046–3060. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Sun, Y. Targeting CDC34 E2 ubiquitin conjugating enzyme for lung cancer therapy. EBioMedicine 2020, 54, 102718. [Google Scholar] [CrossRef]
- Legesse-Miller, A.; Elemento, O.; Pfau, S.J.; Forman, J.J.; Tavazoie, S.; Coller, H.A. let-7 Overexpression leads to an increased fraction of cells in G2/M, direct down-regulation of Cdc34, and stabilization of Wee1 kinase in primary fibroblasts. J. Biol. Chem. 2009, 284, 6605–6609. [Google Scholar] [CrossRef] [PubMed]
- Lim, K.H.; Song, M.H.; Baek, K.H. Decision for cell fate: Deubiquitinating enzymes in cell cycle checkpoint. Cell. Mol. Life Sci. 2016, 73, 1439–1455. [Google Scholar] [CrossRef]
- Wei, X.; You, X.; Zhang, J.; Zhou, C. MicroRNA-1305 Inhibits the Stemness of LCSCs and Tumorigenesis by Repressing the UBE2T-Dependent Akt-Signaling Pathway. Mol. Ther. Nucleic Acids 2019, 16, 721–732. [Google Scholar] [CrossRef]
- Chiu, Y.H.; Sun, Q.; Chen, Z.J. E1-L2 activates both ubiquitin and FAT10. Mol. Cell 2007, 27, 1014–1023. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Lee, Y.K.; Han, K.H.; Jeon, H.; Jeong, I.H.; Kim, S.Y.; Lee, J.B.; Lee, P.C.W. BRC-mediated RNAi targeting of USE1 inhibits tumor growth in vitro and in vivo. Biomaterials 2020, 230, 119630. [Google Scholar] [CrossRef]
- Pierce, N.W.; Kleiger, G.; Shan, S.O.; Deshaies, R.J. Detection of sequential polyubiquitylation on a millisecond timescale. Nature 2009, 462, 615–619. [Google Scholar] [CrossRef] [PubMed]
- Ceccarelli, D.F.; Tang, X.; Pelletier, B.; Orlicky, S.; Xie, W.; Plantevin, V.; Neculai, D.; Chou, Y.C.; Ogunjimi, A.; Al-Hakim, A.; et al. An allosteric inhibitor of the human Cdc34 ubiquitin-conjugating enzyme. Cell 2011, 145, 1075–1087. [Google Scholar] [CrossRef]
- Tsukamoto, S.; Takeuchi, T.; Rotinsulu, H.; Mangindaan, R.E.; van Soest, R.W.; Ukai, K.; Kobayashi, H.; Namikoshi, M.; Ohta, T.; Yokosawa, H. Leucettamol A: A new inhibitor of Ubc13-Uev1A interaction isolated from a marine sponge, Leucetta aff. microrhaphis. Bioorg. Med. Chem. Lett. 2008, 18, 6319–6320. [Google Scholar] [CrossRef]
- Ushiyama, S.; Umaoka, H.; Kato, H.; Suwa, Y.; Morioka, H.; Rotinsulu, H.; Losung, F.; Mangindaan, R.E.; de Voogd, N.J.; Yokosawa, H.; et al. Manadosterols A and B, sulfonated sterol dimers inhibiting the Ubc13-Uev1A interaction, isolated from the marine sponge Lissodendryx fibrosa. J. Nat. Prod. 2012, 75, 1495–1499. [Google Scholar] [CrossRef]
- Strickson, S.; Campbell, D.G.; Emmerich, C.H.; Knebel, A.; Plater, L.; Ritorto, M.S.; Shpiro, N.; Cohen, P. The anti-inflammatory drug BAY 11-7082 suppresses the MyD88-dependent signalling network by targeting the ubiquitin system. Biochem. J. 2013, 451, 427–437. [Google Scholar] [CrossRef] [PubMed]
- Uprichard, S.L. The therapeutic potential of RNA interference. FEBS Lett. 2005, 579, 5996–6007. [Google Scholar] [CrossRef]
- Lee, J.H.; Ku, S.H.; Kim, M.J.; Lee, S.J.; Kim, H.C.; Kim, K.; Kim, S.H.; Kwon, I.C. Rolling circle transcription-based polymeric siRNA nanoparticles for tumor-targeted delivery. J. Control. Release off. J. Control. Release Soc. 2017, 263, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Jeong, J.; Kim, D.; Kwak, G.; Kim, S.H.; Lee, J.B. Bubbled RNA-Based Cargo for Boosting RNA Interference. Adv. Sci. 2017, 4, 1600523. [Google Scholar] [CrossRef] [PubMed]
- Cox, D.B.; Platt, R.J.; Zhang, F. Therapeutic genome editing: Prospects and challenges. Nat. Med. 2015, 21, 121–131. [Google Scholar] [CrossRef] [PubMed]
| Name | Synonyms | Class | Regulated Target | Relevant Cancers |
|---|---|---|---|---|
| UBE2A | UBC2, HR6A, HHR6A, RAD6A | Class I | P53 mono- or multi-monoubiquitination, PCNA monoubiquitination, RSG protein ubiquitination | Ovarian cancer [24], breast cancer [25,26,27], melanoma [28] |
| UBE2B | UBC2, HR6B, HHR6B, RAD6B, E2-17K | Class I | P53 mono- or multi-monoubiquitination, PCNA monoubiquitination, RSG protein ubiquitination | Ovarian cancer [24], breast cancer [25,26,27], melanoma [28] |
| UBE2C | UBCH10, DJ447F3.2, EC 6.3.2.19 | Class II | AKT degradation | Breast cancer [29,30,31], esophageal squamous cell carcinoma [30,32], hepatocellular carcinoma [30,33], lung cancer [30,34], thyroid cancers [30,35], pancreas cancer [30,36], nasopharyngeal carcinoma [37] |
| UBE2D1 | SFT, UBCH5, UBC4/5, UBCH5A | Class I | P53 degradation, β-TrCP1 degradation, RIP1 ubiquitination, BRCA1 ubiquitination | Lung cancer [38], hepatocellular carcinoma [39], bladder cancer [40] |
| UBE2F | NCE2 | Class II | NOXA degradation | Lung cancer [41,42] |
| UBE2I | UBC9, UBCH9 | Class I | Bcl-2 expression levels | Breast cancer [43], ovarian cancer [44] lung cancer [45] |
| UBE2J2 | UBC6, NCUBE2, PRO2121 | Class III | Phosphorylation of EGFR | Hepatocellular carcinoma [46,47] |
| UBE2L3 | E2-F1, UBCH7, UBCM4 | Class I | p27kip1 degradation | Lung cancer [48] |
| UBE2N | UBCH-BEN, UBC13 | Class I | Interacts with UBE2V1 or UBE2V2 to activate the NF-κB and p38 signaling | B-cell lymphoma [49], neuroblastoma [50], colon cancer [51], breast cancer [52], melanoma [53], cervical carcinoma [54] |
| UBE2O | E2-230K, FLJ12878, KIAA1734 | Class IV | SMAD6 ubiquitination, AMPKα2 ubiquitination | Prostate cancer [55], breast cancer [55,56], head and neck squamous carcinoma [57] |
| UBE2R1 | CDC34, UBCH3, UBC3, E2-CDC34 | Class III | CDK inhibitor degradation | Hepatocellular carcinomas [58], breast cancer [59], lung cancer [60] |
| UBE2S | E2-EPF | Class III | P53 degradation | Colorectal cancer [61], breast cancer [62], endometrial cancer [63], lung cancer [64], liver cancer [65] |
| UBE2T | PIG50, HSPC150, FANCT | Class III | Inhibits SOX6 expression and facilitates the nuclear translocation of β-catenin | Breast cancer [66], nasopharyngeal carcinoma [67], bladder cancer [68], hepatocellular carcinoma [69], gastric cancer [70], aggressive clear cell renal cell carcinoma [71], lung cancer [72] |
| UBE2Z | HOYS7, FLJ13855, USE1 | Class IV | RSG protein ubiquitination | Lung cancer [73] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bui, Q.T.; Hong, J.H.; Kwak, M.; Lee, J.Y.; Lee, P.C.-W. Ubiquitin-Conjugating Enzymes in Cancer. Cells 2021, 10, 1383. https://doi.org/10.3390/cells10061383
Bui QT, Hong JH, Kwak M, Lee JY, Lee PC-W. Ubiquitin-Conjugating Enzymes in Cancer. Cells. 2021; 10(6):1383. https://doi.org/10.3390/cells10061383
Chicago/Turabian StyleBui, Quyen Thu, Jeong Hee Hong, Minseok Kwak, Ji Yeon Lee, and Peter Chang-Whan Lee. 2021. "Ubiquitin-Conjugating Enzymes in Cancer" Cells 10, no. 6: 1383. https://doi.org/10.3390/cells10061383
APA StyleBui, Q. T., Hong, J. H., Kwak, M., Lee, J. Y., & Lee, P. C.-W. (2021). Ubiquitin-Conjugating Enzymes in Cancer. Cells, 10(6), 1383. https://doi.org/10.3390/cells10061383







