Paraburkholderia phymatum Homocitrate Synthase NifV Plays a Key Role for Nitrogenase Activity during Symbiosis with Papilionoids and in Free-Living Growth Conditions
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Media and Cultivation
2.2. Phylogenetic Analysis
2.3. Construction of P. phymatum STM815 Mutant Strains
2.4. Nitrogen-Fixation In Vitro Test
2.5. Plant Growth Condition, Inoculation and Plant Harvesting
2.6. Microscopy and Image Analysis
2.7. Metabolite Extraction and Metabolomics Data Analysis
2.8. Statistical Analysis
3. Results
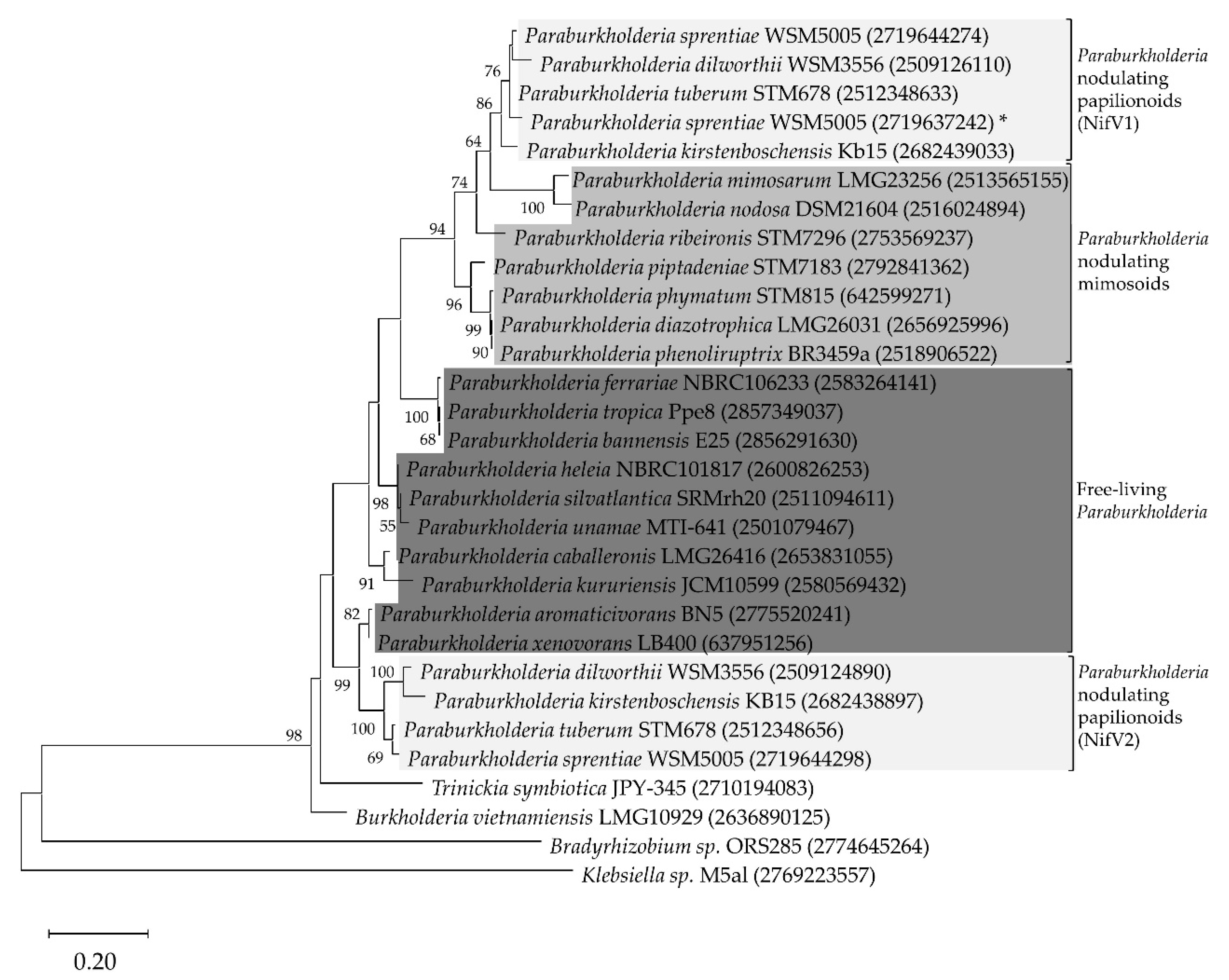
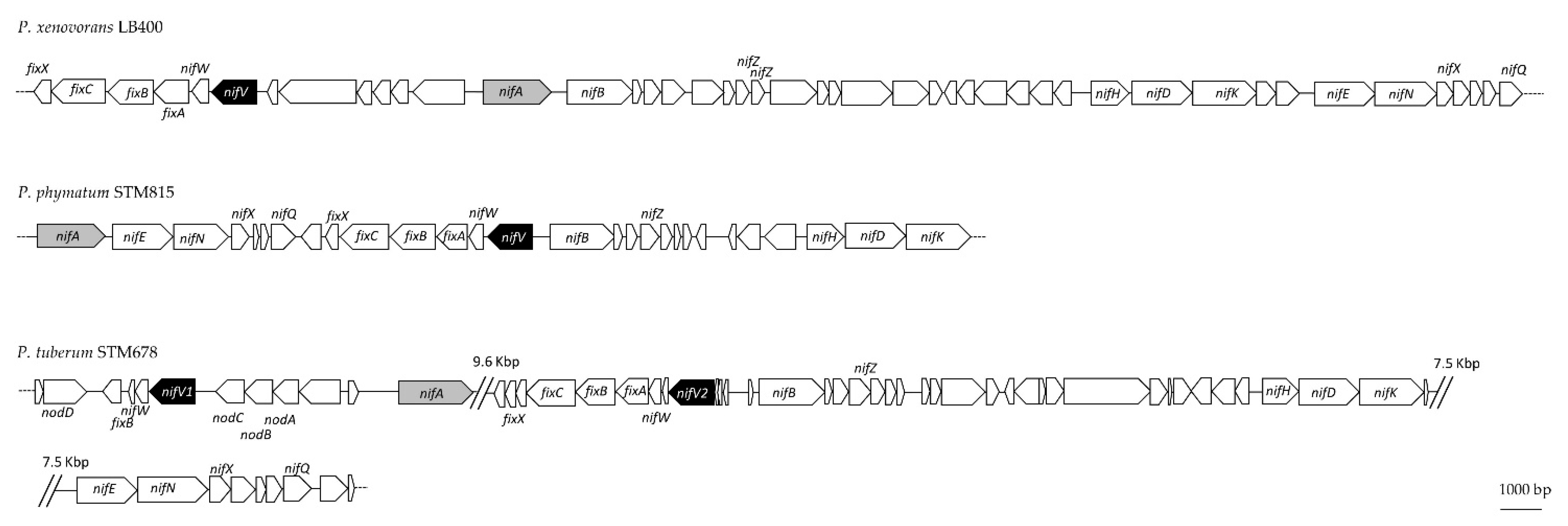
3.1. NifV Is Widely Distributed in Paraburkholderia Strains
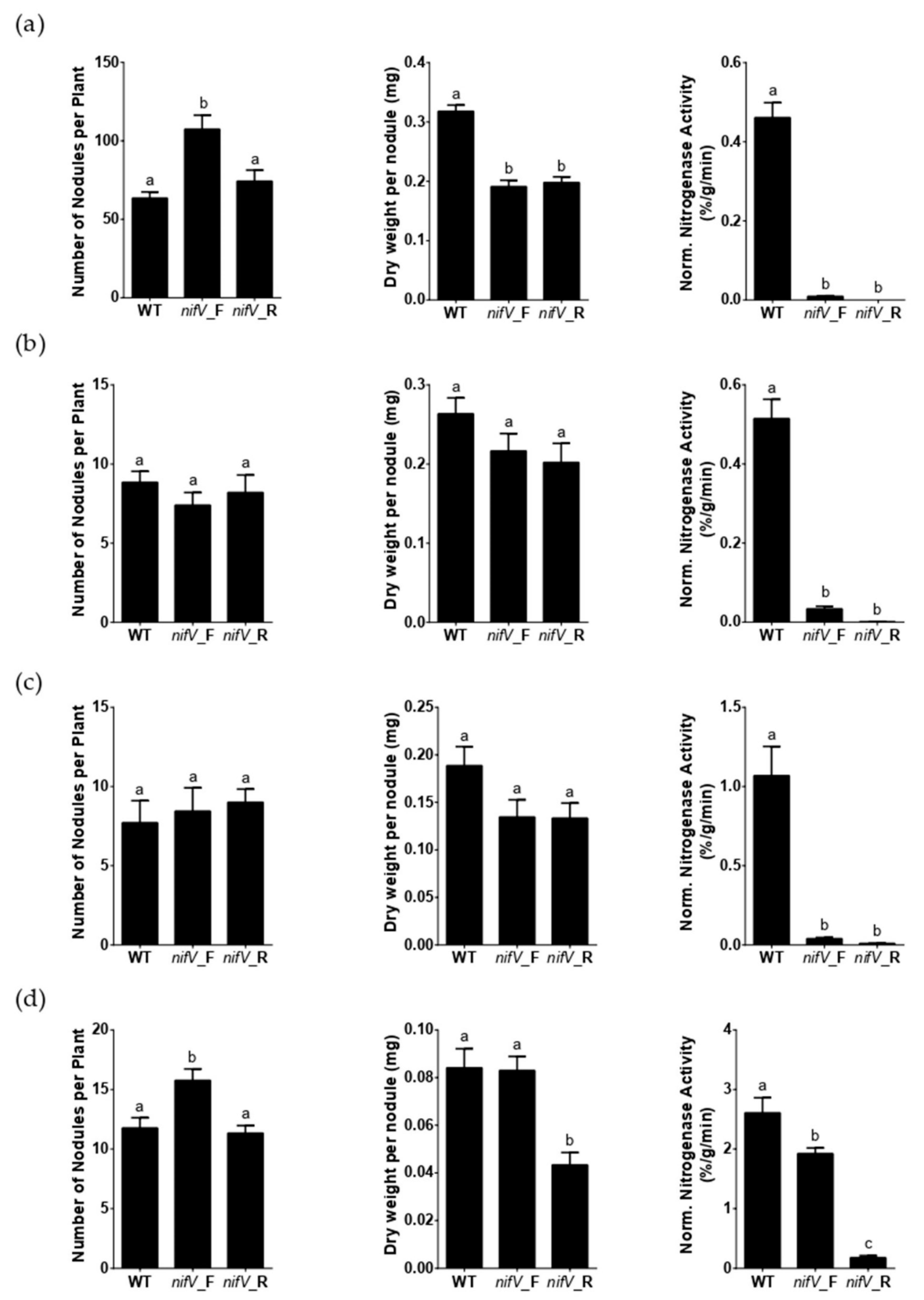
3.2. P. phymatum nifV Is Essential for Nitrogenase Activity in Symbiosis with Papilionoid Host Plants
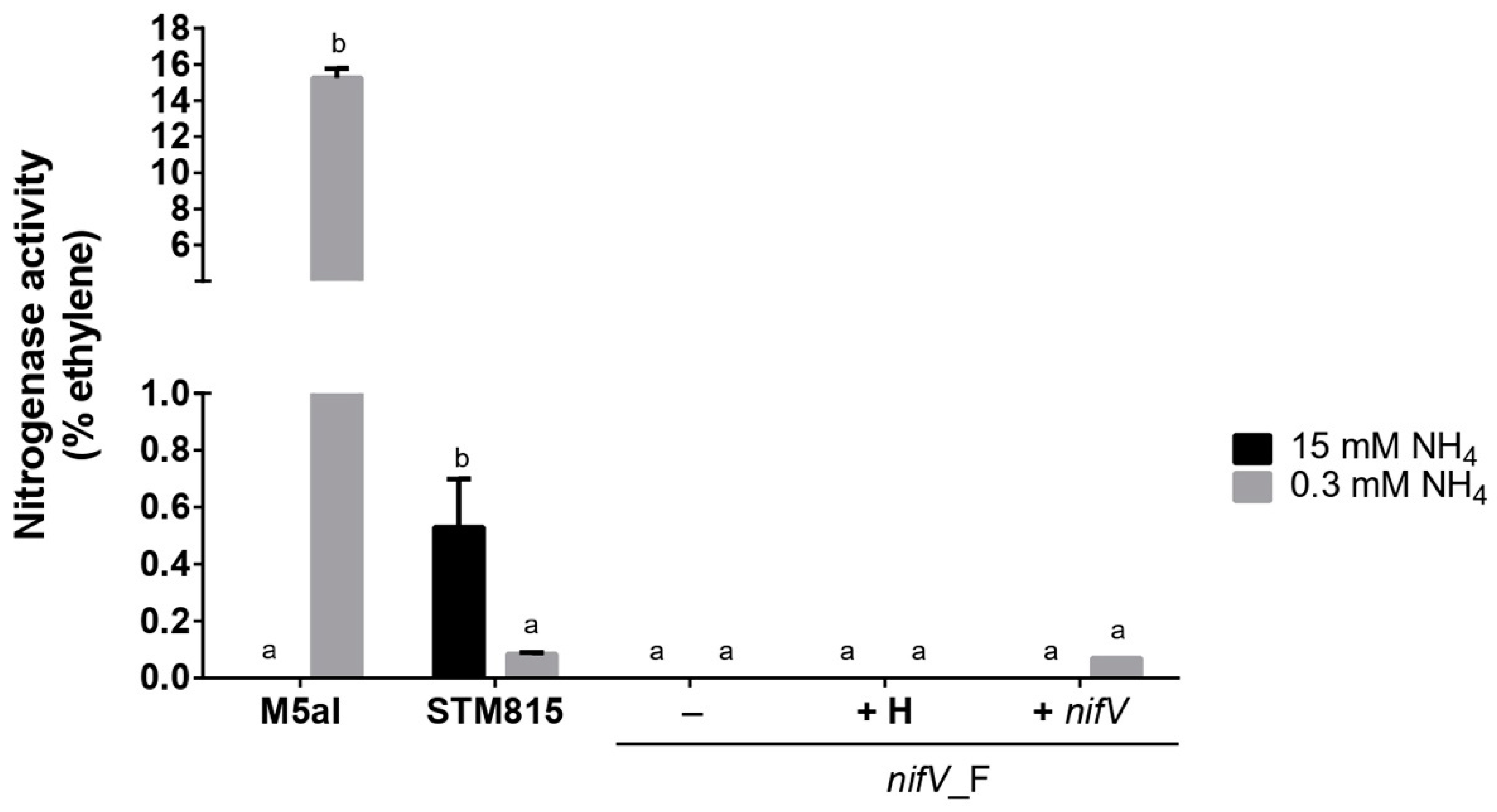
3.3. P. phymatum nifV Is Essential for Nitrogenase Activity in Free-Living Conditions
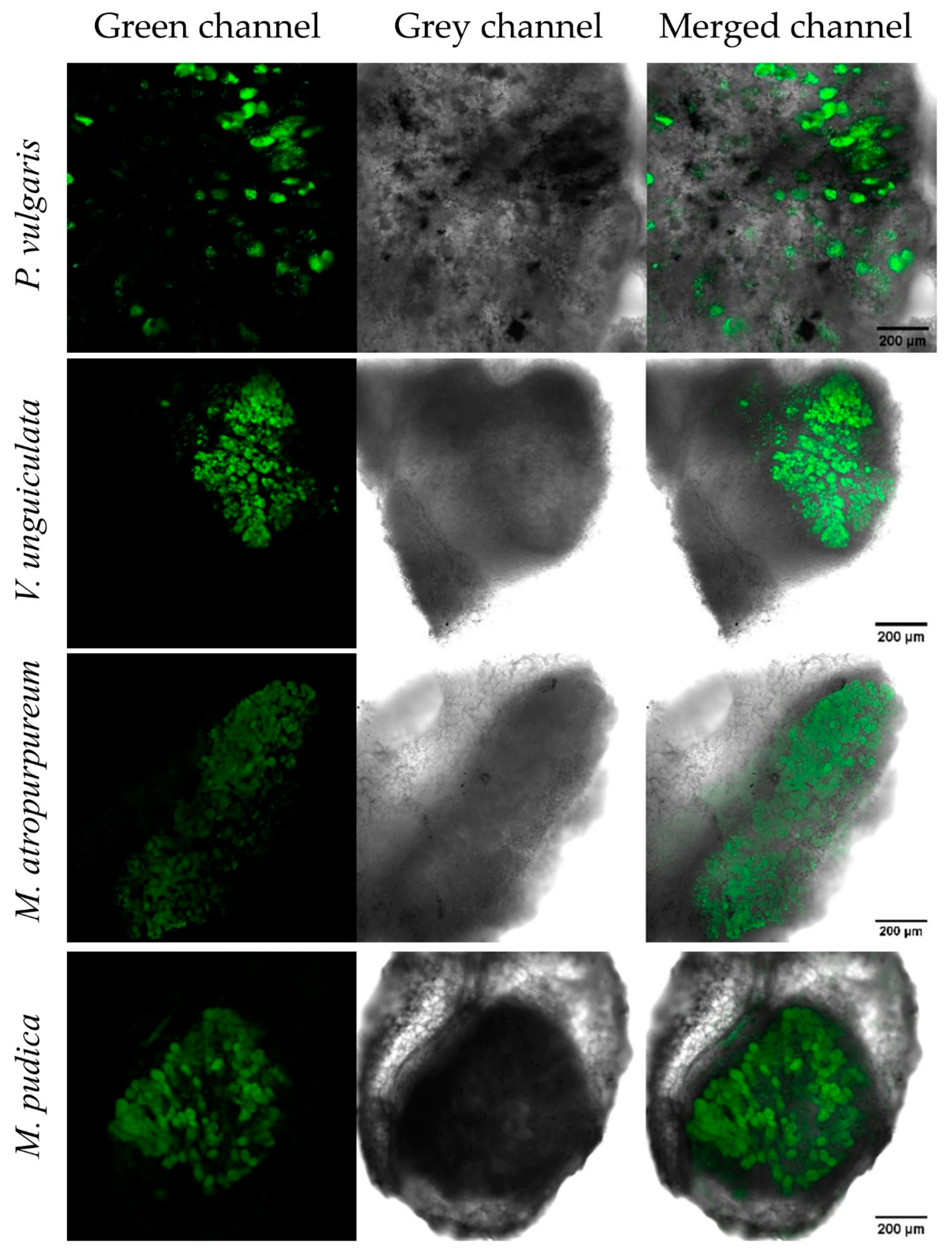
3.4. P. phymatum nifV Is Expressed in Free-Living Microoxic Conditions and in Symbiosis with Papilionoids and Mimosoids
3.5. Metabolic Comparison of P. phymatum STM815 Wild Type in Nodules of Different Host Plants
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bloom, A.J. The increasing importance of distinguishing among plant nitrogen sources. Curr. Opin. Plant Biol. 2015, 25, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Gruber, N.; Galloway, J.N. An Earth-system perspective of the global nitrogen cycle. Nature 2008, 451, 293–296. [Google Scholar] [CrossRef] [PubMed]
- Bohlool, B.B.; Ladha, J.K.; Garrity, D.P.; George, T. Biological nitrogen fixation for sustainable agriculture: A perspective. Plant Soil 1992, 141, 1–11. [Google Scholar] [CrossRef]
- Zahran, H.H. Rhizobium-legume symbiosis and nitrogen fixation under severe conditions and in an arid climate. Microbiol. Mol. Biol. Rev. 1999, 63, 968–989. [Google Scholar] [CrossRef] [Green Version]
- Tilman, D.; Cassman, K.G.; Matson, P.A.; Naylor, R.; Polasky, S. Agricultural sustainability and intensive production practices. Nature 2002, 418, 671–677. [Google Scholar] [CrossRef]
- Santos, M.S.; Nogueira, M.A.; Hungria, M. Microbial inoculants: Reviewing the past, discussing the present and previewing an outstanding future for the use of beneficial bacteria in agriculture. AMB Express 2019, 9. [Google Scholar] [CrossRef]
- Willems, A. The taxonomy of rhizobia: An overview. Plant Soil 2006, 287, 3–14. [Google Scholar] [CrossRef]
- Beukes, C.W.; Palmer, M.; Manyaka, P.; Chan, W.Y.; Avontuur, J.R.; van Zyl, E.; Huntemann, M.; Clum, A.; Pillay, M.; Palaniappan, K.; et al. Genome data provides high support for generic boundaries in Burkholderia sensu lato. Front. Microbiol. 2017, 8, 1–12. [Google Scholar] [CrossRef]
- Poole, P.; Ramachandran, V.; Terpolilli, J. Rhizobia: From saprophytes to endosymbionts. Nat. Rev. Microbiol. 2018, 16, 291–303. [Google Scholar] [CrossRef]
- Moulin, L.; Munive, A.; Dreyfus, B.; Boivin-Masson, C. Nodulation of legumes by members of the beta-subclass of Proteobacteria. Nature 2001, 411, 948. [Google Scholar] [CrossRef]
- Chen, W.M.; Laevens, S.; Lee, T.M.; Coenye, T.; De Vos, P.; Mergeay, M.; Vandamme, P. Ralstonia taiwanensis sp. nov., isolated from root nodules of Mimosa species and sputum of a cystic fibrosis patient. Int. J. Syst. Evol. Microbiol. 2001, 51, 1729–1735. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.M.; Moulin, L.; Bontemps, C.; Vandamme, P.; Béna, G.; Boivin-Masson, C. Legume symbiotic nitrogen fixation by β-Proteobacteria is widespread in nature. J. Bacteriol. 2003, 185, 7266–7272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masson-Boivin, C.; Giraud, E.; Perret, X.; Batut, J. Establishing nitrogen-fixing symbiosis with legumes: How many rhizobium recipes? Trends Microbiol. 2009, 17, 458–466. [Google Scholar] [CrossRef] [PubMed]
- Bontemps, C.; Elliott, G.N.; Simon, M.F.; Dos Reis Júnior, F.B.; Gross, E.; Lawton, R.C.; Neto, N.E.; De Fátima Loureiro, M.; De Faria, S.M.; Sprent, J.I.; et al. Burkholderia species are ancient symbionts of legumes. Mol. Ecol. 2010, 19, 44–52. [Google Scholar] [CrossRef] [PubMed]
- Gyaneshwar, P.; Hirsch, A.M.; Moulin, L.; Chen, W.M.; Elliott, G.N.; Bontemps, C.; De Los Santos, P.E.; Gross, E.; Dos Reis, F.B.; Janet, I.S.; et al. Legume-nodulating betaproteobacteria: Diversity, host range, and future prospects. Mol. Plant-Microbe Interact. 2011, 24, 1276–1288. [Google Scholar] [CrossRef] [Green Version]
- Sawana, A.; Adeolu, M.; Gupta, R.S. Molecular signatures and phylogenomic analysis of the genus Burkholderia: Proposal for division of this genus into the emended genus Burkholderia containing pathogenic organisms and a new genus Paraburkholderia gen. nov. harboring environmental species. Front. Genet. 2014, 5, 1–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Estrada-de los Santos, P.; Palmer, M.; Chávez-Ramírez, B.; Beukes, C.; Steenkamp, E.T.; Briscoe, L.; Khan, N.; Maluk, M.; Lafos, M.; Humm, E.; et al. Whole genome analyses suggests that Burkholderia sensu lato contains two additional novel genera (Mycetohabitans gen. nov., and Trinickia gen. nov.): Implications for the evolution of diazotrophy and nodulation in the Burkholderiaceae. Genes 2018, 9, 389. [Google Scholar] [CrossRef] [Green Version]
- De Meyer, S.E.; Briscoe, L.; Martínez-Hidalgo, P.; Agapakis, C.M.; De-Los Santos, P.E.; Seshadri, R.; Reeve, W.; Weinstock, G.; O’Hara, G.; Howieson, J.G.; et al. Symbiotic Burkholderia species show diverse arrangements of nif/fix and nod genes and lack typical high-affinity cytochrome cbb3 oxidase genes. Mol. Plant-Microbe Interact. 2016, 29, 609–619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bournaud, C.; de Faria, S.M.; dos Santos, J.M.F.; Tisseyre, P.; Silva, M.; Chaintreuil, C.; Gross, E.; James, E.K.; Prin, Y.; Moulin, L. Burkholderia species are the most common and preferred nodulating symbionts of the Piptadenia group (tribe Mimoseae). PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, W.; De Faria, S.M.; Pitard, R.M.; Simo, J.L.; Prescott, A.R.; Elliott, G.N.; Sprent, J.I.; Young, J.P.W.; James, E.K. Proof that Burkholderia strains form effective symbioses with legumes: A study of novel Mimosa-nodulating strains from South America. Appl. Environ. Microbiol. 2005, 71, 7461–7471. [Google Scholar] [CrossRef] [Green Version]
- Barrett, C.F.; Parker, M.A. Coexistence of Burkholderia, Cupriavidus, and Rhizobium sp. nodule bacteria on two Mimosa spp. in Costa Rica craig. Appl. Environ. Microbiol. 2005, 72, 1198–1206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elliott, G.N.; Chen, W.M.; Chou, J.H.; Wang, H.C.; Sheu, S.Y.; Perin, L.; Reis, V.M.; Moulin, L.; Simon, M.F.; Bontemps, C.; et al. Burkholderia phymatum is a highly effective nitrogen-fixing symbiont of Mimosa spp. and fixes nitrogen ex planta. New Phytol. 2006, 173, 168–180. [Google Scholar] [CrossRef]
- Bueno dos Reis, F.; Simon, M.F.; Gross, E.; Boddey, R.M.; Elliott, G.N.; Neto, N.E.; de Fatima Loureiro, M.; de Queiroz, L.P.; Scotti, M.R.; Chen, W.M.; et al. Nodulation and nitrogen fixation by Mimosa spp. in the Cerrado and Caatinga biomes of Brazil. New Phytol. 2010, 186, 934–946. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Wei, S.; Wang, F.; James, E.K.; Guo, X.; Zagar, C.; Xia, L.G.; Dong, X.; Wang, Y.P. Burkholderia and Cupriavidus spp. are the preferred symbionts of Mimosa spp. in Southern China. FEMS Microbiol. Ecol. 2012, 80, 417–426. [Google Scholar] [CrossRef] [PubMed]
- Mishra, R.P.N.; Tisseyre, P.; Melkonian, R.; Chaintreuil, C.; Miché, L.; Klonowska, A.; Gonzalez, S.; Bena, G.; Laguerre, G.; Moulin, L. Genetic diversity of Mimosa pudica rhizobial symbionts in soils of French Guiana: Investigating the origin and diversity of Burkholderia phymatum and other beta-rhizobia. FEMS Microbiol. Ecol. 2012, 79, 487–503. [Google Scholar] [CrossRef] [PubMed]
- Talbi, C.; Delgado, M.J.; Girard, L.; Ramírez-Trujillo, A.; Caballero-Mellado, J.; Bedmar, E.J. Burkholderia phymatum strains capable of nodulating Phaseolus vulgaris are present in moroccan soils. Appl. Environ. Microbiol. 2010, 76, 4587–4591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, W.Y.Y.; Ridgway, H.J.; James, T.K.; James, E.K.; Chen, W.M.; Sprent, J.I.; Young, J.P.W.; Andrews, M. Burkholderia sp. induces functional nodules on the South African invasive legume Dipogon lignosus (Phaseoleae) in New Zealand soils. Microb. Ecol. 2014, 68, 542–555. [Google Scholar] [CrossRef]
- Dixon, R.; Kahn, D. Genetic regulation of biological nitrogen fixation. Nat. Rev. Microbiol. 2004, 2, 621–631. [Google Scholar] [CrossRef]
- Hoover, T.R.; Imperial, J.; Ludden, P.W.; Shah, V.K. Homocitrate is a component of the iron-molybdenum cofactor of nitrogenase. Biochemistry 1989, 28, 2768–2771. [Google Scholar] [CrossRef]
- Burén, S.; Jiménez-Vicente, E.; Echavarri-Erasun, C.; Rubio, L.M. Biosynthesis of nitrogenase cofactors. Chem. Rev. 2020, 120, 4921–4968. [Google Scholar] [CrossRef]
- Yu, Z.; Li, S.; Li, Y.; Jiang, Z.; Zhou, J.; An, Q. Complete genome sequence of N2-fixing model strain Klebsiella sp. nov. M5al, which produces plant cell wall-degrading enzymes and siderophores. Biotechnol. Rep. 2018, 17, 6–9. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; White, R.H.; Dean, D.R. Purification of the Azotobacter vinelandii nifV-encoded homocitrate synthase. J. Bacteriol. 1997, 179, 5963–5966. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hakoyama, T.; Niimi, K.; Watanabe, H.; Tabata, R.; Matsubara, J.; Sato, S.; Nakamura, Y.; Tabata, S.; Jichun, L.; Matsumoto, T.; et al. Host plant genome overcomes the lack of a bacterial gene for symbiotic nitrogen fixation. Nature 2009, 462, 514–517. [Google Scholar] [CrossRef] [Green Version]
- Nouwen, N.; Arrighi, J.F.; Cartieaux, F.; Chaintreuil, C.; Gully, D.; Klopp, C.; Giraud, E. The role of rhizobial (NifV) and plant (FEN1) homocitrate synthases in Aeschynomene/photosynthetic Bradyrhizobium symbiosis. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Lardi, M.; Liu, Y.; Purtschert, G.; de Campos, S.B.; Pessi, G. Transcriptome analysis of Paraburkholderia phymatum under nitrogen starvation and during symbiosis with Phaseolus vulgaris. Genes 2017, 8, 389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, J.H. Experiments in Molecular Genetics; Cold Spring Harbor Lab Press: Cold Spring Harbor, NY, USA, 1972; pp. 352–355. ISBN 0879691069. [Google Scholar]
- Clark, D.J.; Maaløe, O. DNA replication and the division cycle in Escherichia coli. J. Mol. Biol. 1967, 23, 99–112. [Google Scholar] [CrossRef]
- Liu, Y.; Bellich, B.; Hug, S.; Eberl, L.; Cescutti, P.; Pessi, G. The exopolysaccharide cepacian plays a role in the establishment of the Paraburkholderia phymatum—Phaseolus vulgaris symbiosis. Front. Microbiol. 2020, 11, 1–13. [Google Scholar] [CrossRef]
- Chen, I.M.A.; Chu, K.; Palaniappan, K.; Pillay, M.; Ratner, A.; Huang, J.; Huntemann, M.; Varghese, N.; White, J.R.; Seshadri, R.; et al. IMG/M v.5.0: An integrated data management and comparative analysis system for microbial genomes and microbiomes. Nucleic Acids Res. 2019, 47, D666–D677. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Vincent, J.M. A Manual for the Practical Study of the Root-Nodule Bacteria; Blackwell Scientific Publications: Oxford, UK; Edinburgh, UK, 1970; ISBN 0632064102. [Google Scholar]
- Lardi, M.; de Campos, S.B.; Purtschert, G.; Eberl, L.; Pessi, G. Competition experiments for legume infection identify Burkholderia phymatum as a highly competitive β-rhizobium. Front. Microbiol. 2017, 8, 1–12. [Google Scholar] [CrossRef] [Green Version]
- de Campos, S.B.; Lardi, M.; Gandolfi, A.; Eberl, L.; Pessi, G. Mutations in two Paraburkholderia phymatum type VI secretion systems cause reduced fitness in interbacterial competition. Front. Microbiol. 2017, 8, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Rueden, C.T.; Schindelin, J.; Hiner, M.C.; DeZonia, B.E.; Walter, A.E.; Arena, E.T.; Eliceiri, K.W. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinform. 2017, 18, 1–26. [Google Scholar] [CrossRef]
- Lardi, M.; Murset, V.; Fischer, H.M.; Mesa, S.; Ahrens, C.H.; Zamboni, N.; Pessi, G. Metabolomic profiling of Bradyrhizobium diazoefficiens -induced root nodules reveals both host plant-specific and developmental signatures. Int. J. Mol. Sci. 2016, 17, 815. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fuhrer, T.; Heer, D.; Begemann, B.; Zamboni, N. High-throughput, accurate mass metabolome profiling of cellular extracts by flow injection-time-of-flight mass spectrometry. Anal. Chem. 2011, 83, 7074–7080. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Storey, J.D.; Tibshirani, R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 2003, 100, 9440–9445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmitz, R.A.; Klopprogge, K.; Grabbe, R. Regulation of nitrogen fixation in Klebsiella pneumoniae and Azotobacter vinelandii: NifL, transducing two environmental signals to the nif transcriptional activator NifA. J. Mol. Microbiol. Biotechnol. 2002, 4, 235–242. [Google Scholar]
- Lardi, M.; Liu, Y.; Giudice, G.; Ahrens, C.H.; Zamboni, N.; Pessi, G. Metabolomics and transcriptomics identify multiple downstream targets of Paraburkholderia phymatum σ54 during symbiosis with Phaseolus vulgaris. Int. J. Mol. Sci. 2018, 19, 1049. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christinet, L.; Burdet, F.X.; Zaiko, M.; Hinz, U.; Zrÿd, J.P. Characterization and functional identification of a novel plant 4,5-extradiol dioxygenase involved in betalain pigment biosynthesis in Portulaca grandiflora. Plant Physiol. 2004, 134, 265–274. [Google Scholar] [CrossRef] [Green Version]
- Masukawa, H.; Inoue, K.; Sakurai, H. Effects of disruption of homocitrate synthase genes on Nostoc sp. strain PCC 7120 photobiological hydrogen production and nitrogenase. Appl. Environ. Microbiol. 2007, 73, 7562–7570. [Google Scholar] [CrossRef] [Green Version]
- Fernández, N.; Cabrera, J.J.; Varadarajan, A.R.; Lutz, S.; Ledermann, R.; Roschitzki, B.; Eberl, L.; Bedmar, E.J.; Fischer, H.M.; Pessi, G.; et al. An integrated systems approach unveils new aspects of microoxia-mediated regulation in Bradyrhizobium diazoefficiens. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef] [Green Version]
- Hashimoto, S.; Wongdee, J.; Songwattana, P.; Greetatorn, T.; Goto, K.; Tittabutr, P.; Boonkerd, N.; Teaumroong, N.; Uchiumi, T. Homocitrate synthase genes of two wide-host-range Bradyrhizobium strains are differently required for symbiosis depending on host plants. Microbes Environ. 2019, 34, 393–401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mayer, S.M.; Gormal, C.A.; Smith, B.E.; Lawson, D.M. Crystallographic analysis of the MoFe protein of nitrogenase from a nifV mutant of Klebsiella pneumoniae identifies citrate as a ligand to the molybdenum of iron molybdenum cofactor (FeMoco). J. Biol. Chem. 2002, 277, 35263–35266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lardi, M.; Pessi, G. Functional genomics approaches to studying symbioses between legumes and nitrogen-fixing rhizobia. High-Throughput 2018, 7, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishida, H.; Nishiyama, M.; Kobashi, N.; Kosuge, T.; Hoshino, T.; Yamane, H. A prokaryotic gene cluster involved in synthesis of lysine through the amino adipate pathway: A key to the evolution of amino acid biosynthesis. Genome Res. 1999, 9, 1175–1183. [Google Scholar] [CrossRef] [Green Version]
- Ferraioli, S.; Tatè, R.; Cermola, M.; Favre, R.; Iaccarino, M.; Patriarca, E.J. Auxotrophic mutant strains of Rhizobium etli reveal new nodule development phenotypes. Mol. Plant-Microbe Interact. 2002, 15, 501–510. [Google Scholar] [CrossRef] [Green Version]
- Das, S.K.; Gautam, U.S.; Sandhu, K.V.; Bandyopadhyay, S.; Chakrabartty, P.K.; Singh, A. Mutation in the lysA gene impairs the symbiotic properties of Mesorhizobium ciceri. Arch. Microbiol. 2010, 192, 69–77. [Google Scholar] [CrossRef]
- Dunn, M.F. Key roles of microsymbiont amino acid metabolism in rhizobia-legume interactions. Crit. Rev. Microbiol. 2015, 41, 411–451. [Google Scholar] [CrossRef]
- Flores-Tinoco, C.E.; Tschan, F.; Fuhrer, T.; Margot, C.; Sauer, U.; Christen, M.; Christen, B. Co-catabolism of arginine and succinate drives symbiotic nitrogen fixation. Mol. Syst. Biol. 2020, 16, 1–18. [Google Scholar] [CrossRef]
- Liu, A.; Contador, C.A.; Fan, K.; Lam, H.M. Interaction and regulation of carbon, nitrogen, and phosphorus metabolisms in root nodules of legumes. Front. Plant Sci. 2018, 871, 1–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dixon, R.A.; Paiva, N.L. Stress-induced phenylpropanoid metabolism. Plant Cell 1995, 7, 1085–1097. [Google Scholar] [CrossRef]
- Oldroyd, G.E.D.; Downie, J.A. Calcium, kinases and nodulation signalling in legumes. Nat. Rev. Mol. Cell Biol. 2004, 5, 566–576. [Google Scholar] [CrossRef]
- Mierziak, J.; Kostyn, K.; Kulma, A. Flavonoids as important molecules of plant interactions with the environment. Molecules 2014, 19, 16240–16265. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Pandey, A.K. Chemistry and biological activities of flavonoids: An overview. Sci. World J. 2013, 2013. [Google Scholar] [CrossRef] [Green Version]
- Shaw, L.J.; Morris, P.; Hooker, J.E. Perception and modification of plant flavonoid signals by rhizosphere microorganisms. Environ. Microbiol. 2006, 8, 1867–1880. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.; Ma, C.; Dong, S.; Xu, Y.; Gong, Z. Effects of nitrogen concentrations on nodulation and nitrogenase activity in dual root systems of soybean plants. Soil Sci. Plant Nutr. 2017, 63, 470–482. [Google Scholar] [CrossRef]

| Metabolites 1 | ID 1 | log2FC (Bean wt nod vs. Root) 2 | log2FC (Cowpea wt nod vs. Root) 2 | log2FC (Siratro wt nod vs. Root) 2 | log2FC (Mimosa wt nod vs. Root) 2 |
|---|---|---|---|---|---|
| Higher Abundance in Nodules Infected by P. phymatum Wild-Type | |||||
| Diaminopimelate | C00666 | 0.7 | 1.1 | 1.9 | 1.2 |
| Aminoadipate | C00956 | 0.8 | 0.8 | 1.7 | 1.3 |
| 4-aminobutyraldehyde | C00555 | 0.7 | 0.9 | 1.6 | 1.0 |
| Alanine | C00041 | 0.6 | 1.1 | 1.6 | 1.0 |
| Homocitrate | C01251 | 0.6 | 1.3 | 1.4 | 1.2 |
| 4,5-seco-Dopa | C17758 | 0.5 | 0.7 | 1.3 | 1.0 |
| Acetyl-Glu-semialdehyde | C01250 | 1.2 | 1.1 | 1.0 | 1.6 |
| Pipecolate | C00408 | 0.8 | 0.9 | 0.8 | 0.9 |
| Higher Abundance in Uninfected Roots | |||||
| Urocanate | C00785 | −0.7 | −0.7 | −1.4 | −0.5 |
| sn-glycerol 3-phosphate | C00093 | −1.5 | −0.6 | −1.5 | −0.9 |
| 4-hydroxybenzoate | C00156 | −0.5 | −0.6 | −0.7 | −0.8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bellés-Sancho, P.; Lardi, M.; Liu, Y.; Hug, S.; Pinto-Carbó, M.A.; Zamboni, N.; Pessi, G. Paraburkholderia phymatum Homocitrate Synthase NifV Plays a Key Role for Nitrogenase Activity during Symbiosis with Papilionoids and in Free-Living Growth Conditions. Cells 2021, 10, 952. https://doi.org/10.3390/cells10040952
Bellés-Sancho P, Lardi M, Liu Y, Hug S, Pinto-Carbó MA, Zamboni N, Pessi G. Paraburkholderia phymatum Homocitrate Synthase NifV Plays a Key Role for Nitrogenase Activity during Symbiosis with Papilionoids and in Free-Living Growth Conditions. Cells. 2021; 10(4):952. https://doi.org/10.3390/cells10040952
Chicago/Turabian StyleBellés-Sancho, Paula, Martina Lardi, Yilei Liu, Sebastian Hug, Marta Adriana Pinto-Carbó, Nicola Zamboni, and Gabriella Pessi. 2021. "Paraburkholderia phymatum Homocitrate Synthase NifV Plays a Key Role for Nitrogenase Activity during Symbiosis with Papilionoids and in Free-Living Growth Conditions" Cells 10, no. 4: 952. https://doi.org/10.3390/cells10040952
APA StyleBellés-Sancho, P., Lardi, M., Liu, Y., Hug, S., Pinto-Carbó, M. A., Zamboni, N., & Pessi, G. (2021). Paraburkholderia phymatum Homocitrate Synthase NifV Plays a Key Role for Nitrogenase Activity during Symbiosis with Papilionoids and in Free-Living Growth Conditions. Cells, 10(4), 952. https://doi.org/10.3390/cells10040952







