p53 Protein Isoform Profiles in AML: Correlation with Distinct Differentiation Stages and Response to Epigenetic Differentiation Therapy
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines and AML Patient Material
2.2. Compounds
2.3. Western Blotting
2.4. Flow Cytometry
2.5. Cell Proliferation Assay
2.6. Gene Expression Profiling
2.7. Correlation Method and Statistical Analysis
3. Results
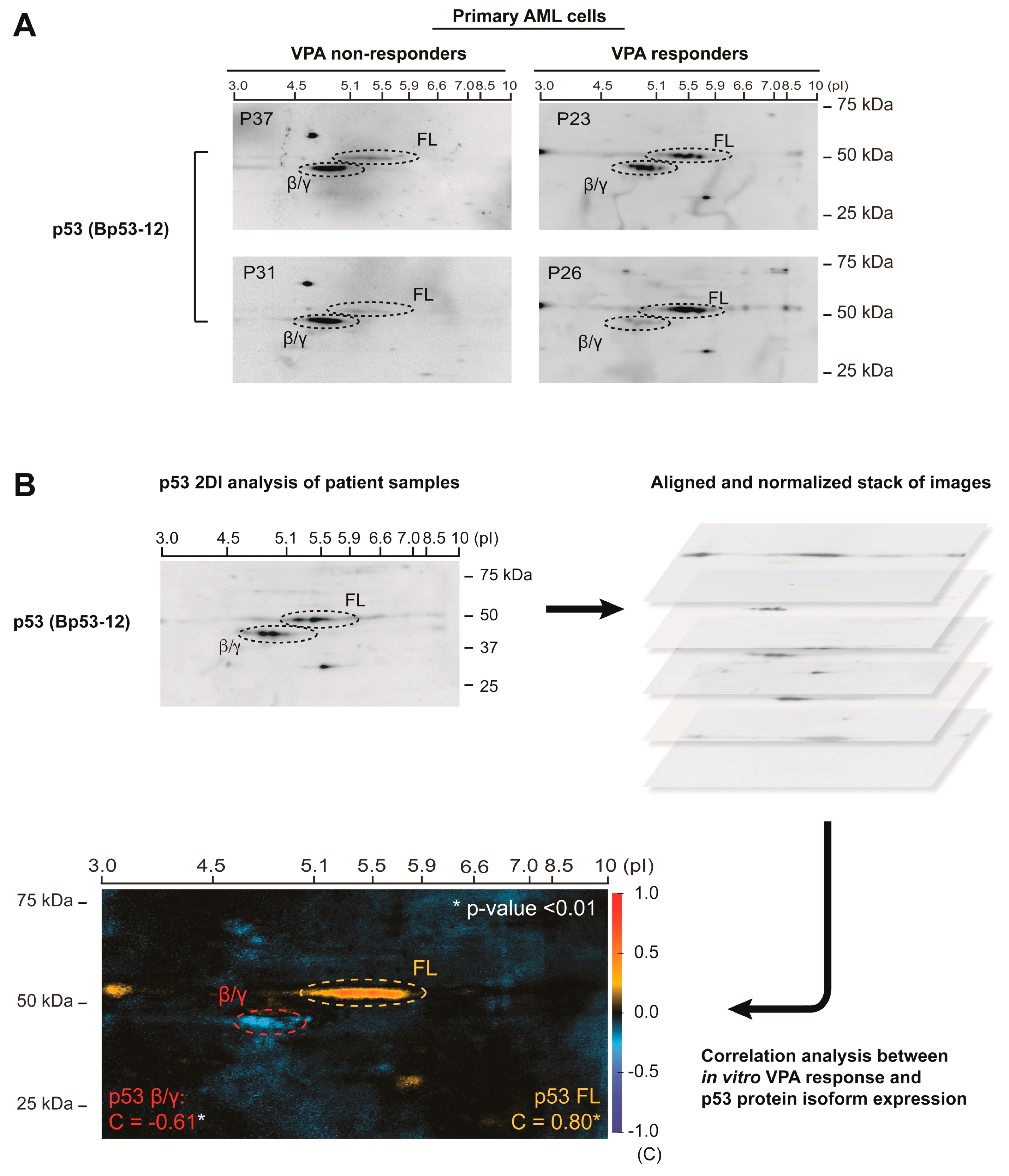
3.1. p53 Protein Isoform Expression Correlates with In Vitro Sensitivity to Valproic Acid in Primary AML Cells
3.2. p53 Protein Isoform Expression Correlates with the Differentiation Stage of AML Blasts
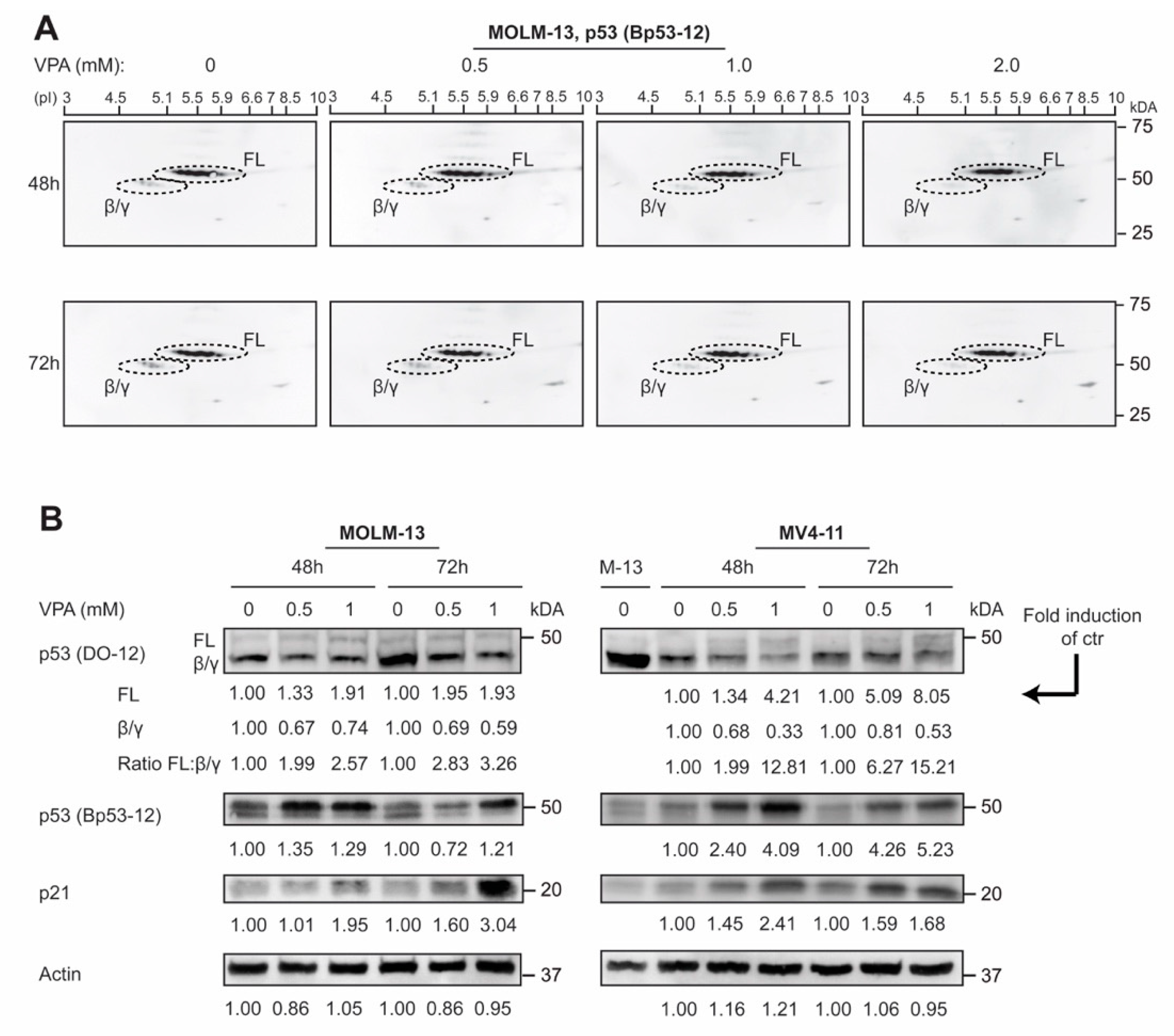
3.3. p53 Protein Isoforms Are Modulated by Valproic Acid in AML Cell Lines
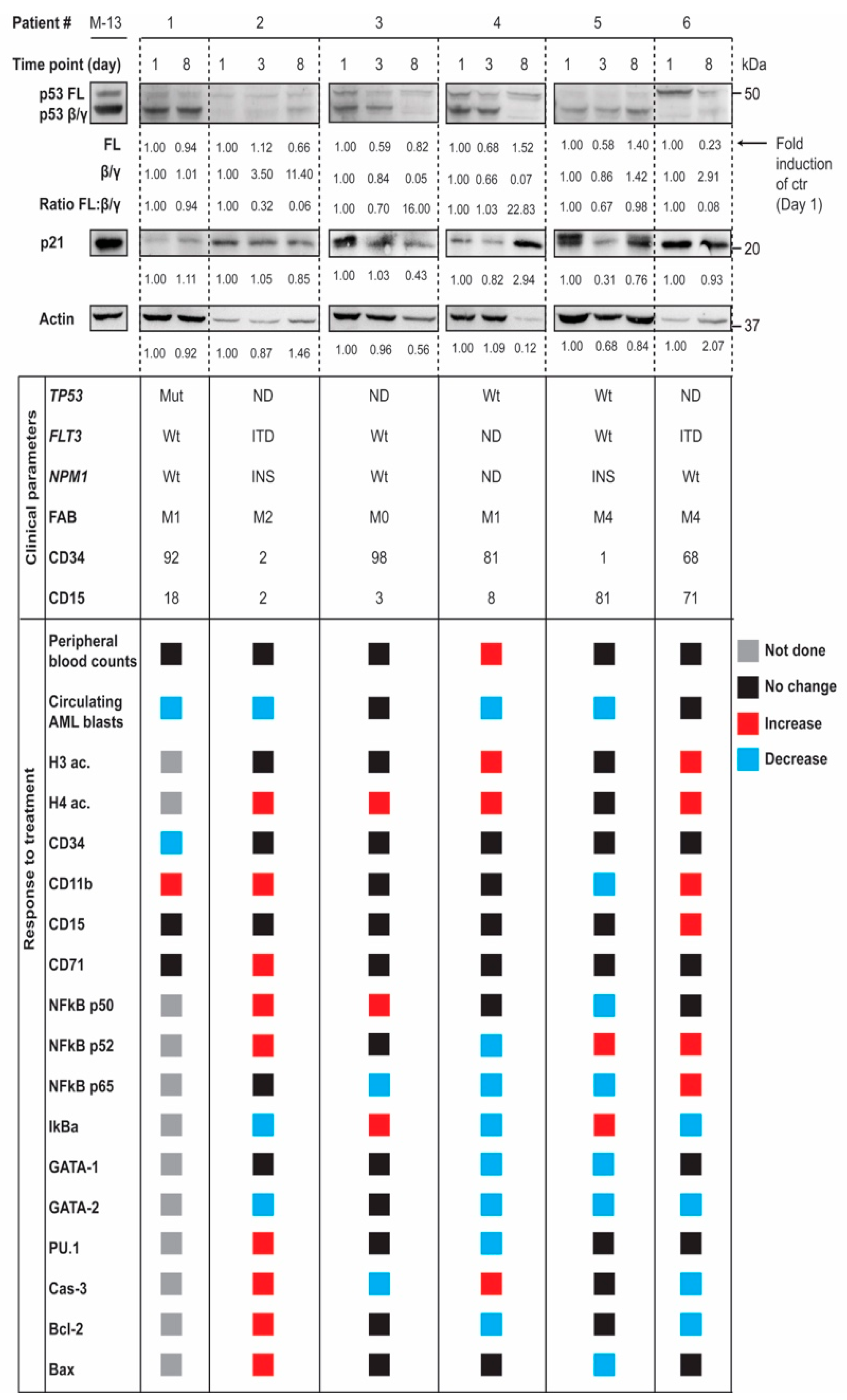
3.4. In Vivo Modulation of p53 Protein Isoforms during Differentiation Therapy of AML
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Anbarasan, T.; Bourdon, J.-C. The Emerging Landscape of p53 Isoforms in Physiology, Cancer and Degenerative Diseases. Int. J. Mol. Sci. 2019, 20, 6257. [Google Scholar] [CrossRef]
- Beck, J.; Turnquist, C.; Horikawa, I.; Harris, C.C. Targeting cellular senescence in cancer and aging: Roles of p53 and its isoforms. Carcinogenesis 2020, 41, 1017–1029. [Google Scholar] [CrossRef]
- Hayman, L.; Chaudhry, W.R.; Revin, V.V.; Zhelev, N.; Bourdon, J.C. What is the potential of p53 isoforms as a predictive biomarker in the treatment of cancer? Expert Rev. Mol. Diagn. 2019, 19, 149–159. [Google Scholar] [CrossRef]
- Bourdon, J.-C.; Surget, S.; Khoury, M.P. Uncovering the role of p53 splice variants in human malignancy: A clinical perspective. OncoTargets Ther. 2013, 7, 57–68. [Google Scholar] [CrossRef]
- Bourdon, J.-C.; Fernandes, K.; Murray-Zmijewski, F.; Liu, G.; Diot, A.; Xirodimas, D.P.; Saville, M.K.; Lane, D.P. p53 isoforms can regulate p53 transcriptional activity. Genes Dev. 2005, 19, 2122–2137. [Google Scholar] [CrossRef]
- Murray-Zmijewski, F.; Lane, D.P.; Bourdon, J.-C. p53/p63/p73 isoforms: An orchestra of isoforms to harmonise cell differentiation and response to stress. Cell Death Differ. 2006, 13, 962–972. [Google Scholar] [CrossRef]
- Marcel, V.; Dichtel-Danjoy, M.-L.; Sagne, C.; Hafsi, H.; Ma, D.; Ortizcuaran, S.; Olivier, M.; Hall, J.F.; Mollereau, B.; Hainaut, P.; et al. Biological functions of p53 isoforms through evolution: Lessons from animal and cellular models. Cell Death Differ. 2011, 18, 1815–1824. [Google Scholar] [CrossRef] [PubMed]
- Joruiz, S.M.; Bourdon, J.-C. p53 Isoforms: Key Regulators of the Cell Fate Decision. Cold Spring Harb. Perspect. Med. 2016, 6, a026039. [Google Scholar] [CrossRef] [PubMed]
- Tu, Q.; Gong, H.; Yuan, C.; Liu, G.; Huang, J.; Li, Z.; Luo, J. Δ133p53/FLp53 Predicts Poor Clinical Outcome in Esophageal Squamous Cell Carcinoma. Cancer Manag. Res. 2020, 12, 7405–7417. [Google Scholar] [CrossRef] [PubMed]
- Kazantseva, M.; Mehta, S.; Eiholzer, R.A.; Gimenez, G.; Bowie, S.; Campbell, H.; Reily-Bell, A.L.; Roth, I.; Ray, S.; Drummond, C.J.; et al. The Δ133p53β isoform promotes an immunosuppressive environment leading to aggressive prostate cancer. Cell Death Dis. 2019, 10, 1–17. [Google Scholar] [CrossRef]
- Hofstetter, G.; Berger, A.; Schuster, E.; Wolf, A.; Hager, G.; Vergote, I.; Cadron, I.; Sehouli, J.; Braicu, E.I.; Mahner, S.; et al. Δ133p53 is an independent prognostic marker in p53 mutant advanced serous ovarian cancer. Br. J. Cancer 2011, 105, 1593–1599. [Google Scholar] [CrossRef]
- Bischof, K.; Knappskog, S.; Hjelle, S.M.; Stefansson, I.; Woie, K.; Salvesen, H.B.; Gjertsen, B.T.; Bjorge, L. Influence of p53 Isoform Expression on Survival in High-Grade Serous Ovarian Cancers. Sci. Rep. 2019, 9, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhao, Y.; Sun, P.; Zhao, M.; Su, Z.; Jin, X.; Song, W. p53β: A new prognostic marker for patients with clear-cell renal cell carcinoma from 5.3 years of median follow-up. Carcinogenesis 2018, 39, 368–374. [Google Scholar] [CrossRef]
- Avery-Kiejda, K.A.; Morten, B.; Wong-Brown, M.W.; Mathe, A.; Scott, R.J. The relative mRNA expression of p53 isoforms in breast cancer is associated with clinical features and outcome. Carcinogenesis 2013, 35, 586–596. [Google Scholar] [CrossRef] [PubMed]
- Bourdon, J.-C.; Khoury, M.P.; Diot, A.; Baker, L.; Fernandes, K.; Aoubala, M.; Quinlan, P.; Purdie, C.A.; Jordan, L.B.; Prats, A.-C.; et al. p53 mutant breast cancer patients expressing p53γ have as good a prognosis as wild-type p53 breast cancer patients. Breast Cancer Res. 2011, 13, R7. [Google Scholar] [CrossRef] [PubMed]
- Ånensen, N.; Hjelle, S.M.; Van Belle, W.; Haaland, I.; Silden, E.; Bourdon, J.-C.; Hovland, R.; Taskén, K.; Knappskog, S.; Lonning, P.E.; et al. Correlation analysis of p53 protein isoforms with NPM1/FLT3 mutations and therapy response in acute myeloid leukemia. Oncogene 2011, 31, 1533–1545. [Google Scholar] [CrossRef] [PubMed]
- Prokocimer, M.; Molchadsky, A.; Rotter, V. Dysfunctional diversity of p53 proteins in adult acute myeloid leukemia: Projections on diagnostic workup and therapy. Blood 2017, 130, 699–712. [Google Scholar] [CrossRef] [PubMed]
- Cleven, A.H.G.; Nardi, V.; Ok, C.Y.; Goswami, M.; Cin, P.D.; Zheng, Z.; Iafrate, A.J.; Hamid, M.A.A.; Wang, S.A.; Hasserjian, R.P. High p53 protein expression in therapy-related myeloid neoplasms is associated with adverse karyotype and poor outcome. Mod. Pathol. 2014, 28, 552–563. [Google Scholar] [CrossRef]
- Anensen, N.; Oyan, A.M.; Bourdon, J.-C.; Kalland, K.H.; Bruserud, O.; Gjertsen, B.T. A Distinct p53 Protein Isoform Signature Reflects the Onset of Induction Chemotherapy for Acute Myeloid Leukemia. Clin. Cancer Res. 2006, 12, 3985–3992. [Google Scholar] [CrossRef]
- Madan, V.; Koeffler, H.P. Differentiation therapy of myeloid leukemia: Four decades of development. Haematologica 2020, 106, 1–13. [Google Scholar] [CrossRef]
- Lübbert, M.; Grishina, O.; Schmoor, C.; Schlenk, R.F.; Jost, E.; Crysandt, M.; Heuser, M.; Thol, F.; Salih, H.R.; Schittenhelm, M.M.; et al. Valproate and Retinoic Acid in Combination with Decitabine in Elderly Nonfit Patients With Acute Myeloid Leukemia: Results of a Multicenter, Randomized, 2 × 2, Phase II Trial. J. Clin. Oncol. 2020, 38, 257–270. [Google Scholar] [CrossRef] [PubMed]
- Tassara, M.; Döhner, K.; Brossart, P.; Held, G.; Götze, K.; Horst, H.-A.; Ringhoffer, M.; Köhne, C.-H.; Kremers, S.; Raghavachar, A.; et al. Valproic acid in combination with all-trans retinoic acid and intensive therapy for acute myeloid leukemia in older patients. Blood 2014, 123, 4027–4036. [Google Scholar] [CrossRef] [PubMed]
- Raffoux, E.; Cras, A.; Recher, C.; Boëlle, P.-Y.; De Labarthe, A.; Turlure, P.; Marolleau, J.-P.; Reman, O.; Gardin, C.; Victor, M.; et al. Phase 2 clinical trial of 5-azacitidine, valproic acid, and all-trans retinoic acid in patients with high-risk acute myeloid leukemia or myelodysplastic syndrome. Oncotarget 2010, 1, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Gjertsen, B.T.; Øyan, A.M.; Marzolf, B.; Hovland, R.; Gausdal, G.; Døskeland, S.-O.; Dimitrov, K.; Golden, A.; Kalland, K.-H.; Hood, L.; et al. Analysis of Acute Myelogenous Leukemia: Preparation of Samples for Genomic and Proteomic Analyses. J. Hematother. 2002, 11, 469–481. [Google Scholar] [CrossRef] [PubMed]
- Chrisanthar, R.; Knappskog, S.; Løkkevik, E.; Anker, G.; Østenstad, B.; Lundgren, S.; Berge, E.O.; Risberg, T.; Mjaaland, I.; Mæhle, L.; et al. CHEK2 Mutations Affecting Kinase Activity Together with Mutations in TP53 Indicate a Functional Pathway Associated with Resistance to Epirubicin in Primary Breast Cancer. PLoS ONE 2008, 3, e3062. [Google Scholar] [CrossRef] [PubMed]
- McCormack, E.; Haaland, I.; Venås, G.; Forthun, R.B.; Huseby, S.; Gausdal, G.; Knappskog, S.; Micklem, D.R.; Lorens, J.B.; Bruserud, Ø.; et al. Synergistic induction of p53 mediated apoptosis by valproic acid and nutlin-3 in acute myeloid leukemia. Leukemia 2011, 26, 910–917. [Google Scholar] [CrossRef]
- Ryningen, A.; Stapnes, C.; Lassalle, P.; Corbascio, M.; Gjertsen, B.-T.; Bruserud, Ø. A subset of patients with high-risk acute myelogenous leukemia shows improved peripheral blood cell counts when treated with the combination of valproic acid, theophylline and all-trans retinoic acid. Leuk. Res. 2009, 33, 779–787. [Google Scholar] [CrossRef]
- Reikvam, H.; Hovland, R.; Forthun, R.B.; Erdal, S.; Gjertsen, B.T.; Fredly, H.; Bruserud, Ø. Disease-stabilizing treatment based on all-trans retinoic acid and valproic acid in acute myeloid leukemia—Identification of responders by gene expression profiling of pretreatment leukemic cells. BMC Cancer 2017, 17, 630. [Google Scholar] [CrossRef] [PubMed]
- Van Belle, W.; Anensen, N.; Haaland, I.; Bruserud, Ø.; Høgda, K.-A.; Gjertsen, B.T. Correlation analysis of two-dimensional gel electrophoretic protein patterns and biological variables. BMC Bioinform. 2006, 7, 198. [Google Scholar] [CrossRef][Green Version]
- Xu, J.; Morris, G.F. p53-Mediated Regulation of Proliferating Cell Nuclear Antigen Expression in Cells Exposed to Ionizing Radiation. Mol. Cell. Biol. 1999, 19, 12–20. [Google Scholar] [CrossRef]
- Masuda, Y.; Kanao, R.; Kawai, H.; Kukimoto, I.; Masutani, C. Preferential digestion of PCNA-ubiquitin and p53-ubiquitin linkages by USP7 to remove polyubiquitin chains from substrates. J. Biol. Chem. 2019, 294, 4177–4187. [Google Scholar] [CrossRef]
- Mayo, L.D.; Dixon, J.E.; Durden, D.L.; Tonks, N.K.; Donner, D.B. PTEN Protects p53 from Mdm2 and Sensitizes Cancer Cells to Chemotherapy. J. Biol. Chem. 2002, 277, 5484–5489. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Eng, C. PTEN Autoregulates Its Expression by Stabilization of p53 in a Phosphatase-Independent Manner. Cancer Res. 2006, 66, 736–742. [Google Scholar] [CrossRef]
- Zheng, H.; Ying, H.; Yan, H.; Kimmelman, A.C.; Hiller, D.J.; Chen, A.-J.; Perry, S.R.; Tonon, G.; Chu, G.C.; Ding, Z.; et al. p53 and Pten control neural and glioma stem/progenitor cell renewal and differentiation. Nature 2008, 455, 1129–1133. [Google Scholar] [CrossRef] [PubMed]
- Rivlin, N.; Koifman, G.; Rotter, V. p53 orchestrates between normal differentiation and cancer. Semin. Cancer Biol. 2015, 32, 10–17. [Google Scholar] [CrossRef]
- Marcel, V.; Fernandes, K.; Terrier, O.; Lane, D.P.; Bourdon, J.-C. Modulation of p53β and p53γ expression by regulating the alternative splicing of TP53 gene modifies cellular response. Cell Death Differ. 2014, 21, 1377–1387. [Google Scholar] [CrossRef] [PubMed]
- Fujita, K.; Mondal, A.M.; Horikawa, I.; Nguyen, G.H.; Kumamoto, K.; Sohn, J.J.; Bowman, E.D.; Mathe, E.A.; Schetter, A.J.; Pine, S.R.; et al. p53 isoforms Δ133p53 and p53β are endogenous regulators of replicative cellular senescence. Nat. Cell Biol. 2009, 11, 1135–1142. [Google Scholar] [CrossRef]
- Silden, E.; Hjelle, S.M.; Wergeland, L.; Sulen, A.; Andresen, V.; Bourdon, J.-C.; Micklem, D.R.; McCormack, E.; Gjertsen, B.T. Expression of TP53 Isoforms p53β or p53γ Enhances Chemosensitivity in TP53null Cell Lines. PLoS ONE 2013, 8, e56276. [Google Scholar] [CrossRef] [PubMed]
- De Thé, H. Differentiation therapy revisited. Nat. Rev. Cancer 2018, 18, 117–127. [Google Scholar] [CrossRef]

| Age and Gender | Fab Classification | ||
|---|---|---|---|
| Median age (years) | 63 | M0/1 | 6 |
| Range age (years) | 29/82 | M2 | 4 |
| Female | 11 | M3 | 1 |
| Male | 12 | M4/5 | 10 |
| Total | 21 | ||
| Previous Malignancies | Cytogenetics | ||
| MDS | 2 | Adverse | 3 |
| CML, recidiv | 1 | Favorable | 2 |
| PVR | 1 | Intermediate | 11 |
| MDS, AML relapse | 1 | Unknown | 5 |
| Cancer ovarii | 1 | ||
| Mutations | CD-markers | ||
| NPM1 mutations | CD13 neg | 1 | |
| Mut | 4 | pos | 16 |
| WT | 6 | CD14 neg | 15 |
| n.d. | 11 | pos | 2 |
| FLT3 mutations | CD15 neg | 12 | |
| ITD | 4 | pos | 5 |
| WT | 14 | CD33 neg | 0 |
| n.d. | 3 | pos | 17 |
| TP53 mutations | CD34 neg | 6 | |
| Mut (del) | 2 | pos | 11 |
| WT | 17 | n.d. | 3 |
| n.d. | 2 | ||
| Disease res. | Survival (months) | ||
| Yes | 7 | Median survival | 4 |
| No | 4 | Survival range | 0–36 |
| n.d. | 10 | ||
| Valproic acid response (%) | |||
| Median response | 8.5 | ||
| Range response | −55.6–48.6 | ||
| Age and Gender | Fab Classification | ||
|---|---|---|---|
| Median age (years) | 64 | M0/1 | 11 |
| Range age (years) | 34-82 | M2 | 5 |
| Female | 13 | M3 | 0 |
| Male | 16 | M4/5 | 12 |
| Total | 29 | M6 | 1 |
| Previous Malignancies | Cytogenetics | ||
| MDS | 6 | Adverse | 5 |
| CML, recidiv | 1 | Favorable | 1 |
| AML, recidiv | 1 | Intermediate | 11 |
| PVR | 1 | Unknown | 12 |
| MDS, AML relapse | 1 | ||
| Mutations | CD-markers | ||
| NPM1 mutations | CD13 neg | 3 | |
| Mut | 3 | pos | 22 |
| WT | 14 | CD14 neg | 18 |
| n.d. | 12 | pos | 6 |
| FLT3 mutations | CD15 neg | 10 | |
| ITD | 10 | pos | 11 |
| WT | 15 | CD33 neg | 6 |
| n.d. | 4 | pos | 19 |
| TP53 mutations | CD34 neg | 8 | |
| Mut (del) | 2 | pos | 13 |
| WT | 22 | n.d. | 4 |
| n.d. | 5 | ||
| Disease res. | Survival (months) | ||
| Yes | 5 | Median survival | 5 |
| No | 7 | Survival range | 0–72 |
| n.d. | 17 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Haaland, I.; Hjelle, S.M.; Reikvam, H.; Sulen, A.; Ryningen, A.; McCormack, E.; Bruserud, Ø.; Gjertsen, B.T. p53 Protein Isoform Profiles in AML: Correlation with Distinct Differentiation Stages and Response to Epigenetic Differentiation Therapy. Cells 2021, 10, 833. https://doi.org/10.3390/cells10040833
Haaland I, Hjelle SM, Reikvam H, Sulen A, Ryningen A, McCormack E, Bruserud Ø, Gjertsen BT. p53 Protein Isoform Profiles in AML: Correlation with Distinct Differentiation Stages and Response to Epigenetic Differentiation Therapy. Cells. 2021; 10(4):833. https://doi.org/10.3390/cells10040833
Chicago/Turabian StyleHaaland, Ingvild, Sigrun M. Hjelle, Håkon Reikvam, André Sulen, Anita Ryningen, Emmet McCormack, Øystein Bruserud, and Bjørn Tore Gjertsen. 2021. "p53 Protein Isoform Profiles in AML: Correlation with Distinct Differentiation Stages and Response to Epigenetic Differentiation Therapy" Cells 10, no. 4: 833. https://doi.org/10.3390/cells10040833
APA StyleHaaland, I., Hjelle, S. M., Reikvam, H., Sulen, A., Ryningen, A., McCormack, E., Bruserud, Ø., & Gjertsen, B. T. (2021). p53 Protein Isoform Profiles in AML: Correlation with Distinct Differentiation Stages and Response to Epigenetic Differentiation Therapy. Cells, 10(4), 833. https://doi.org/10.3390/cells10040833








