The Expression and Function of Metastases Associated Lung Adenocarcinoma Transcript-1 Long Non-Coding RNA in Subchondral Bone and Osteoblasts from Patients with Osteoarthritis
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Recruitment
2.2. Isolation and Culture of Primary Osteoblasts
2.3. Analysis of Serum Cytokines
2.4. MALAT1 Functional Studies in Human OA Osteoblasts
2.5. Pathway Analysis
2.6. Statistical Analysis
3. Results
3.1. Expression of MALAT1 in OA Subchondral Bone and Relationship to Patient Characteristics and OA Disease Severity
3.2. MALAT Expression in OA Osteoblasts Is Induced by Inflammatory Challenge
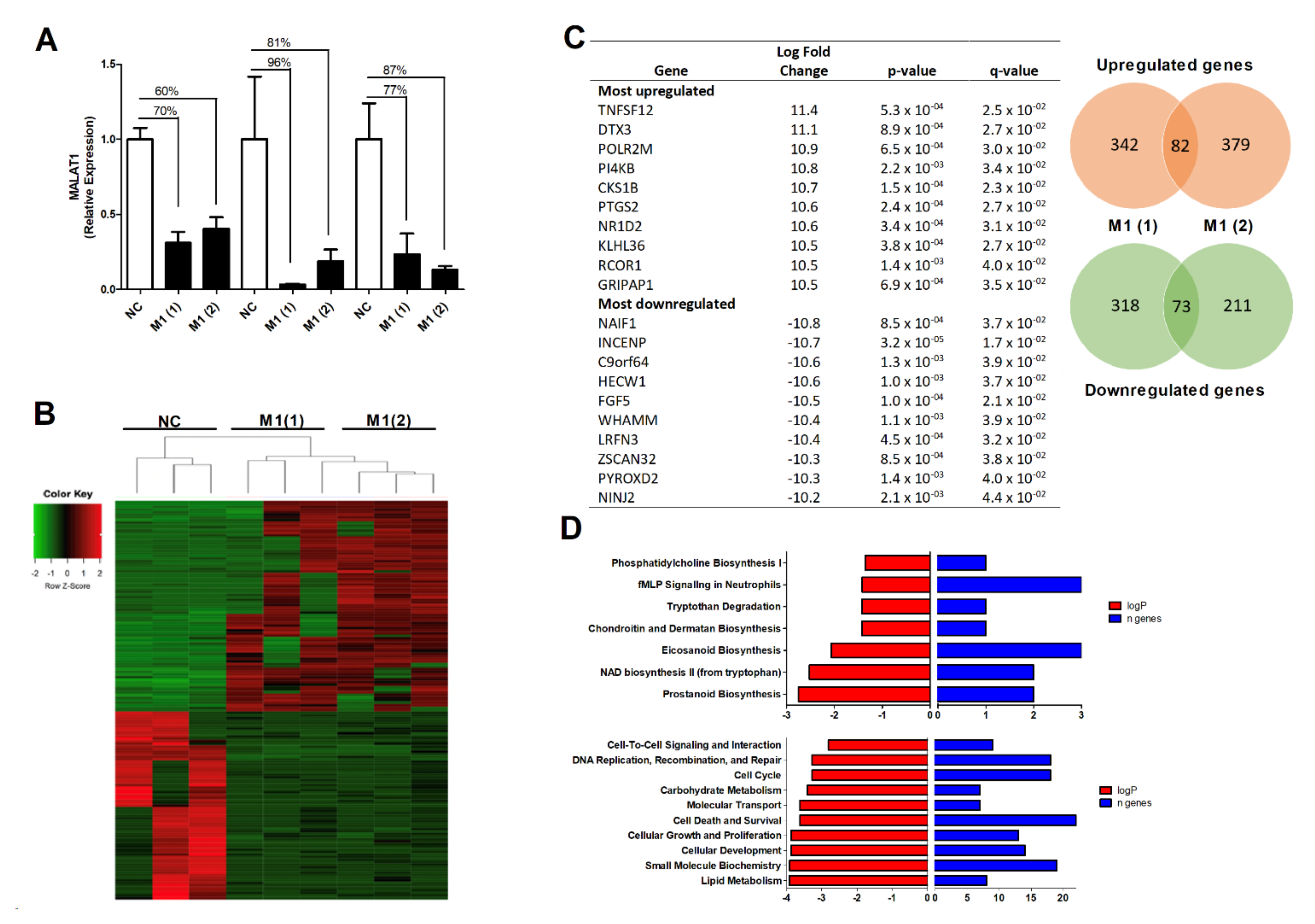
3.3. The Effect of MALAT1 Knockdown on the OA Osteoblast Transcriptome
3.4. Modulation of MALAT1 Expression in Osteoblasts Induced PGE2 Secretion but Did Not Impair Osteoblast OPG Secretion or Their Innate Ability to Mineralise
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Philp, A.M.; Davis, E.T.; Jones, S.W. Developing anti-inflammatory therapeutics for patients with osteoarthritis. Rheumatology 2017, 56, 869–881. [Google Scholar] [CrossRef] [PubMed]
- Tonge, D.P.; Pearson, M.J.; Jones, S.W. The hallmarks of osteoarthritis and the potential to develop personalised disease-modifying pharmacological therapeutics. Osteoarthr. Cartil. 2014, 22, 609–621. [Google Scholar] [CrossRef] [PubMed]
- Hawker, G.A. Osteoarthritis is a serious disease. Clin. Exp. Rheumatol. 2019, 37, 3–6. [Google Scholar] [PubMed]
- Primorac, D.; Molnar, V.; Rod, E.; Jelec, Z.; Cukelj, F.; Matisic, V.; Vrdoljak, T.; Hudetz, D.; Hajsok, H.; Boric, I. Knee Osteoarthritis: A Review of Pathogenesis and State-Of-The-Art Non-Operative Therapeutic Considerations. Genes 2020, 11. [Google Scholar] [CrossRef]
- Dibonaventura, M.; Gupta, S.; McDonald, M.; Sadosky, A. Evaluating the health and economic impact of osteoarthritis pain in the workforce: Results from the National Health and Wellness Survey. BMC Musculoskelet. Disord. 2011, 12, 83. [Google Scholar] [CrossRef]
- Hunter, D.J.; Gerstenfeld, L.; Bishop, G.; Davis, A.D.; Mason, Z.D.; Einhorn, T.A.; Maciewicz, R.A.; Newham, P.; Foster, M.; Jackson, S.; et al. Bone marrow lesions from osteoarthritis knees are characterized by sclerotic bone that is less well mineralized. Arthritis Res. Ther. 2009, 11, R11. [Google Scholar] [CrossRef]
- Philp, A.M.; Collier, R.L.; Grover, L.M.; Davis, E.T.; Jones, S.W. Resistin promotes the abnormal Type I collagen phenotype of subchondral bone in obese patients with end stage hip osteoarthritis. Sci. Rep. 2017, 7, 4042. [Google Scholar] [CrossRef]
- Bailey, A.J.; Sims, T.J.; Knott, L. Phenotypic expression of osteoblast collagen in osteoarthritic bone: Production of type I homotrimer. Int. J. Biochem. Cell Biol. 2002, 34, 176–182. [Google Scholar] [CrossRef]
- Burr, D.B.; Gallant, M.A. Bone remodelling in osteoarthritis. Nat. Rev. Rheumatol. 2012, 8, 665–673. [Google Scholar] [CrossRef]
- Chang, J.; Jackson, S.G.; Wardale, J.; Jones, S.W. Hypoxia modulates the phenotype of osteoblasts isolated from knee osteoarthritis patients, leading to undermineralized bone nodule formation. Arthritis Rheumatol. 2014, 66, 1789–1799. [Google Scholar] [CrossRef]
- Quasnichka, H.L.; Anderson-MacKenzie, J.M.; Bailey, A.J. Subchondral bone and ligament changes precede cartilage degradation in guinea pig osteoarthritis. Biorheology 2006, 43, 389–397. [Google Scholar]
- Radin, E.L.; Rose, R.M. Role of subchondral bone in the initiation and progression of cartilage damage. Clin. Orthop. Relat. Res. 1986, 213, 34–40. [Google Scholar] [CrossRef]
- Khalil, A.M.; Guttman, M.; Huarte, M.; Garber, M.; Raj, A.; Rivea Morales, D.; Thomas, K.; Presser, A.; Bernstein, B.E.; van Oudenaarden, A.; et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. USA 2009, 106, 11667–11672. [Google Scholar] [CrossRef]
- Wang, K.C.; Chang, H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell 2011, 43, 904–914. [Google Scholar] [CrossRef]
- Ilott, N.E.; Heward, J.A.; Roux, B.; Tsitsiou, E.; Fenwick, P.S.; Lenzi, L.; Goodhead, I.; Hertz-Fowler, C.; Heger, A.; Hall, N.; et al. Corrigendum: Long non-coding RNAs and enhancer RNAs regulate the lipopolysaccharide-induced inflammatory response in human monocytes. Nat. Commun. 2015, 6, 6814. [Google Scholar] [CrossRef]
- Roux, B.T.; Heward, J.A.; Donnelly, L.E.; Jones, S.W.; Lindsay, M.A. Catalog of Differentially Expressed Long Non-Coding RNA following Activation of Human and Mouse Innate Immune Response. Front. Immunol. 2017, 8, 1038. [Google Scholar] [CrossRef]
- Wijesinghe, S.N.; Nicholson, T.; Tsintzas, K.; Jones, S.W. Involvements of long noncoding RNAs in obesity-associated inflammatory diseases. Obes. Rev. 2020. [Google Scholar] [CrossRef]
- Hadjicharalambous, M.R.; Roux, B.T.; Feghali-Bostwick, C.A.; Murray, L.A.; Clarke, D.L.; Lindsay, M.A. Long Non-coding RNAs Are Central Regulators of the IL-1beta-Induced Inflammatory Response in Normal and Idiopathic Pulmonary Lung Fibroblasts. Front. Immunol. 2018, 9, 2906. [Google Scholar] [CrossRef]
- Hadjicharalambous, M.R.; Lindsay, M.A. Idiopathic Pulmonary Fibrosis: Pathogenesis and the Emerging Role of Long Non-Coding RNAs. Int. J. Mol. Sci. 2020, 21, 524. [Google Scholar] [CrossRef]
- Hamann, P.D.; Roux, B.T.; Heward, J.A.; Love, S.; McHugh, N.J.; Jones, S.W.; Lindsay, M.A. Transcriptional profiling identifies differential expression of long non-coding RNAs in Jo-1 associated and inclusion body myositis. Sci. Rep. 2017, 7, 8024. [Google Scholar] [CrossRef]
- Pearson, M.J.; Jones, S.W. Review: Long Noncoding RNAs in the Regulation of Inflammatory Pathways in Rheumatoid Arthritis and Osteoarthritis. Arthritis Rheumatol. 2016, 68, 2575–2583. [Google Scholar] [CrossRef]
- Pearson, M.J.; Philp, A.M.; Heward, J.A.; Roux, B.T.; Walsh, D.A.; Davis, E.T.; Lindsay, M.A.; Jones, S.W. Long Intergenic Noncoding RNAs Mediate the Human Chondrocyte Inflammatory Response and Are Differentially Expressed in Osteoarthritis Cartilage. Arthritis Rheumatol. 2016, 68, 845–856. [Google Scholar] [CrossRef]
- Nanus, D.E.; Wijesinghe, S.N.; Pearson, M.J.; Hadjicharalambous, M.R.; Rosser, A.; Davis, E.T.; Lindsay, M.A.; Jones, S.W. Regulation of the Inflammatory Synovial Fibroblast Phenotype by Metastasis-Associated Lung Adenocarcinoma Transcript 1 Long Noncoding RNA in Obese Patients With Osteoarthritis. Arthritis Rheumatol. 2020, 72, 609–619. [Google Scholar] [CrossRef]
- Yang, X.; Zhang, Y.; Li, Y.; Wen, T. MALAT1 enhanced the proliferation of human osteoblasts treated with ultrahigh molecular weight polyethylene by targeting VEGF via miR225p. Int. J. Mol. Med. 2018, 41, 1536–1546. [Google Scholar] [CrossRef]
- Zhu, J.; Zhang, X.; Gao, W.; Hu, H.; Wang, X.; Hao, D. lncRNA/circRNAmiRNAmRNA ceRNA network in lumbar intervertebral disc degeneration. Mol. Med. Rep. 2019, 20, 3160–3174. [Google Scholar] [CrossRef]
- Yi, J.; Liu, D.; Xiao, J. LncRNA MALAT1 sponges miR-30 to promote osteoblast differentiation of adipose-derived mesenchymal stem cells by promotion of Runx2 expression. Cell Tissue Res. 2019, 376, 113–121. [Google Scholar] [CrossRef]
- Wang, Q.; Yang, Q.; Chen, G.; Du, Z.; Ren, M.; Wang, A.; Zhao, H.; Li, Z.; Zhang, G.; Song, Y. LncRNA expression profiling of BMSCs in osteonecrosis of the femoral head associated with increased adipogenic and decreased osteogenic differentiation. Sci. Rep. 2018, 8, 9127. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Qiang, Y.W.; Barlogie, B.; Rudikoff, S.; Shaughnessy, J.D., Jr. Dkk1-induced inhibition of Wnt signaling in osteoblast differentiation is an underlying mechanism of bone loss in multiple myeloma. Bone 2008, 42, 669–680. [Google Scholar] [CrossRef] [PubMed]
- Hurson, C.J.; Butler, J.S.; Keating, D.T.; Murray, D.W.; Sadlier, D.M.; O’Byrne, J.M.; Doran, P.P. Gene expression analysis in human osteoblasts exposed to dexamethasone identifies altered developmental pathways as putative drivers of osteoporosis. BMC Musculoskelet. Disord. 2007, 8, 12. [Google Scholar] [CrossRef] [PubMed]
- Butler, J.S.; Murray, D.W.; Hurson, C.J.; O’Brien, J.; Doran, P.P.; O’Byrne, J.M. The role of Dkk1 in bone mass regulation: Correlating serum Dkk1 expression with bone mineral density. J. Orthop. Res. 2011, 29, 414–418. [Google Scholar] [CrossRef] [PubMed]
- Andersen, H.; Jensen, O.N.; Moiseeva, E.P.; Eriksen, E.F. A proteome study of secreted prostatic factors affecting osteoblastic activity: Galectin-1 is involved in differentiation of human bone marrow stromal cells. J. Bone Miner. Res. 2003, 18, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Muller, J.; Duray, E.; Lejeune, M.; Dubois, S.; Plougonven, E.; Leonard, A.; Storti, P.; Giuliani, N.; Cohen-Solal, M.; Hempel, U.; et al. Loss of Stromal Galectin-1 Enhances Multiple Myeloma Development: Emphasis on a Role in Osteoclasts. Cancers 2019, 11, 261. [Google Scholar] [CrossRef]
- Vincent, C.; Findlay, D.M.; Welldon, K.J.; Wijenayaka, A.R.; Zheng, T.S.; Haynes, D.R.; Fazzalari, N.L.; Evdokiou, A.; Atkins, G.J. Pro-inflammatory cytokines TNF-related weak inducer of apoptosis (TWEAK) and TNFalpha induce the mitogen-activated protein kinase (MAPK)-dependent expression of sclerostin in human osteoblasts. J. Bone Miner. Res. 2009, 24, 1434–1449. [Google Scholar] [CrossRef]
- Du, Y.Y.; Zhao, Y.X.; Liu, Y.P.; Liu, W.; Wang, M.M.; Yuan, C.M. Regulatory Tweak/Fn14 signaling pathway as a potent target for controlling bone loss. Biomed. Pharmacother. 2015, 70, 170–173. [Google Scholar] [CrossRef]
- Yao, C.; Narumiya, S. Prostaglandin-cytokine crosstalk in chronic inflammation. Br. J. Pharmacol. 2019, 176, 337–354. [Google Scholar] [CrossRef]
- Lee, A.S.; Ellman, M.B.; Yan, D.; Kroin, J.S.; Cole, B.J.; van Wijnen, A.J.; Im, H.J. A current review of molecular mechanisms regarding osteoarthritis and pain. Gene 2013, 527, 440–447. [Google Scholar] [CrossRef]
- Li, X.; Ellman, M.; Muddasani, P.; Wang, J.H.; Cs-Szabo, G.; van Wijnen, A.J.; Im, H.J. Prostaglandin E2 and its cognate EP receptors control human adult articular cartilage homeostasis and are linked to the pathophysiology of osteoarthritis. Arthritis Rheum. 2009, 60, 513–523. [Google Scholar] [CrossRef]
- Puljak, L.; Marin, A.; Vrdoljak, D.; Markotic, F.; Utrobicic, A.; Tugwell, P. Celecoxib for osteoarthritis. Cochrane Database Syst. Rev. 2017, 5, CD009865. [Google Scholar] [CrossRef]
- Suri, S.; Gill, S.E.; Massena de Camin, S.; Wilson, D.; McWilliams, D.F.; Walsh, D.A. Neurovascular invasion at the osteochondral junction and in osteophytes in osteoarthritis. Ann. Rheum. Dis. 2007, 66, 1423–1428. [Google Scholar] [CrossRef]
- Barr, A.J.; Campbell, T.M.; Hopkinson, D.; Kingsbury, S.R.; Bowes, M.A.; Conaghan, P.G. A systematic review of the relationship between subchondral bone features, pain and structural pathology in peripheral joint osteoarthritis. Arthritis Res. Ther. 2015, 17, 228. [Google Scholar] [CrossRef]
- Che, W.; Dong, Y.; Quan, H.B. RANKL inhibits cell proliferation by regulating MALAT1 expression in a human osteoblastic cell line hFOB 1.19. Cell Mol. Biol. 2015, 61, 7–14. [Google Scholar]
- Xiao, X.; Zhou, T.; Guo, S.; Guo, C.; Zhang, Q.; Dong, N.; Wang, Y. LncRNA MALAT1 sponges miR-204 to promote osteoblast differentiation of human aortic valve interstitial cells through up-regulating Smad4. Int. J. Cardiol. 2017, 243, 404–412. [Google Scholar] [CrossRef]
- Silva, A.M.; Moura, S.R.; Teixeira, J.H.; Barbosa, M.A.; Santos, S.G.; Almeida, M.I. Long noncoding RNAs: A missing link in osteoporosis. Bone Res. 2019, 7, 10. [Google Scholar] [CrossRef]
- Gao, Y.; Xiao, F.; Wang, C.; Wang, C.; Cui, P.; Zhang, X.; Chen, X. Long noncoding RNA MALAT1 promotes osterix expression to regulate osteogenic differentiation by targeting miRNA-143 in human bone marrow-derived mesenchymal stem cells. J. Cell Biochem. 2018, 119, 6986–6996. [Google Scholar] [CrossRef]


| All OA Patients | Knee OA Patients | Hip OA Patients | |
|---|---|---|---|
| Age (years) | 65 ± 2.2 | 67 ± 3.1 | 63.2 ± 3.2 |
| Gender (m:f) | 9:7 | 4:4 | 5:3 |
| Height (cm) | 167.2 ± 3.2 | 164.8 ± 5.4 | 169.4 ± 3.8 |
| Weight (Kg) | 82.4 ± 4.2 | 84.2 ± 7.9 | 80.8 ± 4.3 |
| BMI (kg/m2) | 29.7 ± 1.6 | 31.1 ± 2.7 | 28.4 ± 1.9 |
| % Fat | 34.8 ± 2.4 | 35.0 ± 5.1 | 34.8 ± 2.3 |
| WHR 1 | 0.92 ± 0.02 | 0.93 ± 0.03 | 0.91 ± 0.03 |
| KL Grade | 4 (3.5–4) | 4 (3.25–4) | 4 (3.5–4) |
| Concentration a | r2 | p-Value | |
|---|---|---|---|
| Visfatin | 2979 ± 355 | 0.11 | 0.23 |
| Resistin | 15441 ± 1394 | 0.17 | 0.18 |
| Leptin | 18365 ± 5865 | 0.03 | 0.55 |
| Adiponectin | 10640 ± 2081 b | <0.01 | 0.94 |
| Chemerin | 6920 ± 937 | <0.01 | 0.67 |
| Dkk1 | 3.7 ± 0.3 b | 0.3 | 0.04 |
| Galectin1 | 41 ± 3 b | 0.44 | 0.009 |
| Eotaxin | 170 ± 44 | 0.15 | 0.16 |
| Amphiregulin | 570 ± 54 | 0.24 | 0.07 |
| Aggrecan | 183 ± 54 | 0.10 | 0.28 |
| FABP4 | 47.4 ± 15.2 b | 0.04 | 0.48 |
| Serpin E1 | 131 ± 19 b | 0.02 | 0.63 |
| IP10 | 32.3 ± 5.9 | 0.03 | 0.56 |
| MIP1a | 400 ± 76 | 0.06 | 0.39 |
| MIP1b | 188 ± 31 | 0.01 | 0.74 |
| MCP1 | 398 ± 38 | <0.01 | 0.94 |
| MIP3a | 37.3 ± 6.8 | 0.07 | 0.38 |
| IL-1β | 17.6 ± 3.1 | 0.007 | 0.78 |
| IL6 | 10.9 ± 4.1 | 0.05 | 0.55 |
| IL7 | 3.4 ± 0.3 | 0.09 | 0.29 |
| IL10 | 4.5 ± 0.3 | 0.01 | 0.69 |
| IL15 | 5.3 ± 1.6 | 0.02 | 0.66 |
| TNF-α | 4.3 ± 0.3 | 0.08 | 0.32 |
| Gp130 | 94 ± 7.1 b | 0.02 | 0.63 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alnajjar, F.A.; Sharma-Oates, A.; Wijesinghe, S.N.; Farah, H.; Nanus, D.E.; Nicholson, T.; Davis, E.T.; Jones, S.W. The Expression and Function of Metastases Associated Lung Adenocarcinoma Transcript-1 Long Non-Coding RNA in Subchondral Bone and Osteoblasts from Patients with Osteoarthritis. Cells 2021, 10, 786. https://doi.org/10.3390/cells10040786
Alnajjar FA, Sharma-Oates A, Wijesinghe SN, Farah H, Nanus DE, Nicholson T, Davis ET, Jones SW. The Expression and Function of Metastases Associated Lung Adenocarcinoma Transcript-1 Long Non-Coding RNA in Subchondral Bone and Osteoblasts from Patients with Osteoarthritis. Cells. 2021; 10(4):786. https://doi.org/10.3390/cells10040786
Chicago/Turabian StyleAlnajjar, Fawzeyah A., Archana Sharma-Oates, Susanne N. Wijesinghe, Hussein Farah, Dominika E. Nanus, Tom Nicholson, Edward T. Davis, and Simon W. Jones. 2021. "The Expression and Function of Metastases Associated Lung Adenocarcinoma Transcript-1 Long Non-Coding RNA in Subchondral Bone and Osteoblasts from Patients with Osteoarthritis" Cells 10, no. 4: 786. https://doi.org/10.3390/cells10040786
APA StyleAlnajjar, F. A., Sharma-Oates, A., Wijesinghe, S. N., Farah, H., Nanus, D. E., Nicholson, T., Davis, E. T., & Jones, S. W. (2021). The Expression and Function of Metastases Associated Lung Adenocarcinoma Transcript-1 Long Non-Coding RNA in Subchondral Bone and Osteoblasts from Patients with Osteoarthritis. Cells, 10(4), 786. https://doi.org/10.3390/cells10040786







