Changes in Biomechanical Properties of A375 Cells Due to the Silencing of TMSB4X Expression Are Not Directly Correlated with Alterations in Their Stemness Features
Abstract
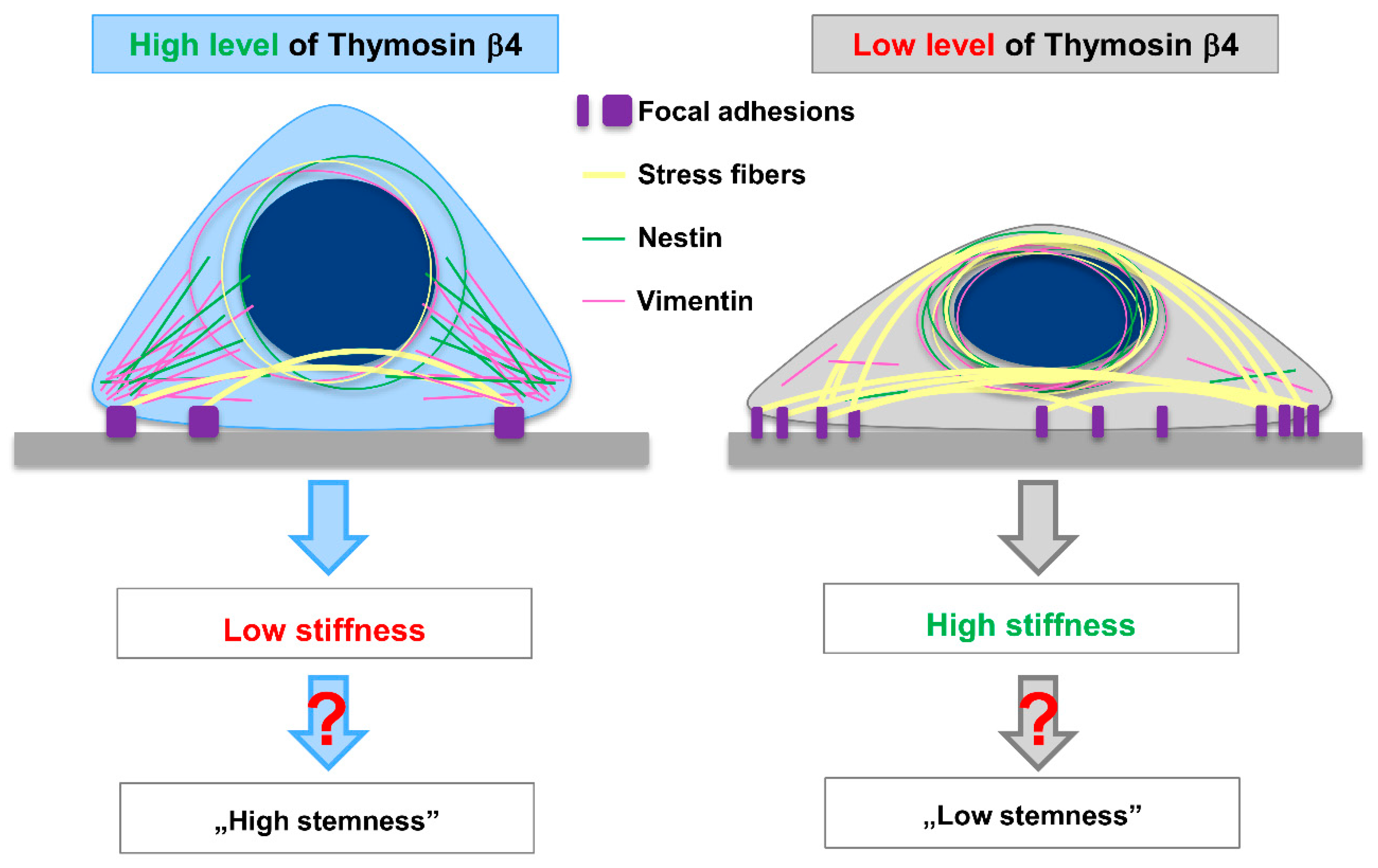
:1. Introduction
2. Materials and Methods
2.1. Cell Lines and Culture Conditions
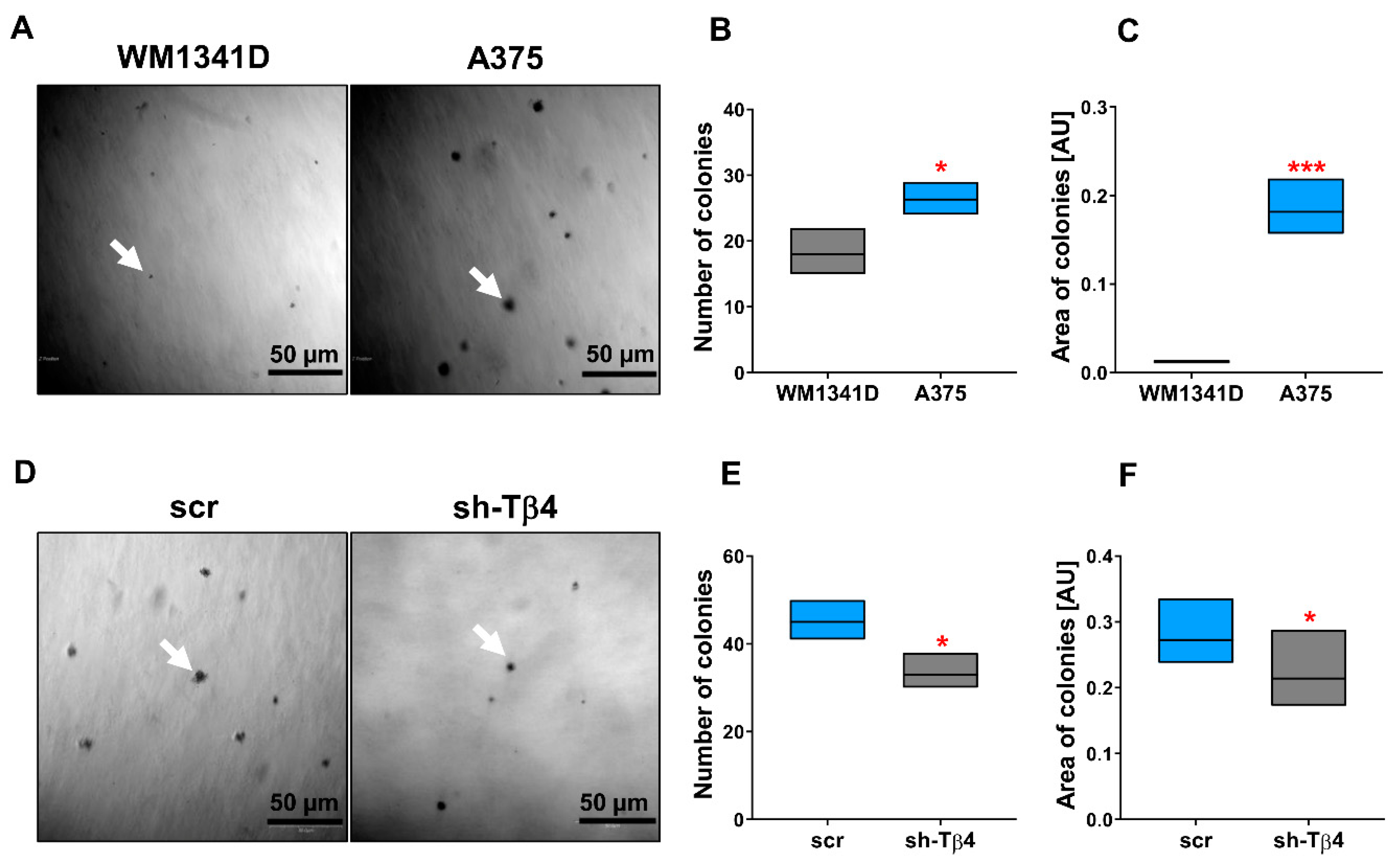
2.2. The Soft Agar Colony Formation Assay
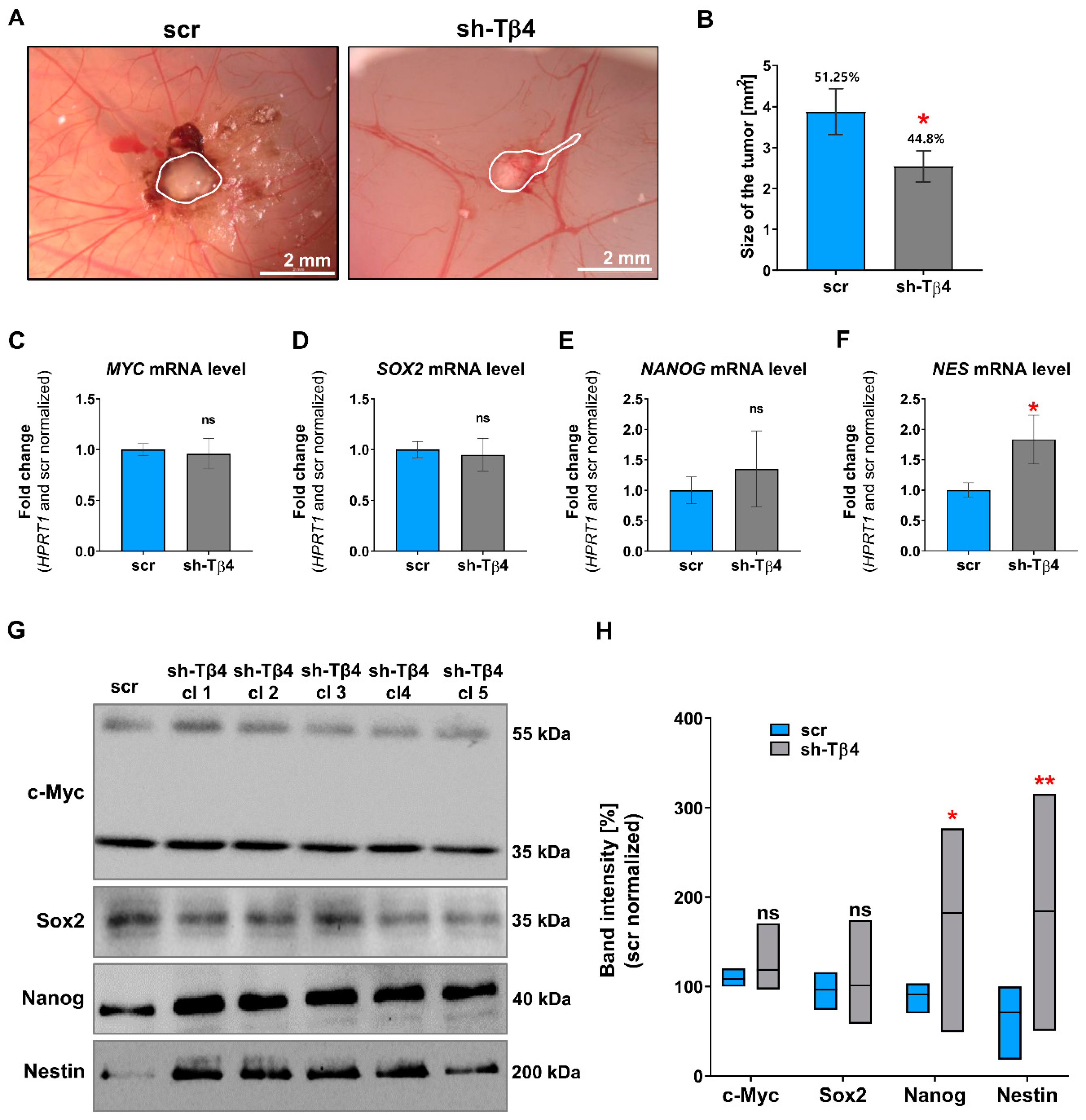
2.3. Chorioallantoic Membrane (CAM) Assay
2.4. Ethics Statement
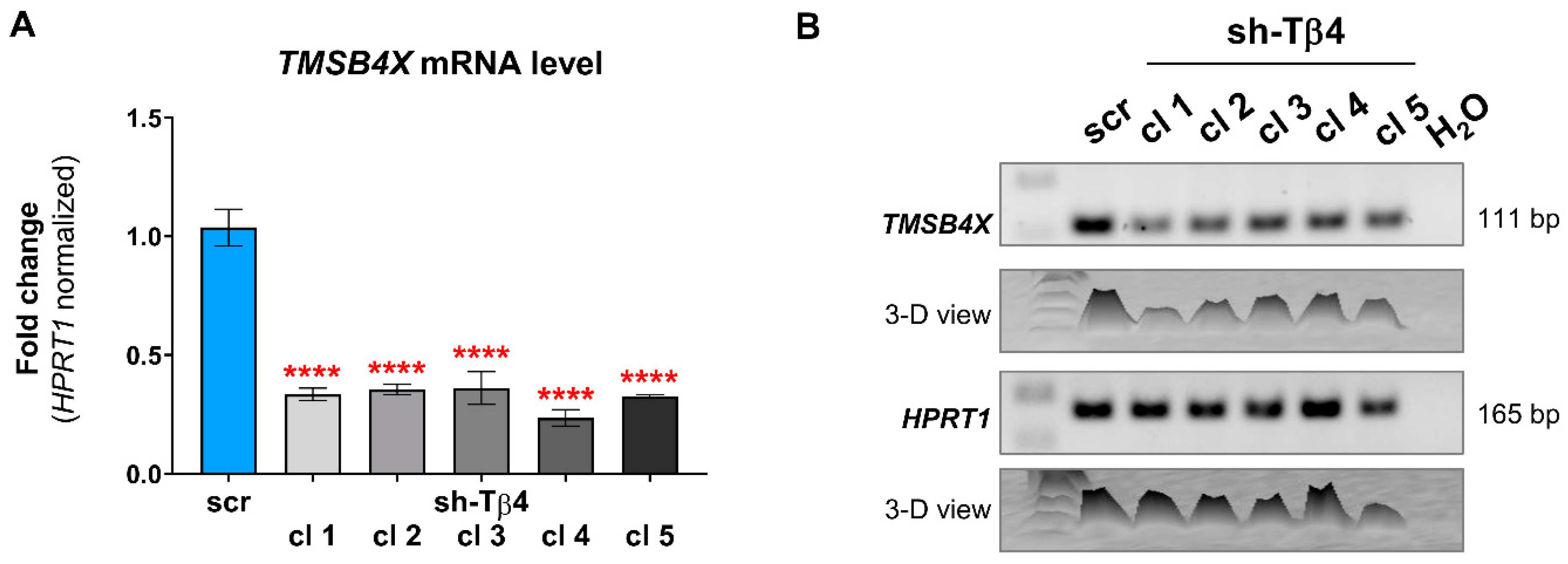
2.5. Quantitative Polymerase Chain Reaction (qPCR) and RT-PCR
2.6. Immunocytochemistry and Confocal Microscopy
2.7. Western Blot Analysis (WB)
2.8. Cell Imaging with Atomic Force Microscopy (AFM)
2.9. Single-Cell Force Spectroscopy (SCFS)
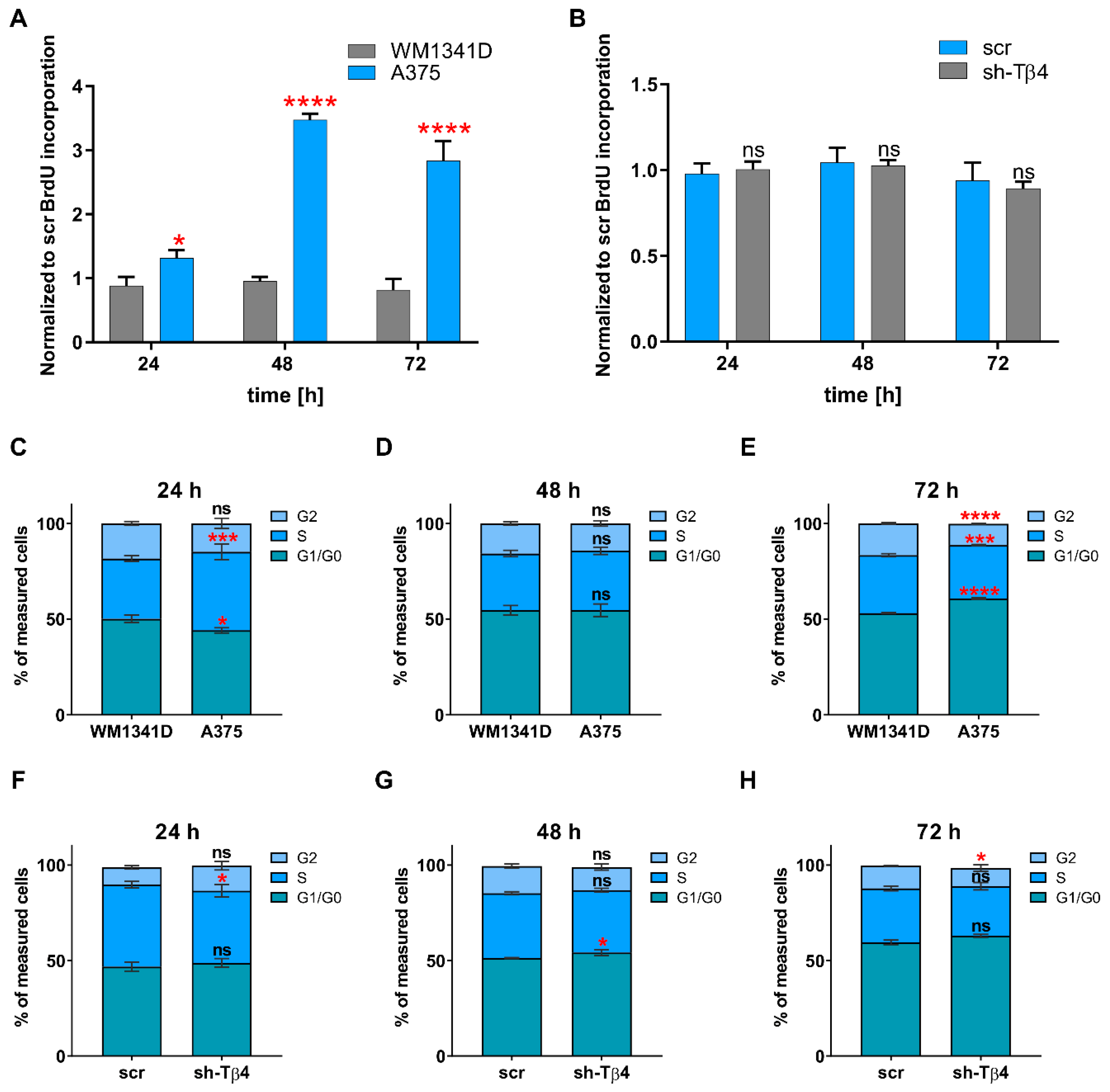
2.10. BrdU Cell Proliferation Assay
2.11. Cell Cycle Analysis
2.12. Statistical Analysis
3. Results
3.1. Some A375 Clones with Low TMSB4X Expression Exhibited an Impaired Ability to Form Colonies in Soft Agar
3.2. Part of A375 Melanoma Cells with Lowered Tβ4 Level Had Diminished Tumorigenic Potential
3.3. Lowering of TMSB4X Expression Did Not Affect Cell Proliferation Rate of A375 Cells
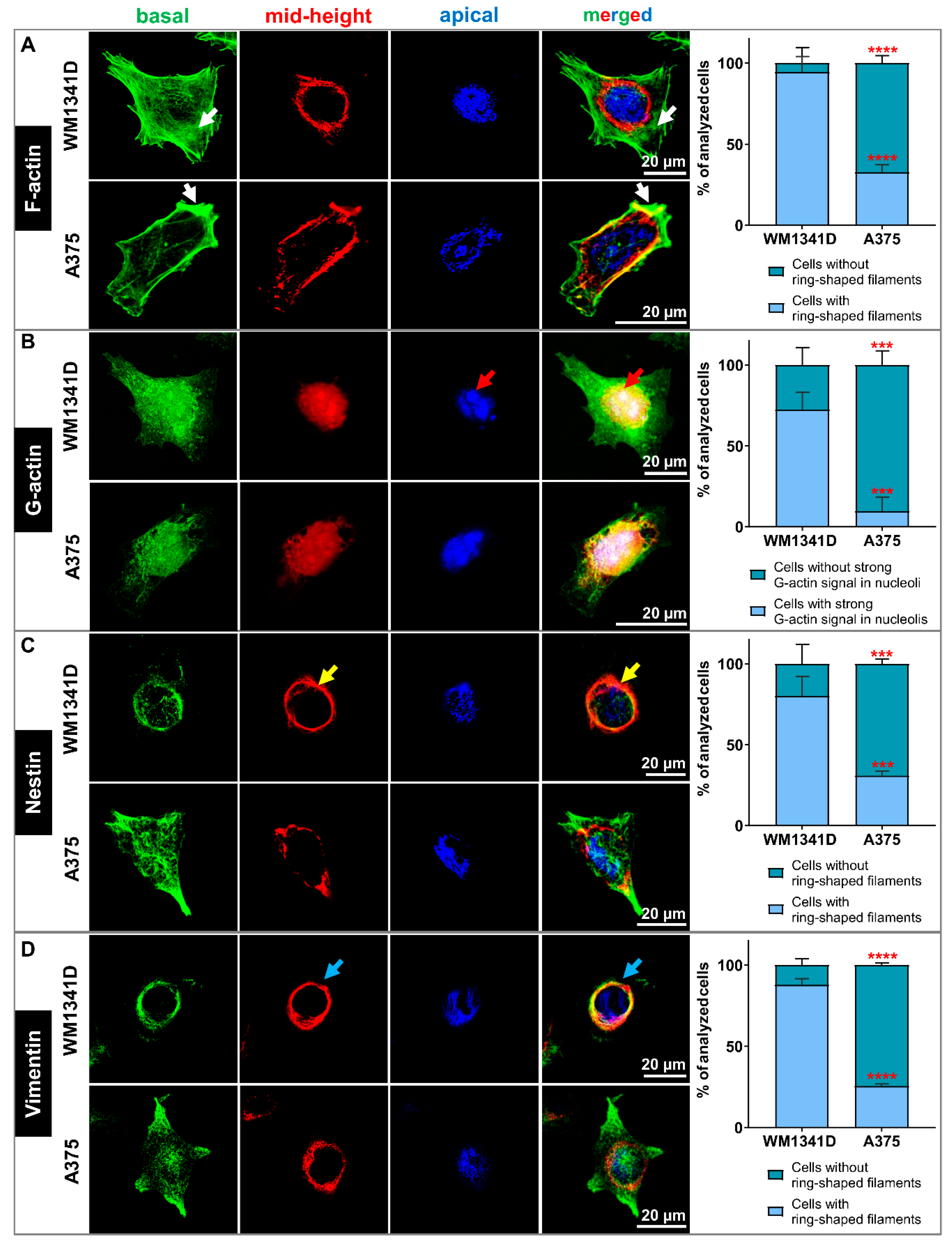
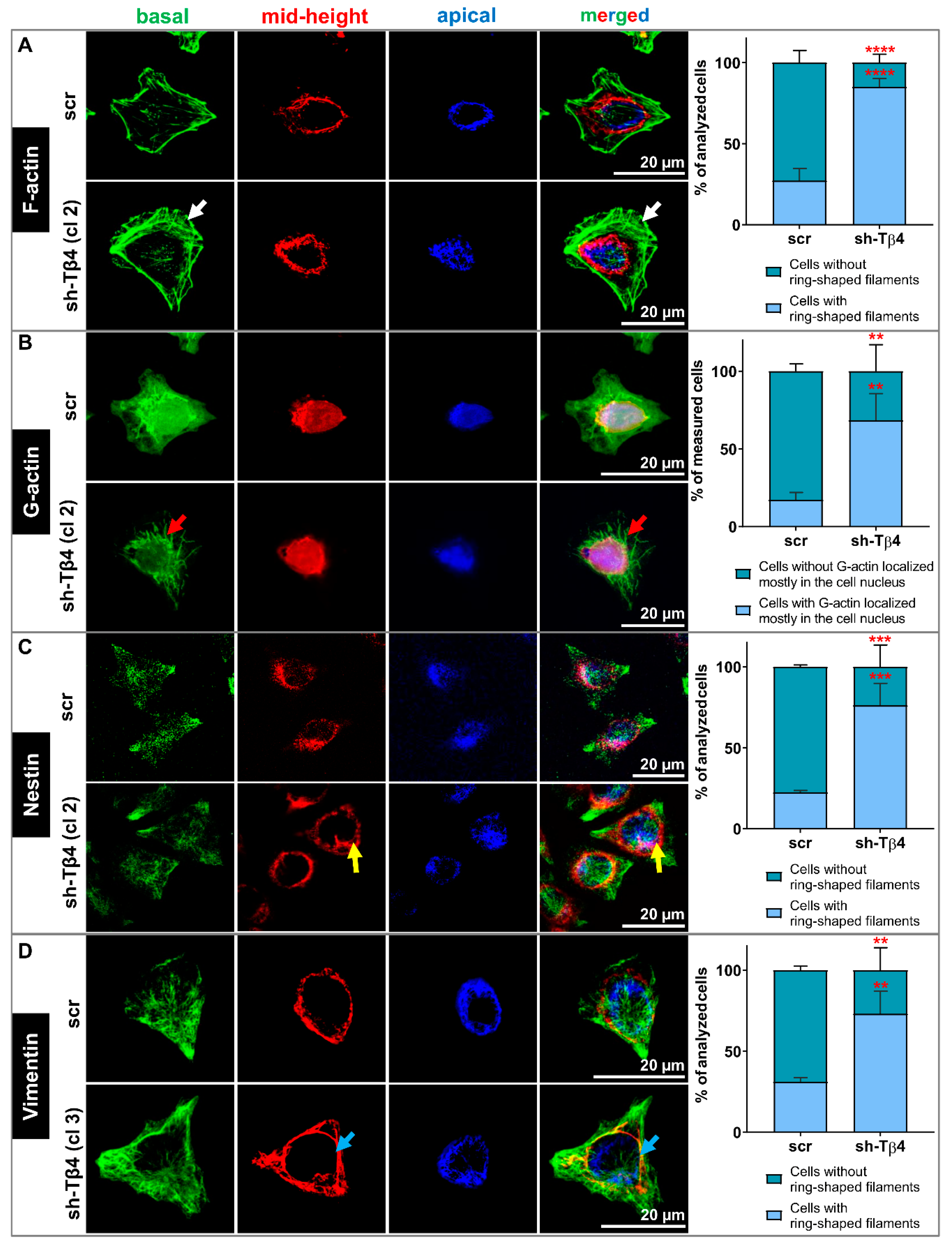
3.4. Lower Tβ4 Level Was Correlated with Alterations in Actin Cytoskeleton and Intermediate Filaments Organization
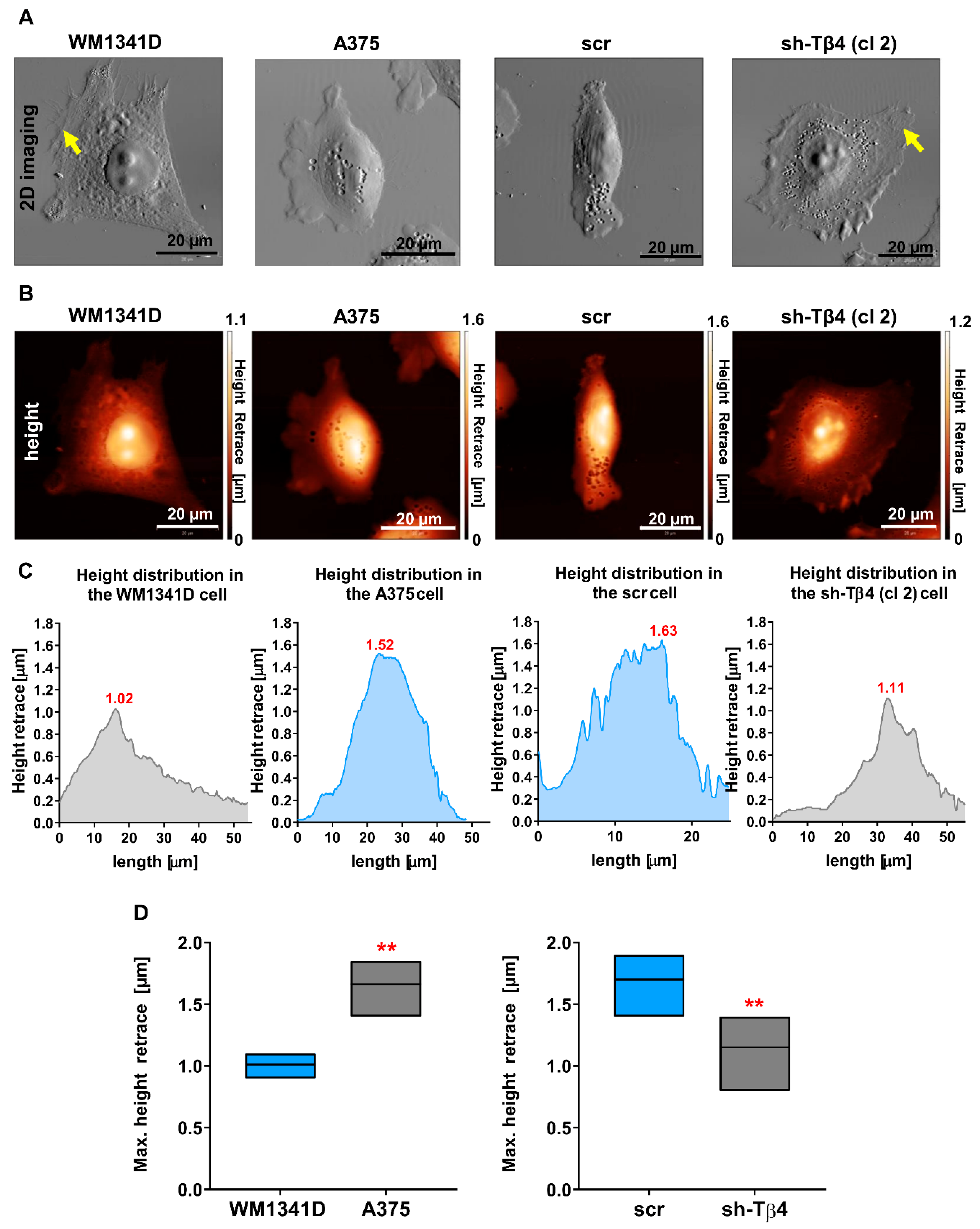
3.5. Tβ4 Influenced the Height of the Melanoma Cells
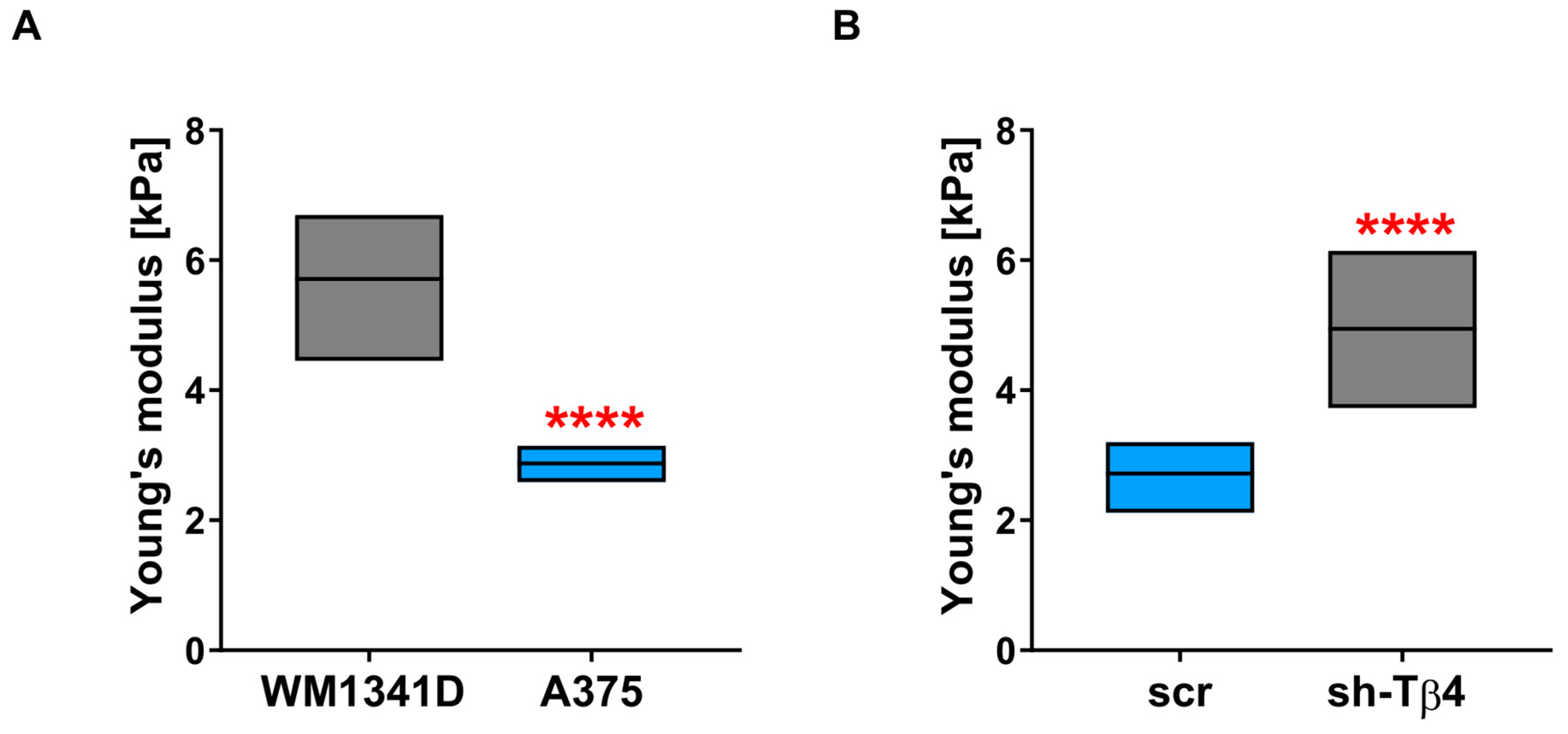
3.6. Lower Level of Tβ4 Was Positively Correlated with Cells Being Stiffer
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AFM | atomic force microscopy |
| CSCs | cancer stem cells |
| DNase | deoxyribonuclease I |
| EMT | epithelial-mesenchymal transition |
| FA | focal adhesion |
| F-actin | filamentous actin |
| G-actin | monomeric actin |
| IFs | intermediate filaments |
| iPSCs | induced pluripotent stem cells |
| mESCs | mouse embryonic stem cells |
| MSCs | mesenchymal stem cells |
| SCFS | single-cell force spectroscopy |
| scr | control A375 cells transfected with scrambled sh-sequence |
| sh-Tβ4 | stable A375 clones with silenced TMSB4X expression |
| Tβ4 | Thymosin β4 |
| TMSB4X | gene coding for Thymosin β4 |
References
- Goldstein, A.; Hannappel, E.; Kleinman, H. Thymosin β: Actin-sequestering protein moonlights to repair injured tissues. Trends Mol. Med. 2005, 11, 421–429. [Google Scholar] [CrossRef]
- Sanders, M.C.; Goldstein, A.L.; Wang, Y.L. Thymosin beta 4 (Fx peptide) is a potent regulator of actin polymerization in living cells. Proc. Natl. Acad. Sci. USA 1992, 89, 4678–4682. [Google Scholar] [CrossRef] [Green Version]
- Malinda, K.M.; Goldstein, A.L.; Kleinman, H.K. Thymosin beta 4 stimulates directional migration of human umbilical vein endothelial cells. FASEB J. 1997, 11, 474–481. [Google Scholar] [CrossRef]
- Crockford, D.; Turjman, N.; Allan, C.; Angel, J. Thymosin β4: Structure, function, and biological properties supporting current and future clinical applications. Ann. N. Y. Acad. Sci. 2010, 1194, 179–189. [Google Scholar] [CrossRef]
- Smart, N.; Risebro, C.A.; Melville, A.A.D.; Moses, K.; Schwartz, R.J.; Chien, K.R.; Riley, P.R. Thymosin β4 induces adult epicardial progenitor mobilization and neovascularization. Nature 2007, 445, 177–182. [Google Scholar] [CrossRef]
- Sosne, G.; Szliter, E.A.; Barrett, R.; Kernacki, K.A.; Kleinman, H.; Hazlett, L.D. Thymosin beta 4 promotes corneal wound healing and decreases inflammation in vivo following alkali injury. Exp. Eye Res. 2002, 74, 293–299. [Google Scholar] [CrossRef]
- Qiu, P.; Wheater, M.K.; Qiu, Y.; Sosne, G. Thymosin beta4 inhibits TNF-alpha-induced NF-kappaB activation, IL-8 expression, and the sensitizing effects by its partners PINCH-1 and ILK. FASEB J. 2011, 25, 1815–1826. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Makowiecka, A.; Malek, N.; Mazurkiewicz, E.; Mrówczyńska, E.; Nowak, D.; Mazur, A.J. Thymosin β4 Regulates Focal Adhesion Formation in Human Melanoma Cells and Affects Their Migration and Invasion. Front. Cell Dev. Biol. 2019, 7, 304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wirsching, H.-G.; Krishnan, S.; Florea, a.-M.; Frei, K.; Krayenbuhl, N.; Hasenbach, K.; Reifenberger, G.; Weller, M.; Tabatabai, G. Thymosin beta 4 gene silencing decreases stemness and invasiveness in glioblastoma. Brain 2014, 137, 433–448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scatena, R. Cancer Stem Cells: A Revisitation of the “Anaplasia” Concept. In Advances in Cancer Stem Cell Biology; Springer: New York, NY, USA, 2012; pp. 1–16. [Google Scholar]
- Kondo, T. Stem Cells and Cancer Stem Cells: New Insights. In Advances in Cancer Stem Cell Biology; Springer: New York, NY, USA, 2012; pp. 17–31. [Google Scholar]
- Fang, D.; Nguyen, T.K.; Leishear, K.; Finko, R.; Kulp, A.N.; Hotz, S.; Van Belle, P.A.; Xu, X.; Elder, D.E.; Herlyn, M. A tumorigenic subpopulation with stem cell properties in melanomas. Cancer Res. 2005, 65, 9328–9337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schatton, T.; Murphy, G.F.; Frank, N.Y.; Yamaura, K.; Waaga-Gasser, A.M.; Gasser, M.; Zhan, Q.; Jordan, S.; Duncan, L.M.; Weishaupt, C.; et al. Identification of cells initiating human melanomas. Nature 2008, 451, 345–349. [Google Scholar] [CrossRef] [Green Version]
- Mollinari, C.; Ricci-Vitiani, L.; Pieri, M.; Lucantoni, C.; Rinaldi, A.M.; Racaniello, M.; De Maria, R.; Zona, C.; Pallini, R.; Merlo, D.; et al. Downregulation of thymosin β4 in neural progenitor grafts promotes spinal cord regeneration. J. Cell Sci. 2009, 122, 4195–4207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ricci-Vitiani, L.; Mollinari, C.; di Martino, S.; Biffoni, M.; Pilozzi, E.; Pagliuca, A.; Chiara de Stefano, M.; Circo, R.; Merlo, D.; De Maria, R.; et al. Thymosin beta4 targeting impairs tumorigenic activity of colon cancer stem cells. FASEB J. 2010, 24, 4291–4301. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.-I.; Lee, B.-Y.; Kang, Y.-J.; Jo, J.-O.; Lee, S.H.; Kim, H.Y.; Kim, Y.-O.; Lee, C.; Koh, S.B.; Kim, A.; et al. Expression Patterns of Thymosin β4 and Cancer Stem Cell Marker CD133 in Ovarian Cancers. Pathol. Oncol. Res. 2013, 19, 237–245. [Google Scholar] [CrossRef]
- Chu, Y.; You, M.; Zhang, J.; Gao, G.; Han, R.; Luo, W.; Liu, T.; Zuo, J.; Wang, F. Adipose-Derived Mesenchymal Stem Cells Enhance Ovarian Cancer Growth and Metastasis by Increasing Thymosin Beta 4X-Linked Expression. Stem Cells Int. 2019, 2019, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Fan, Y.-L.; Zhao, H.-C.; Li, B.; Zhao, Z.-L.; Feng, X.-Q. Mechanical Roles of F-Actin in the Differentiation of Stem Cells: A Review. ACS Biomater. Sci. Eng. 2019, 5, 3788–3801. [Google Scholar] [CrossRef] [PubMed]
- Närvä, E.; Stubb, A.; Guzmán, C.; Blomqvist, M.; Balboa, D.; Lerche, M.; Saari, M.; Otonkoski, T.; Ivaska, J. A Strong Contractile Actin Fence and Large Adhesions Direct Human Pluripotent Colony Morphology and Adhesion. Stem. Cell Rep. 2017, 9, 67–76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pillarisetti, A.; Desai, J.P.; Ladjal, H.; Schiffmacher, A.; Ferreira, A.; Keefer, C.L. Mechanical Phenotyping of Mouse Embryonic Stem Cells: Increase in Stiffness with Differentiation. Cell. Reprogram. 2011, 13, 371–380. [Google Scholar] [CrossRef] [Green Version]
- Xu, W.; Mezencev, R.; Kim, B.; Wang, L.; McDonald, J.; Sulchek, T. Cell Stiffness Is a Biomarker of the Metastatic Potential of Ovarian Cancer Cells. PLoS ONE 2012, 7, e46609. [Google Scholar] [CrossRef] [Green Version]
- Andolfi, L.; Bourkoula, E.; Migliorini, E.; Palma, A.; Pucer, A.; Skrap, M.; Scoles, G.; Beltrami, A.P.; Cesselli, D.; Lazzarino, M. Investigation of Adhesion and Mechanical Properties of Human Glioma Cells by Single Cell Force Spectroscopy and Atomic Force Microscopy. PLoS ONE 2014, 9, e112582. [Google Scholar] [CrossRef] [Green Version]
- Watanabe, T.; Kuramochi, H.; Takahashi, A.; Imai, K.; Katsuta, N.; Nakayama, T.; Fujiki, H.; Suganuma, M. Higher cell stiffness indicating lower metastatic potential in B16 melanoma cell variants and in (-)-epigallocatechin gallate-treated cells. J. Cancer Res. Clin. Oncol. 2012, 138, 859–866. [Google Scholar] [CrossRef]
- Lathia, J.D.; Liu, H. Overview of Cancer Stem Cells and Stemness for Community Oncologists. Target. Oncol. 2017, 12, 387–399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herlyn, M.; Clark, W.H.; Mastrangelo, M.J.; Guerry, D.P.; Elder, D.E.; LaRossa, D.; Hamilton, R.; Bondi, E.; Tuthill, R.; Steplewski, Z.; et al. Specific immunoreactivity of hybridoma-secreted monoclonal anti-melanoma antibodies to cultured cells and freshly derived human cells. Cancer Res. 1980, 40, 3602–3609. [Google Scholar] [PubMed]
- Borowicz, S.; Van Scoyk, M.; Avasarala, S.; Karuppusamy Rathinam, M.K.; Tauler, J.; Bikkavilli, R.K.; Winn, R.A. The Soft Agar Colony Formation Assay. J. Vis. Exp. 2014, 92, 51998. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crespo, P.; Casar, B. The Chick Embryo Chorioallantoic Membrane as an in vivo Model to Study Metastasis. Bio-Protocol 2016, 6, 6. [Google Scholar] [CrossRef] [Green Version]
- Schindelin, J.; Arganda-Carrera, I.; Frise, E.; Verena, K.; Mark, L.; Tobias, P.; Stephan, P.; Curtis, R.; Stephan, S.; Benjamin, S.; et al. Fiji—An Open platform for biological image analysis. Nat. Methods 2009, 9, 676–682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malek, N.; Mrówczyńska, E.; Michrowska, A.; Mazurkiewicz, E.; Pavlyk, I.; Mazur, A.J. Knockout of ACTB and ACTG1 with CRISPR/Cas9(D10A) Technique Shows that Non-Muscle β and γ Actin Are Not Equal in Relation to Human Melanoma Cells’ Motility and Focal Adhesion Formation. Int. J. Mol. Sci. 2020, 21, 2746. [Google Scholar] [CrossRef] [Green Version]
- te Riet, J.; Katan, A.J.; Rankl, C.; Stahl, S.W.; van Buul, A.M.; Phang, I.Y.; Gomez-Casado, A.; Schön, P.; Gerritsen, J.W.; Cambi, A.; et al. Interlaboratory round robin on cantilever calibration for AFM force spectroscopy. Ultramicroscopy 2011, 111, 1659–1669. [Google Scholar] [CrossRef]
- Sneddon, I.N. The relation between load and penetration in the axisymmetric boussinesq problem for a punch of arbitrary profile. Int. J. Eng. Sci. 1965, 3, 47–57. [Google Scholar] [CrossRef]
- Farahani, E.; Patra, H.K.; Jangamreddy, J.R.; Rashedi, I.; Kawalec, M.; Rao Pariti, R.K.; Batakis, P.; Wiechec, E. Cell adhesion molecules and their relation to (cancer) cell stemness. Carcinogenesis 2014, 35, 747–759. [Google Scholar] [CrossRef] [Green Version]
- Ribatti, D.; Tamma, R. The chick embryo chorioallantoic membrane as an in vivo experimental model to study human neuroblastoma. J. Cell. Physiol. 2019, 234, 152–157. [Google Scholar] [CrossRef] [Green Version]
- Hadjimichael, C.; Chanoumidou, K.; Papadopoulou, N.; Arampatzi, P.; Papamatheakis, J.; Kretsovali, A. Common stemness regulators of embryonic and cancer stem cells. World J. Stem. Cells 2015, 7, 1150–1184. [Google Scholar] [PubMed]
- Mazur, A.J.; Morosan-Puopolo, G.; Makowiecka, A.; Malicka-Błaszkiewicz, M.; Nowak, D.; Brand-Saberi, B. Analysis of gelsolin expression pattern in developing chicken embryo reveals high GSN expression level in tissues of neural crest origin. Brain Struct. Funct. 2016, 221, 515–534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dittmer, A.; Dittmer, J. Beta-actin is not a reliable loading control in Western blot analysis. Electrophoresis 2006, 27, 2844–2845. [Google Scholar] [CrossRef] [PubMed]
- Eaton, S.L.; Roche, S.L.; Llavero Hurtado, M.; Oldknow, K.J.; Farquharson, C.; Gillingwater, T.H.; Wishart, T.M. Total protein analysis as a reliable loading control for quantitative fluorescent Western blotting. PLoS ONE 2013, 8, e72457. [Google Scholar] [CrossRef] [PubMed]
- Hann, S.R.; Eisenman, R.N. Proteins encoded by the human c-myc oncogene: Differential expression in neoplastic cells. Mol. Cell. Biol. 1984, 4, 2486–2497. [Google Scholar] [CrossRef]
- Spotts, G.D.; Patel, S.V.; Xiao, Q.; Hann, S.R. Identification of downstream-initiated c-Myc proteins which are dominant-negative inhibitors of transactivation by full-length c-Myc proteins. Mol. Cell. Biol. 1997, 17, 1459–1468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cavanagh, B.L.; Walker, T.; Norazit, A.; Meedeniya, A.C.B. Thymidine analogues for tracking DNA synthesis. Molecules 2011, 16, 7980–7993. [Google Scholar] [CrossRef] [Green Version]
- Huang, W.Q.; Wang, B.H.; Wang, Q.R. Thymosin β4 and AcSDKP inhibit the proliferation of HL-60 cells and induce their differentiation and apoptosis. Cell Biol. Int. 2006, 30, 514–520. [Google Scholar] [CrossRef]
- Moon, E.Y.; Song, J.H.; Yang, K.H. Actin-sequestering protein, thymosin-beta-4 (TB4), inhibits caspase-3 activation in paclitaxel-induced tumor cell death. Oncol. Res. 2007, 16, 507–516. [Google Scholar] [CrossRef] [PubMed]
- Pozarowski, P.; Darzynkiewicz, Z. Analysis of cell cycle by flow cytometry. Methods Mol. Biol. 2004, 281, 301–311. [Google Scholar]
- Bernal, A.; Arranz, L. Nestin-expressing progenitor cells: Function, identity and therapeutic implications. Cell. Mol. Life Sci. 2018, 75, 2177–2195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, D.; Xiang, A.P.; Mao, F.F.; Zhang, L.; Di, C.-G.; Liu, X.-M.; Shao, Y.; Ma, B.-F.; Lee, J.-H.; Ha, K.-S.; et al. Nestin Is Required for the Proper Self-Renewal of Neural Stem Cells. Stem Cells 2010, 28, 2162–2171. [Google Scholar] [CrossRef] [PubMed]
- Khatau, S.B.; Hale, C.M.; Stewart-Hutchinson, P.J.; Patel, M.S.; Stewart, C.L.; Searson, P.C.; Hodzic, D.; Wirtz, D. A perinuclear actin cap regulates nuclear shape. Proc. Natl. Acad. Sci. USA 2009, 106, 19017–19022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiou, Y.-W.; Lin, H.-K.; Tang, M.-J.; Lin, H.-H.; Yeh, M.-L. The influence of physical and physiological cues on atomic force microscopy-based cell stiffness assessment. PLoS ONE 2013, 8, e77384. [Google Scholar]
- Zemła, J.; Bobrowska, J.; Kubiak, A.; Zieliński, T.; Pabijan, J.; Pogoda, K.; Bobrowski, P.; Lekka, M. Indenting soft samples (hydrogels and cells) with cantilevers possessing various shapes of probing tip. Eur. Biophys. J. 2020, 49, 485–495. [Google Scholar] [CrossRef]
- Quintana, E.; Shackleton, M.; Sabel, M.S.; Fullen, D.R.; Johnson, T.M.; Morrison, S.J. Efficient tumour formation by single human melanoma cells. Nature 2008, 456, 593–598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.; Li, M.; Yang, H.; Chai, H.; Fisher, W.; Yao, Q. Roles of Thymosins in Cancers and Other Organ Systems. World J. Surg. 2005, 29, 264–270. [Google Scholar] [CrossRef]
- Clark, E.A.; Golub, T.R.; Lander, E.S.; Hynes, R.O. Genomic analysis of metastasis reveals an essential role for RhoC. Nature 2000, 406, 532–535. [Google Scholar] [CrossRef]
- Zhao, W.; Li, Y.; Zhang, X. Stemness-Related Markers in Cancer. Cancer Transl. Med. 2017, 3, 87. [Google Scholar]
- Perego, M.; Tortoreto, M.; Tragni, G.; Mariani, L.; Deho, P.; Carbone, A.; Santinami, M.; Patuzzo, R.; Della Mina, P.; Villa, A.; et al. Heterogeneous Phenotype of Human Melanoma Cells with In Vitro and In Vivo Features of Tumor-Initiating Cells. J. Investig. Dermatol. 2010, 130, 1877–1886. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dang, C.V. MYC on the Path to Cancer. Cell 2012, 149, 22–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, S.; Li, Y.; Cao, X.; Du, J.; Huang, X. NANOG regulates epithelial–mesenchymal transition and chemoresistance in ovarian cancer. Biosci. Rep. 2017, 37, BSR20160247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ellis, P.; Fagan, B.M.; Magness, S.T.; Hutton, S.; Taranova, O.; Hayashi, S.; McMahon, A.; Rao, M.; Pevny, L. SOX2, a Persistent Marker for Multipotential Neural Stem Cells Derived from Embryonic Stem Cells, the Embryo or the Adult. Dev. Neurosci. 2004, 26, 148–165. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.S.; Tan, J.; Tien, J. Mechanotransduction at cell-matrix and cell-cell contacts. Annu. Rev. Biomed. Eng. 2004, 6, 275–302. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.-Y.; Lin, H.-H.; Tang, M.-J.; Wang, Y.-K. Vimentin contributes to epithelial-mesenchymal transition cancer cell mechanics by mediating cytoskeletal organization and focal adhesion maturation. Oncotarget 2015, 6, 15966–15983. [Google Scholar] [CrossRef] [Green Version]
- Seetharaman, S.; Etienne-Manneville, S. Cytoskeletal Crosstalk in Cell Migration. Trends Cell Biol. 2020, 30, 720–735. [Google Scholar] [CrossRef]
- Serres, M.P.; Samwer, M.; Truong Quang, B.A.; Lavoie, G.; Perera, U.; Görlich, D.; Charras, G.; Petronczki, M.; Roux, P.P.; Paluch, E.K. F-Actin Interactome Reveals Vimentin as a Key Regulator of Actin Organization and Cell Mechanics in Mitosis. Dev. Cell 2020, 52, 210–222.e7. [Google Scholar] [CrossRef] [Green Version]
- Jiu, Y.; Lehtimäki, J.; Tojkander, S.; Cheng, F.; Jäälinoja, H.; Liu, X.; Varjosalo, M.; Eriksson, J.E.; Lappalainen, P. Bidirectional Interplay between Vimentin Intermediate Filaments and Contractile Actin Stress Fibers. Cell Rep. 2015, 11, 1511–1518. [Google Scholar] [CrossRef] [Green Version]
- Yamagishi, A.; Susaki, M.; Takano, Y.; Mizusawa, M.; Mishima, M.; Iijima, M.; Kuroda, S.; Okada, T.; Nakamura, C. The Structural Function of Nestin in Cell Body Softening is Correlated with Cancer Cell Metastasis. Int. J. Biol. Sci. 2019, 15, 1546. [Google Scholar] [CrossRef] [Green Version]
- Ho, J.H.-C.; Ma, W.-H.; Su, Y.; Tseng, K.-C.; Kuo, T.K.-C.; Lee, O.K.-S. Thymosin beta-4 directs cell fate determination of human mesenchymal stem cells through biophysical effects. J. Orthop. Res. 2009, 28, 131–138. [Google Scholar]
- D’Angelo, M.; Benedetti, E.; Tupone, M.G.; Catanesi, M.; Castelli, V.; Antonosante, A.; Cimini, A. The Role of Stiffness in Cell Reprogramming: A Potential Role for Biomaterials in Inducing Tissue Regeneration. Cells 2019, 8, 1036. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gerardo, H.; Lima, A.; Carvalho, J.; Ramos, J.R.D.; Couceiro, S.; Travasso, R.D.M.; Pires das Neves, R.; Grãos, M. Soft culture substrates favor stem-like cellular phenotype and facilitate reprogramming of human mesenchymal stem/stromal cells (hMSCs) through mechanotransduction. Sci. Rep. 2019, 9, 9086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rotsch, C.; Radmacher, M. Drug-induced changes of cytoskeletal structure and mechanics in fibroblasts: An atomic force microscopy study. Biophys. J. 2000, 78, 520–535. [Google Scholar] [CrossRef] [Green Version]
- Wagner, O.I.; Rammensee, S.; Korde, N.; Wen, Q.; Leterrier, J.-F.; Janmey, P.A. Softness, strength and self-repair in intermediate filament networks. Exp. Cell Res. 2007, 313, 2228–2235. [Google Scholar] [CrossRef] [Green Version]
- Sang, X.-B.; Sun, K.-X.; Wang, L.-L.; Chen, S.; Wu, D.-D.; Zong, Z.-H.; Zhao, Y. Effects and mechanism of RhoC downregulation in suppressing ovarian cancer stem cell proliferation, drug resistance, invasion and metastasis. Oncol. Rep. 2016, 36, 3267–3274. [Google Scholar] [CrossRef] [Green Version]
- Rosenthal, D.T.; Zhang, J.; Bao, L.; Zhu, L.; Wu, Z.; Toy, K.; Kleer, C.G.; Merajver, S.D. RhoC impacts the metastatic potential and abundance of breast cancer stem cells. PLoS ONE 2012, 7, e40979. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.; Sharma, S.; Teknos, T.N. RhoC Regulates Cancer Stem Cells in Head and Neck Squamous Cell Carcinoma by Overexpressing IL-6 and Phosphorylation of STAT3. PLoS ONE 2014, 9, e88527. [Google Scholar] [CrossRef]
- Cooper, J.; Giancotti, F.G. Integrin Signaling in Cancer: Mechanotransduction, Stemness, Epithelial Plasticity, and Therapeutic Resistance. Cancer Cell 2019, 35, 347–367. [Google Scholar] [CrossRef] [PubMed]
- Jansen, K.A.; Atherton, P.; Ballestrem, C. Mechanotransduction at the cell-matrix interface. Semin. Cell Dev. Biol. 2017, 71, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Xie, K.; Yang, Y.; Jiang, H. Controlling Cellular Volume via Mechanical and Physical Properties of Substrate. Biophys. J. 2018, 114, 675–687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burridge, K.; Guilluy, C. Focal adhesions, stress fibers and mechanical tension. Exp. Cell Res. 2016, 343, 14–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fusco, S.; Panzetta, V.; Embrione, V.; Netti, P.A. Crosstalk between focal adhesions and material mechanical properties governs cell mechanics and functions. Acta Biomater. 2015, 23, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Kemper, K.; de Goeje, P.L.; Peeper, D.S.; van Amerongen, R. Phenotype switching: Tumor cell plasticity as a resistance mechanism and target for therapy. Cancer Res. 2014, 74, 5937–5941. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pietraszek-Gremplewicz, K.; Simiczyjew, A.; Dratkiewicz, E.; Podgórska, M.; Styczeń, I.; Matkowski, R.; Ziętek, M.; Nowak, D. Expression level of EGFR and MET receptors regulates invasiveness of melanoma cells. J. Cell. Mol. Med. 2019, 23, 8453–8463. [Google Scholar] [CrossRef] [PubMed]
- Mandicourt, G.; Iden, S.; Ebnet, K.; Aurrand-Lions, M.; Imhof, B.A. JAM-C regulates tight junctions and integrin-mediated cell adhesion and migration. J. Biol. Chem. 2007, 282, 1830–1837. [Google Scholar] [CrossRef] [Green Version]
- García-Bartolomé, A.; Peñas, A.; Marín-Buera, L.; Lobo-Jarne, T.; Pérez-Pérez, R.; Morán, M.; Arenas, J.; Martín, M.A.; Ugalde, C. Respiratory chain enzyme deficiency induces mitochondrial location of actin-binding gelsolin to modulate the oligomerization of VDAC complexes and cell survival. Hum. Mol. Genet. 2017, 26, 2493–2506. [Google Scholar] [CrossRef]
- Shen, Y.; Schmidt, B.U.S.; Kubitschke, H.; Morawetz, E.W.; Wolf, B.; Käs, J.A.; Losert, W. Detecting heterogeneity in and between breast cancer cell lines. Cancer Converg. 2020, 4, 1–11. [Google Scholar] [CrossRef]
- Islam, M.; Mezencev, R.; McFarland, B.; Brink, H.; Campbell, B.; Tasadduq, B.; Waller, E.K.; Lam, W.; Alexeev, A.; Sulchek, T. Microfluidic cell sorting by stiffness to examine heterogenic responses of cancer cells to chemotherapy. Cell Death Dis. 2018, 9, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Sato, S.; Rancourt, A.; Sato, Y.; Satoh, M.S. Single-cell lineage tracking analysis reveals that an established cell line comprises putative cancer stem cells and their heterogeneous progeny. Sci. Rep. 2016, 6, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Snyder, D.; Wang, Y.; Kaetzel, D.M. A rare subpopulation of melanoma cells with low expression of metastasis suppressor NME1 is highly metastatic in vivo. Sci. Rep. 2020, 10, 1971. [Google Scholar] [CrossRef] [PubMed]
- Velazquez-Villarreal, E.I.; Maheshwari, S.; Sorenson, J.; Fiddes, I.T.; Kumar, V.; Yin, Y.; Webb, M.G.; Catalanotti, C.; Grigorova, M.; Edwards, P.A.; et al. Single-cell sequencing of genomic DNA resolves sub-clonal heterogeneity in a melanoma cell line. Commun. Biol. 2020, 3, 1–8. [Google Scholar] [CrossRef]
- Freitas, Z.F.O.; Rodrigues, E.G.; Oliveira, V.; Carmona, A.K.; Travassos, L.R. Melanoma heterogeneity: Differential, invasive, metastatic properties and profiles of cathepsin B, D and L activities in subclones of the B16F10-NEX2 cell line. Melanoma Res. 2004, 14, 333–344. [Google Scholar] [CrossRef]
- Pucciarelli, D.; Lengger, N.; Takáčová, M.; Csaderova, L.; Bartosova, M.; Breiteneder, H.; Pastorekova, S.; Hafner, C. Hypoxia increases the heterogeneity of melanoma cell populations and affects the response to vemurafenib. Mol. Med. Rep. 2016, 13, 3281–3288. [Google Scholar] [CrossRef]
- Malek, N.; Michrowska, A.; Mazurkiewicz, E.; Mrówczyńska, E.; Mackiewicz, P.; Mazur, A.J. The origin of the expressed retrotransposed gene ACTBL2 and its influence on human melanoma cells’ motility and focal adhesion formation. Sci. Rep. 2021, 11, 3329. [Google Scholar] [CrossRef]
- Raser, J.M.; O’Shea, E. REVIEW Noise in Gene Expression. Science 2005, 309, 2010–2014. [Google Scholar] [CrossRef] [Green Version]
- Bonny, A.R.; Fonseca, J.P.; Park, J.E.; El-Samad, H. Orthogonal control of mean and variability of endogenous genes in a human cell line. Nat. Commun. 2021, 12, 1–9. [Google Scholar] [CrossRef]
- Ruscetti, M.; Dadashian, E.L.; Guo, W.; Quach, B.; Mulholland, D.J.; Park, J.W.; Tran, L.M.; Kobayashi, N.; Bianchi-Frias, D.; Xing, Y.; et al. HDAC inhibition impedes epithelial-mesenchymal plasticity and suppresses metastatic, castration-resistant prostate cancer. Oncogene 2016, 35, 3781–3795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tripathi, S.; Chakraborty, P.; Levine, H.; Jolly, M.K. A mechanism for epithelial-mesenchymal heterogeneity in a population of cancer cells. PLoS Comput. Biol. 2020, 16, 1–27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fraser, D.; Kærn, M. A chance at survival: Gene expression noise and phenotypic diversification strategies. Mol. Microbiol. 2009, 71, 1333–1340. [Google Scholar] [CrossRef] [PubMed]
- Capp, J.P. Stochastic gene expression stabilization as a new therapeutic strategy for cancer. BioEssays 2012, 34, 170–173. [Google Scholar] [CrossRef] [PubMed]
- Brock, A.; Krause, S.; Ingber, D.E. Control of cancer formation by intrinsic genetic noise and microenvironmental cues. Nat. Rev. Cancer 2015, 15, 499–509. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Makowiecka, A.; Mazurkiewicz, E.; Mrówczyńska, E.; Malek, N.; Battistella, A.; Lazzarino, M.; Nowak, D.; Mazur, A.J. Changes in Biomechanical Properties of A375 Cells Due to the Silencing of TMSB4X Expression Are Not Directly Correlated with Alterations in Their Stemness Features. Cells 2021, 10, 769. https://doi.org/10.3390/cells10040769
Makowiecka A, Mazurkiewicz E, Mrówczyńska E, Malek N, Battistella A, Lazzarino M, Nowak D, Mazur AJ. Changes in Biomechanical Properties of A375 Cells Due to the Silencing of TMSB4X Expression Are Not Directly Correlated with Alterations in Their Stemness Features. Cells. 2021; 10(4):769. https://doi.org/10.3390/cells10040769
Chicago/Turabian StyleMakowiecka, Aleksandra, Ewa Mazurkiewicz, Ewa Mrówczyńska, Natalia Malek, Alice Battistella, Marco Lazzarino, Dorota Nowak, and Antonina Joanna Mazur. 2021. "Changes in Biomechanical Properties of A375 Cells Due to the Silencing of TMSB4X Expression Are Not Directly Correlated with Alterations in Their Stemness Features" Cells 10, no. 4: 769. https://doi.org/10.3390/cells10040769
APA StyleMakowiecka, A., Mazurkiewicz, E., Mrówczyńska, E., Malek, N., Battistella, A., Lazzarino, M., Nowak, D., & Mazur, A. J. (2021). Changes in Biomechanical Properties of A375 Cells Due to the Silencing of TMSB4X Expression Are Not Directly Correlated with Alterations in Their Stemness Features. Cells, 10(4), 769. https://doi.org/10.3390/cells10040769





