miRNAs Potentially Involved in Post Lung Transplant-Obliterative Bronchiolitis: The Role of miR-21-5p
Abstract
1. Introduction
2. Materials and Methods
2.1. Tissue Samples
2.2. miRNAs Selection
2.3. In Situ Hybridization (ISH)
2.4. RNA Extraction and Reverse Trascriptase-Polymerase Chain Reaction
2.5. miRNA Expression Analyses
2.6. Computational Analysis of miR-21-5p Interactors
2.7. Immunohistochemistry
2.8. Cell Culture and miR-21-5p Inhibition
2.9. Western Blot Analysis
2.10. Statistical Analysis
3. Results
3.1. Patient Data
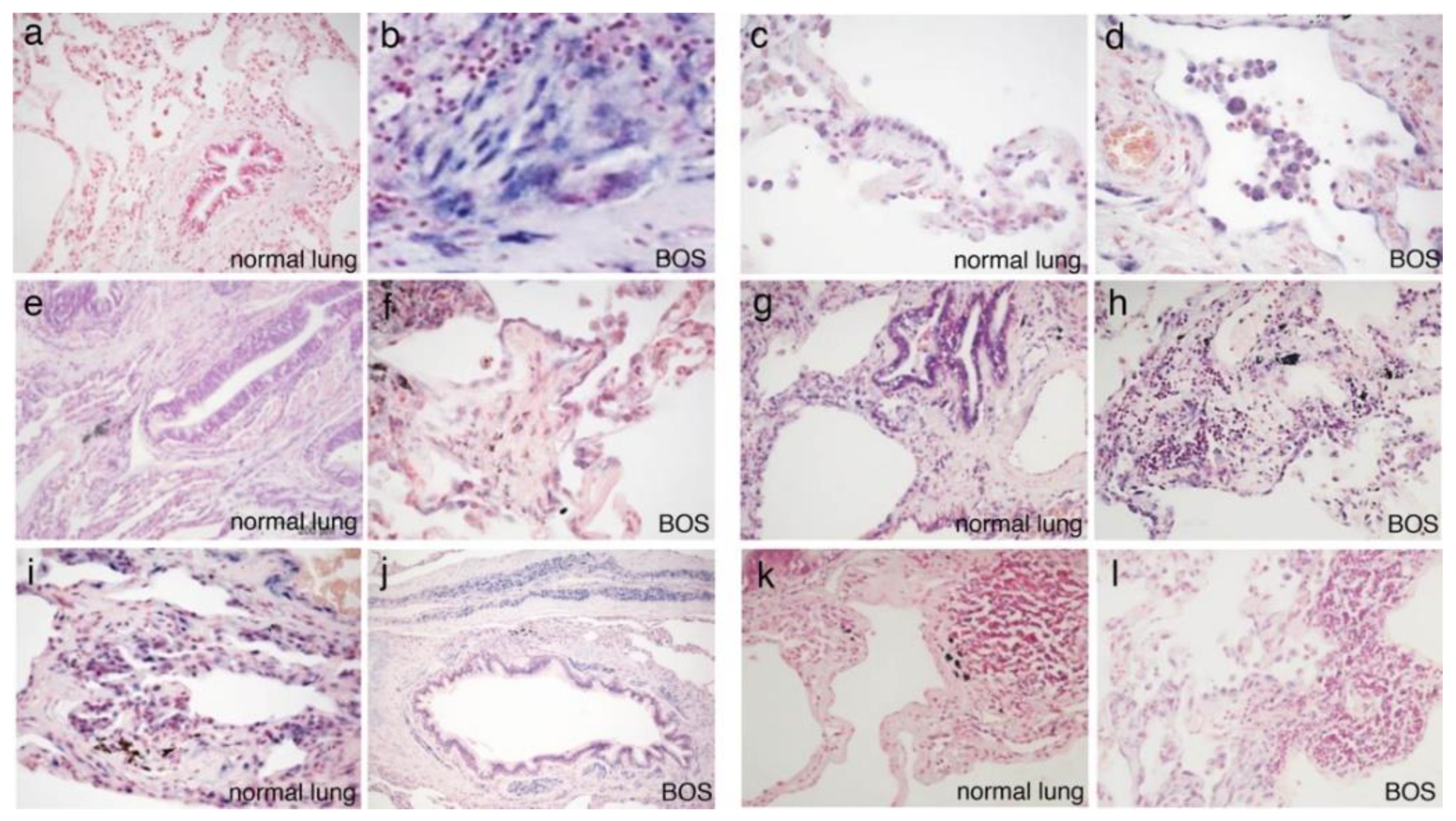
3.2. In Situ Hybridization
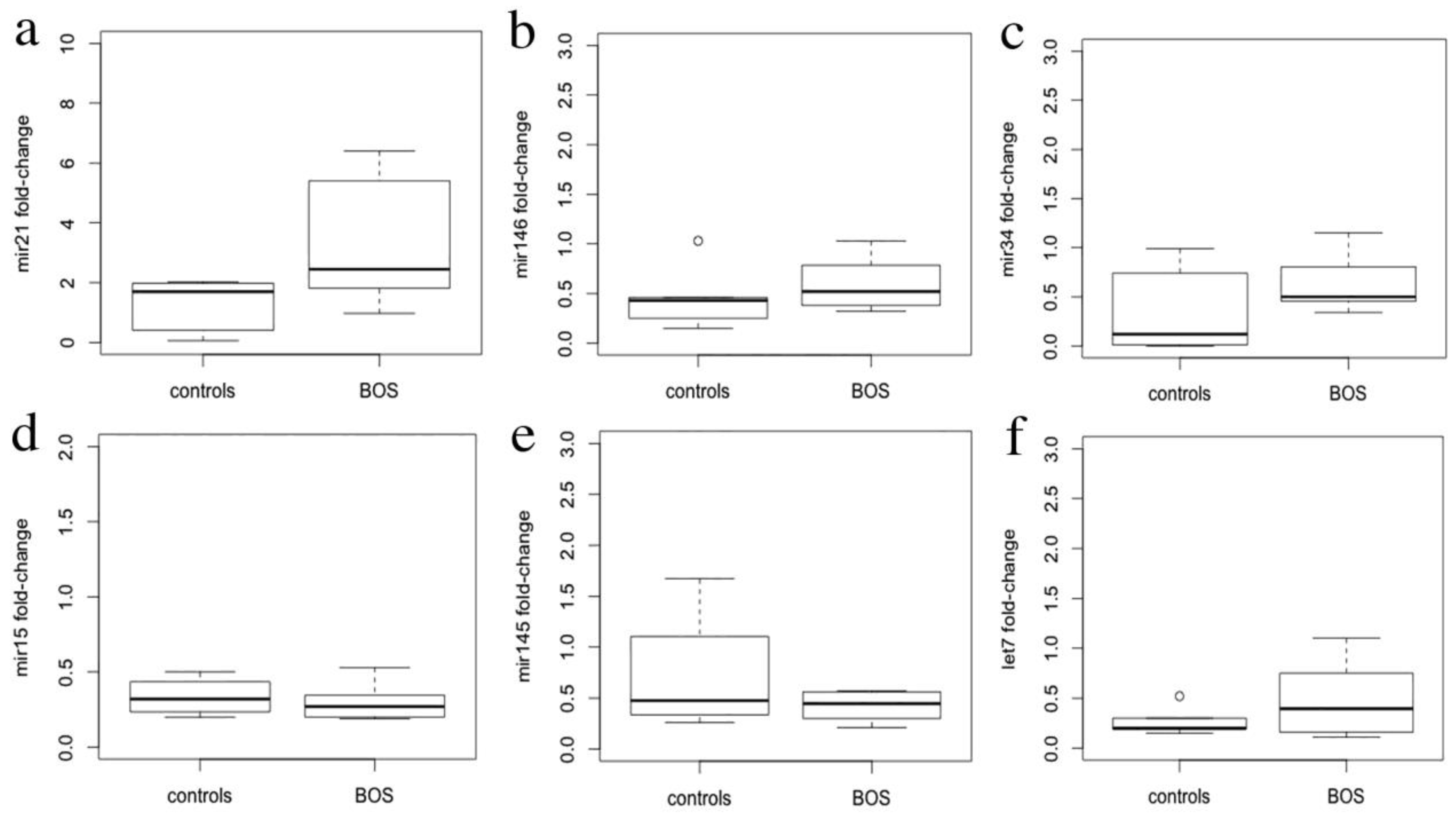
3.3. miRNA Expression Levels
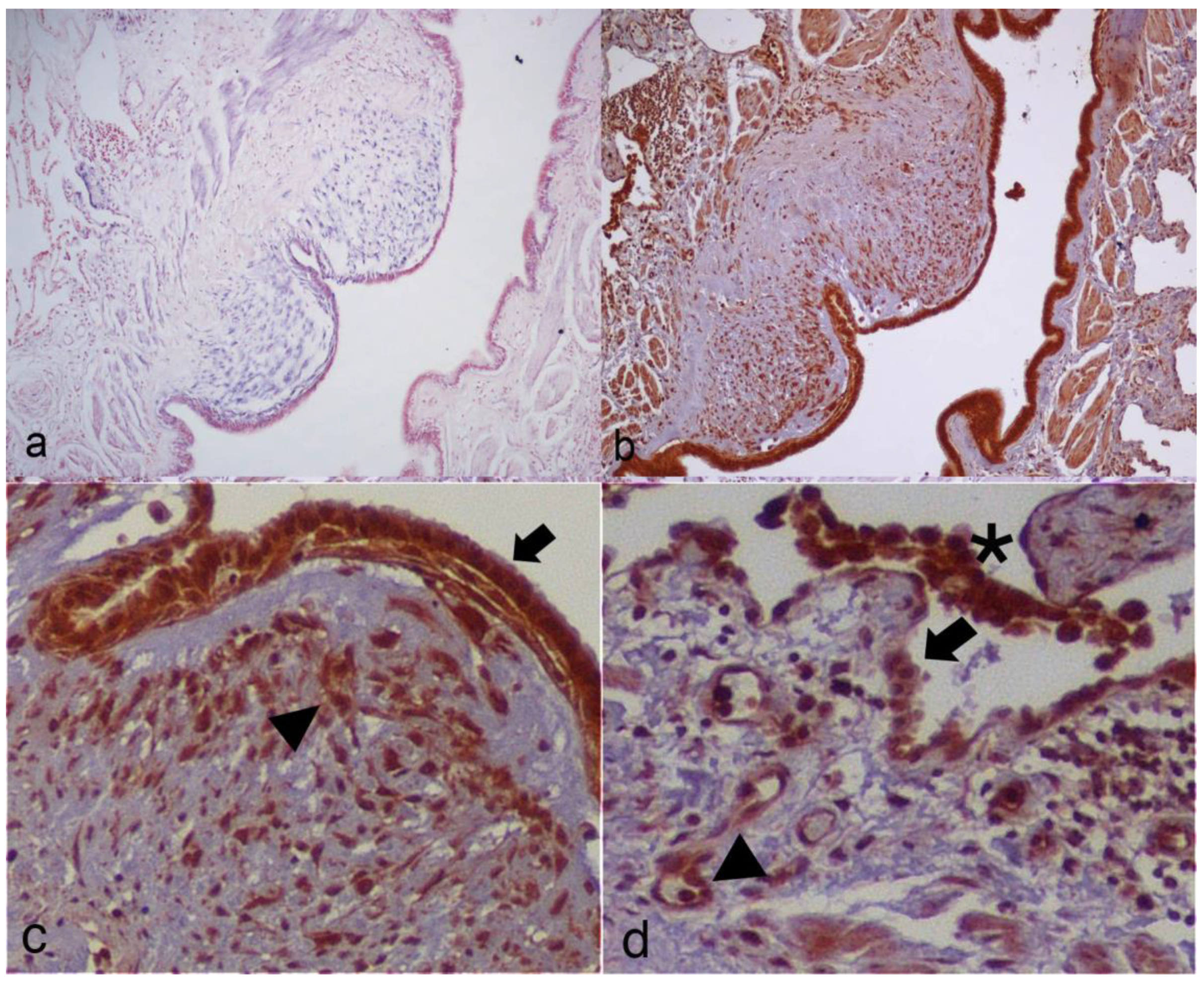
3.4. Computational Identification of miR-21-5p Targets and Immunohistochemistry
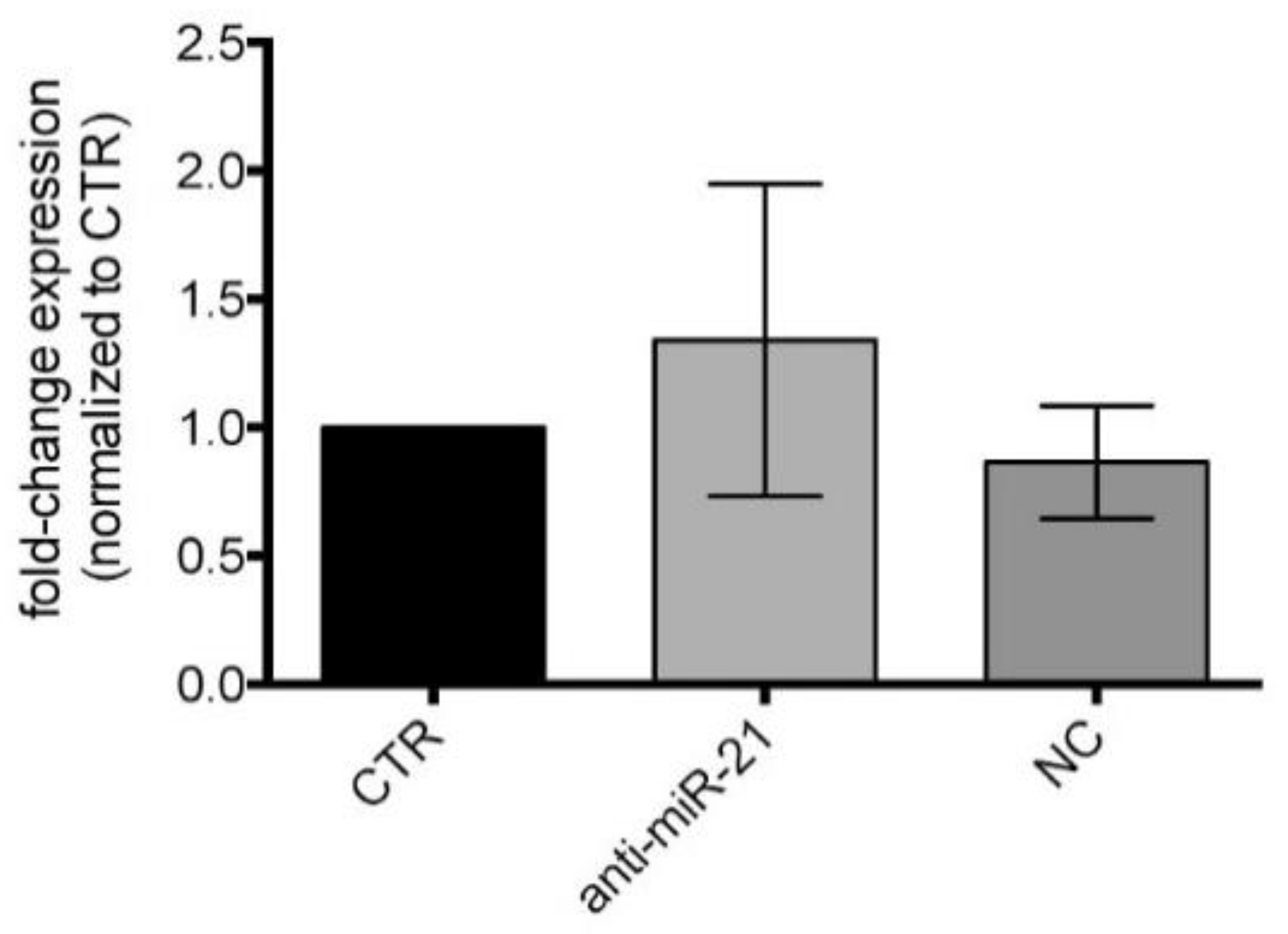
3.5. miR-21-5p Inhibition in Cell Cultures
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Woodrow, J.P.; Shlobin, O.A.; Barnett, S.D.; Burton, N.; Nathan, S.D. Comparison of bronchiolitis obliterans syndrome to other forms of chronic lung allograft dysfunction after lung transplantation. J. Heart Lung Transpl. 2010, 29, 1159–1164. [Google Scholar] [CrossRef]
- Hogg, J.C.; Paré, P.D.; Hackett, T.L. The Contribution of Small Airway Obstruction to the Pathogenesis of Chronic Obstructive Pulmonary Disease. Physiol. Rev. 2017, 97, 529–552. [Google Scholar] [CrossRef]
- Todd, J.; Palmer, S.M. Bronchiolitis obliterans syndrome: The final frontier for lung transplantation. Chest 2011, 140, 502–508. [Google Scholar] [CrossRef] [PubMed]
- Vella, S.; Conaldi, P.G.; Cova, E.; Meloni, F.; Liotta, R.; Cuzzocrea, S.; Martino, L.; Bertani, A.; Luca, A.; Vitulo, P. Lung resident mesenchymal cells isolated from patients with the Bronchiolitis Obliterans Syndrome display a deregulated epigenetic profile. Sci. Rep. 2018, 8, 11167. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C. MicroRNomics: A newly emerging approach for disease biology. Physiol. Genom. 2008, 33, 139–147. [Google Scholar] [CrossRef]
- Poddar, S.; Kesharwani, D.; Datta, M. Interplay between the miRNome and the epigenetic machinery: Implications in health and disease. J. Cell Physiol. 2017, 232, 2938–2945. [Google Scholar] [CrossRef] [PubMed]
- O’Reilly, S. MicroRNAs in fibrosis: Opportunities and challenges. Arthritis Res. Ther. 2016, 18, 11. [Google Scholar] [CrossRef]
- Pandit, K.V.; Milosevic, J.; Kaminski, N. MicroRNAs in idiopathic pulmonary fibrosis. Transl. Res. 2011, 157, 191–199. [Google Scholar] [CrossRef]
- Chen, Z.; Li, Y.; Dian, K.; Rao, L. Modulating microRNAs as Novel Therapeutic Targets in Cardiac Fibrosis. Theranostics 2017, 7, 2287–2288. [Google Scholar] [CrossRef][Green Version]
- Petrovic, N.; Ergun, S. miRNAs as Potential Treatment Targets and Treatment Options in Cancer. Mol. Diagn. Ther. 2018, 22, 157–168. [Google Scholar] [CrossRef]
- Di Carlo, S.; Rossi, E.; Politano, G.; Inghilleri, S.; Morbini, P.; Calabrese, F.; Benso, A.; Savino, A.; Cova, E.; Zampieri, D.; et al. Identification of miRNAs Potentially Involved in Bronchiolitis Obliterans Syndrome: A Computational Study. PLoS ONE 2016, 11, e0161771. [Google Scholar] [CrossRef] [PubMed]
- Yousem, S.A.; Berry, G.J.; Cagle, P.T.; Chamberlain, D.; Husain, A.N.; Hruban, R.H.; Marchevsky, A.; Ohori, N.P.; Ritter, J.; Stewart, S.; et al. Revision of the 1990 working formulation for the classification of pulmonary allograft rejection: Lung Rejection Study Group. J. Heart Lung Transpl. 1996, 15, 1–15. [Google Scholar]
- Verleden, S.E.; Vos, R.; Vanaudenaerde, B.M.; Verleden, G.M. Chronic lung allograft dysfunction phenotypes and treatment. J. Thorac. Dis. 2017, 9, 2650–2659. [Google Scholar] [CrossRef] [PubMed]
- Gauthier, J.M.; Hachem, R.R.; Kreisel, D. Update on Chronic Lung Allograft Dysfunction. Curr. Transpl. Rep. 2016, 3, 185–191. [Google Scholar] [CrossRef]
- Song, X.; Cao, G.; Jing, L.; Lin, S.; Wang, X.; Zhang, J.; Wang, M.; Liu, W.; Lv, C. Analysing the relationship between lncRNA and protein-coding gene and the role of lncRNA as ceRNA in pulmonary fibrosis. J. Cell Mol. Med. 2014, 18, 991–1003. [Google Scholar] [CrossRef]
- Politano, G.; Orso, F.; Raimo, M.; Benso, A.; Savino, A.; Taverna, D.; Di Carlo, S. CyTransfinder: A Cytoscape 3.3 plugin for three-component (TF, gene, miRNA) signal transduction pathway construction. BMC Bioinform. 2016, 17, 157. [Google Scholar] [CrossRef] [PubMed]
- Cova, E.; Colombo, M.; Inghilleri, S.; Morosini, M.; Miserere, S.; Peñaranda-Avila, J.; Santini, B.; Piloni, D.; Magni, S.; Gramatica, F.; et al. Antibody-engineered nanoparticles selectively inhibit mesenchymal cells isolated from patients with chronic lung allograft dysfunction. Nanomedicine 2015, 10, 9–23. [Google Scholar] [CrossRef] [PubMed]
- Bauer-Mehren, A.; Rautschka, M.; Sanz, F.; Furlong, L.I. DisGeNET: A Cytoscape plugin to visualize, integrate, search and analyze gene-disease networks. Bioinformatics 2010, 26, 2924–2926. [Google Scholar] [CrossRef]
- Kasembeli, M.M.; Bharadwaj, U.; Robinson, P.; Tweardy, D.J. Contribution of STAT3 to Inflammatory and Fibrotic Diseases and Prospects for its Targeting for Treatment. Int. J. Mol. Sci. 2018, 19, 2299. [Google Scholar] [CrossRef] [PubMed]
- Honeyman, L.; Bazett, M.; Tomko, T.G.; Haston, C.K. MicroRNA profiling implicates the insulin-like growth factor pathway in bleomycin-induced pulmonary fibrosis in mice. Fibrogenesis Tissue Repair 2013, 6, 16. [Google Scholar] [CrossRef] [PubMed]
- Prêle, C.M.; Yao, E.; O’Donoghue, R.J.; Mutsaers, S.E.; Knight, D.A. STAT3: A central mediator of pulmonary fibrosis? Proc. Am. Thorac. Soc. 2012, 9, 177–182. [Google Scholar] [CrossRef]
- Iliopoulos, D.; Jaeger, S.A.; Hirsch, H.A.; Bulyk, M.L.; Struhl, K. STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Mol. Cell. 2010, 39, 493–506. [Google Scholar] [CrossRef]
- Yamada, M.; Kubo, H.; Ota, C.; Takahashi, T.; Tando, Y.; Suzuki, T.; Fujino, N.; Makiguchi, T.; Takagi, K.; Suzuki, T.; et al. The increase of microRNA-21 during lung fibrosis and its contribution to epithelial-mesenchymal transition in pulmonary epithelial cells. Respir. Res. 2013, 14, 95. [Google Scholar] [CrossRef]
- Liu, G.; Friggeri, A.; Yang, Y.; Milosevic, J.; Ding, Q.; Thannickal, V.J.; Kaminski, N.; Abraham, E. miR-21 mediates fibrogenic activation of pulmonary fibroblasts and lung fibrosis. J. Exp. Med. 2010, 207, 1589–1597. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Zhao, G.Q.; Chen, T.F.; Chang, J.X.; Wang, H.Q.; Chen, S.S.; Zhang, G.J. Serum miR-21 and miR-155 expression in idiopathic pulmonary fibrosis. J. Asthma 2013, 50, 960–964. [Google Scholar] [CrossRef]
- Makiguchi, T.; Yamada, M.; Yoshioka, Y.; Sugiura, H.; Koarai, A.; Chiba, S.; Fujino, N.; Tojo, Y.; Ota, C.; Kubo, H.; et al. Serum extracellular vesicular miR-21-5p is a predictor of the prognosis in idiopathic pulmonary fibrosis. Respir. Res. 2016, 17, 110. [Google Scholar] [CrossRef] [PubMed]
- Budding, K.; Rossato, M.; van de Graaf, E.A.; Kwakkel-Van Erp, J.M.; Radstake, T.; Otten, H.G. Serum miRNAs as potential biomarkers for the bronchiolitis obliterans syndrome after lung transplantation. Transpl. Immunol. 2017, 42, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Gong, Q.; Lin, Z.; Wang, Y.; Li, M.; Wang, L.; Ding, H.; Li, P. Emerging Roles of SRSF3 as a Therapeutic Target for Cancer. Front. Oncol. 2020, 10, 577636. [Google Scholar] [CrossRef]
- Zhou, F.; Wang, Y.; Liu, H.; Ready, N.; Han, Y.; Hung, R.J.; Brhane, Y.; McLaughlin, J.; Brennan, P.; Bickeböller, H.; et al. Susceptibility loci of CNOT6 in the general mRNA degradation pathway and lung cancer risk-A re-analysis of eight GWASs. Mol. Carcinog. 2017, 56, 1227–1238. [Google Scholar] [CrossRef]
- Sutterlüty, H.; Mayer, C.E.; Setinek, U.; Attems, J.; Ovtcharov, S.; Mikula, M.; Mikulits, W.; Micksche, M.; Berger, W. Down-regulation of Sprouty2 in non-small cell lung cancer contributes to tumor malignancy via extracellular signal-regulated kinase pathway-dependent and -independent mechanisms. Mol. Cancer Res. 2007, 5, 509–520. [Google Scholar] [CrossRef] [PubMed]
- Huffman, N.; Palmieri, D.; Coppola, V. The CTLH Complex in Cancer Cell Plasticity. J. Oncol. 2019, 2019, 4216750. [Google Scholar] [CrossRef]
- Chakraborty, D.; Šumová, B.; Mallano, T.; Chen, C.W.; Distler, A.; Bergmann, C.; Ludolph, I.; Horch, R.E.; Gelse, K.; Ramming, A.; et al. Activation of STAT3 integrates common profibrotic pathways to promote fibroblast activation and tissue fibrosis. Nat. Commun. 2017, 8, 1130. [Google Scholar] [CrossRef]
- Pedroza, M.; To, S.; Assassi, S.; Wu, M.; Tweardy, D.; Agarwal, S.K. Role of STAT3 in skin fibrosis and transforming growth factor beta signalling. Rheumatology 2018, 57, 1838–1850. [Google Scholar] [CrossRef]
- Seo, H.Y.; Jeon, J.H.; Jung, Y.A.; Jung, G.S.; Lee, E.J.; Choi, Y.K.; Park, K.G.; Choe, M.S.; Jang, B.K.; Kim, M.K.; et al. Fyn deficiency attenuates renal fibrosis by inhibition of phospho-STAT3. Kidney Int. 2016, 90, 1285–1297. [Google Scholar] [CrossRef]
- Milara, J.; Hernandez, G.; Ballester, B.; Morell, A.; Roger, I.; Montero, P.; Escrivá, J.; Lloris, J.M.; Molina-Molina, M.; Morcillo, E.; et al. The JAK2 pathway is activated in idiopathic pulmonary fibrosis. Respir. Res. 2018, 19, 24. [Google Scholar] [CrossRef]
- Pedroza, M.; Le, T.T.; Lewis, K.; Karmouty-Quintana, H.; To, S.; George, A.T.; Blackburn, M.R.; Tweardy, D.J.; Agarwal, S.K. STAT-3 contributes to pulmonary fibrosis through epithelial injury and fibroblast-myofibroblast differentiation. FASEB 2016, 30, 129–140. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Liu, Y.; Pan, H.; Xu, T.; Li, Y.; Yuan, J.; Li, P.; Yao, W.; Yan, W.; Ni, C. Aberrant expression of miR-125a-3p promotes fibroblast activation via Fyn/STAT3 pathway during silica-induced pulmonary fibrosis. Toxicology 2019, 414, 57–67. [Google Scholar] [CrossRef] [PubMed]
- Ryu, M.S.; Zhang, D.; Protchenko, O.; Shakoury-Elizeh, M.; Philpott, C.C. PCBP1 and NCOA4 regulate erythroid iron storage and heme biosynthesis. J. Clin. Investig. 2017, 127, 1786–1797. [Google Scholar] [CrossRef]
- Zhang, X.; Di, C.; Chen, Y.; Wang, J.; Su, R.; Huang, G.; Xu, C.; Chen, X.; Long, F.; Yang, H.; et al. Multilevel regulation and molecular mechanism of poly (rC)-binding protein 1 in cancer. FASEB J. 2020, 34, 15647–15658. [Google Scholar] [CrossRef]
- Rozovski, U.; Calin, G.A.; Setoyama, T.; D’Abundo, L.; Harris, D.M.; Li, P.; Liu, Z.; Grgurevic, S.; Ferrajoli, A.; Faderl, S.; et al. Signal transducer and activator of transcription (STAT)-3 regulates microRNA gene expression in chronic lymphocytic leukemia cells. Mol. Cancer 2013, 12, 50. [Google Scholar] [CrossRef] [PubMed]
- Löffler, D.; Brocke-Heidrich, K.; Pfeifer, G.; Stocsits, C.; Hackermüller, J.; Kretzschmar, A.K.; Burger, R.; Gramatzki, M.; Blumert, C.; Bauer, K.; et al. Interleukin-6 dependent survival of multiple myeloma cells involves the Stat3-mediated induction of microRNA-21 through a highly conserved enhancer. Blood 2007, 110, 1330–1333. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.Y.; Sun, G.; Luo, H.; Wang, X.F.; Lan, F.M.; Yue, X.; Fu, L.S.; Pu, P.Y.; Kang, C.S.; Liu, N.; et al. MiR-21 modulates hTERT through a STAT3-dependent manner on glioblastoma cell growth. CNS Neurosci. Ther. 2019, 18, 722–728. [Google Scholar] [CrossRef] [PubMed]
- Luo, F.; Xu, Y.; Ling, M.; Zhao, Y.; Xu, W.; Liang, X.; Jiang, R.; Wang, B.; Bian, Q.; Liu, Q. Arsenite evokes IL-6 secretion, autocrine regulation of STAT3 signaling, and miR-21 expression, processes involved in the EMT and malignant transformation of human bronchial epithelial cells. Toxicol. Appl. Pharmacol. 2013, 273, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Bi, C.; Chng, W.J. MicroRNA: Important player in the pathobiology of multiple myeloma. Biomed. Res. Int. 2014, 2014, 521586. [Google Scholar] [CrossRef] [PubMed]


| Case | Sex | Age at First LTx | Original Diagnosis | Months to Retransplant | Pathological Pattern of CLAD g |
|---|---|---|---|---|---|
| Str-1 | M | 66 | NSIP a (Sjogren Syndrome) | 28 | OB h, RAS i, HV l |
| Str-2 | M | 51 | UIP b | 27 | OB, HV |
| Str-3 | M | 28 | CF c | 32 | OB, HV |
| Str-4 | F | 27 | CF | 1 | OB |
| Str-5 | M | 33 | BOS d | 115 | OB |
| Str-6 | M | 20 | CF | 43 | OB, RAS |
| Str-7 | M | 18 | CF | 41 | OB, RAS |
| Pav-1 | M | 50 | UIP | 90 | OB, RAS, HV |
| Pav-2 | F | 36 | CF | 100 | OB, LB m |
| Pad-1 | M | 37 | CLL e | 82 | OB, LB |
| Pad-2 | M | 31 | CML f | 104 | OB |
| Pad-3 | M | 26 | CF | 126 | OB |
| Type II Pneumocyte | Bronchial Epithelia | Endothelia | Bronchial Smooth Muscle | Vascular Smooth Muscle | Myofibroblasts | Inflammatory Cells | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NL | TL | NL | TL | NL | TL | NL | TL | NL | TL | TL | NL | TL | |
| miR-21-5p | - | + | - | + | - | + | - | ++ | - | + | +++ | M | M |
| miR-34a-5p | + | + | + | + | - | + | - | - | - | - | + | M | PC |
| miR-15a-5p | +++ | +++ | +++ | +++ | ++ | ++ | ++ | ++ | ++ | ++ | +++ | L, M | L, M |
| miR-145-5p | + | + | ++ | ++ | + | + | +++ | +++ | +++ | +++ | + | - | - |
| miR-146b-5p | + | + | + | - | - | - | - | - | - | - | + | M | PC |
| Let-7d-5p | ++ | +++ | +++ | +++ | ++ | ++ | - | - | - | + | ++ | PC | PC |
| Type II Pneumocytes | Bronchial Epithelia | Endothelia | Bronchial Smooth Muscle | Vascular Smooth Muscle | Myofibro | Inflammatory Cells | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NL | TL | NL | TL | NL | TL | NL | TL | NL | TL | TL | NL | TL | |
| STAT3 | + | ++ | ++ | ++ | - | + | - | ++ | - | + | +++ | M, PC | M, PC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bozzini, S.; Pandolfi, L.; Rossi, E.; Inghilleri, S.; Zorzetto, M.; Ferrario, G.; Di Carlo, S.; Politano, G.; De Silvestri, A.; Frangipane, V.; et al. miRNAs Potentially Involved in Post Lung Transplant-Obliterative Bronchiolitis: The Role of miR-21-5p. Cells 2021, 10, 688. https://doi.org/10.3390/cells10030688
Bozzini S, Pandolfi L, Rossi E, Inghilleri S, Zorzetto M, Ferrario G, Di Carlo S, Politano G, De Silvestri A, Frangipane V, et al. miRNAs Potentially Involved in Post Lung Transplant-Obliterative Bronchiolitis: The Role of miR-21-5p. Cells. 2021; 10(3):688. https://doi.org/10.3390/cells10030688
Chicago/Turabian StyleBozzini, Sara, Laura Pandolfi, Elena Rossi, Simona Inghilleri, Michele Zorzetto, Giuseppina Ferrario, Stefano Di Carlo, Gianfranco Politano, Annalisa De Silvestri, Vanessa Frangipane, and et al. 2021. "miRNAs Potentially Involved in Post Lung Transplant-Obliterative Bronchiolitis: The Role of miR-21-5p" Cells 10, no. 3: 688. https://doi.org/10.3390/cells10030688
APA StyleBozzini, S., Pandolfi, L., Rossi, E., Inghilleri, S., Zorzetto, M., Ferrario, G., Di Carlo, S., Politano, G., De Silvestri, A., Frangipane, V., Porzio, M., Kessler, R., Calabrese, F., Meloni, F., & Morbini, P. (2021). miRNAs Potentially Involved in Post Lung Transplant-Obliterative Bronchiolitis: The Role of miR-21-5p. Cells, 10(3), 688. https://doi.org/10.3390/cells10030688








