Novel Gene Regulation in Normal and Abnormal Spermatogenesis
Abstract
1. Introduction
2. Novel Gene Regulation in the Fate Decisions of Human SSCs
3. Novel Gene Regulation in Fate Determinations of Rodent SSCs
4. Novel Gene Regulation in Human and Rodent Other Germ Cells
5. Novel Gene Regulation in Testicular Microenvironment
6. Novel Gene Regulation in Abnormal Human Spermatogenesis
7. Perspectives and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Garcia, T.X.; Hofmann, M.C. Regulation of germ line stem cell homeostasis. Anim. Reprod. 2015, 12, 35. [Google Scholar]
- Chen, Z.; Niu, M.; Sun, M.; Yuan, Q.; Yao, C.; Hou, J.; Wang, H.; Wen, L.; Fu, H.; Zhou, F. Transdifferentiation of human male germline stem cells to hepatocytes in vivo via the transplantation under renal capsules. Oncotarget 2017, 8, 14576–14592. [Google Scholar] [CrossRef]
- Chen, Z.; Sun, M.; Yuan, Q.; Niu, M.; Yao, C.; Hou, J.; Wang, H.; Wen, L.; Liu, Y.; Li, Z.; et al. Generation of functional hepatocytes from human spermatogonial stem cells. Oncotarget 2016, 7, 8879–8895. [Google Scholar] [CrossRef]
- Guo, Y.; Liu, L.; Sun, M.; Hai, Y.; Li, Z.; He, Z. Expansion and long-term culture of human spermatogonial stem cells via the activation of SMAD3 and AKT pathways. Exp. Biol. Med. 2015, 240, 1112. [Google Scholar] [CrossRef]
- Simon, L.; Ekman, G.C.; Kostereva, N.; Zhang, Z.; Hess, R.A.; Hofmann, M.C.; Cooke, P.S. Direct Transdifferentiation of Stem/Progenitor Spermatogonia Into Reproductive and Nonreproductive Tissues of All Germ Layers. Stem Cells 2010, 27, 1666–1675. [Google Scholar] [CrossRef] [PubMed]
- Kossack, N.; Meneses, J.; Shefi, S.; Nguyen, H.N.; Chavez, S.; Nicholas, C.; Gromoll, J.; Turek, P.J.; Reijo-Pera, R.A. Isolation and Characterization of Pluripotent Human Spermatogonial Stem Cell-Derived Cells. Stem Cells 2009, 27, 138–149. [Google Scholar] [CrossRef] [PubMed]
- Izadyar, F.; Wong, J.; Maki, C.; Pacchiarotti, J.; Ramos, T.; Howerton, K.; Yuen, C.; Greilach, S.; Zhao, H.H.; Chow, M.; et al. Identification and characterization of repopulating spermatogonial stem cells from the adult human testis. Hum. Reprod. 2011, 26, 1296–1306. [Google Scholar] [CrossRef]
- Sohni, A.; Tan, K.; Song, H.W.; Burow, D.; de Rooij, D.G.; Laurent, L.; Hsieh, T.C.; Rabah, R.; Hammoud, S.S.; Vicini, E.; et al. The Neonatal and Adult Human Testis Defined at the Single-Cell Level. Cell Rep. 2019, 26, 1501–1517.e4. [Google Scholar] [CrossRef]
- Wang, M.; Liu, X.; Chang, G.; Chen, Y.; An, G.; Yan, L.; Gao, S.; Xu, Y.; Cui, Y.; Dong, J.; et al. Single-Cell RNA Sequencing Analysis Reveals Sequential Cell Fate Transition during Human Spermatogenesis. Cell Stem Cell 2018, 23, 599–614.e4. [Google Scholar] [CrossRef]
- Guo, J.; Grow, E.J.; Mlcochova, H.; Maher, G.J.; Lindskog, C.; Nie, X.; Guo, Y.; Takei, Y.; Yun, J.; Cai, L.; et al. The adult human testis transcriptional cell atlas. Cell Res. 2018, 28, 1141–1157. [Google Scholar] [CrossRef]
- Guo, J.; Grow, E.J.; Yi, C.; Mlcochova, H.; Maher, G.J.; Lindskog, C.; Murphy, P.J.; Wike, C.L.; Carrell, D.T.; Goriely, A.; et al. Chromatin and Single-Cell RNA- Seq Profiling Reveal Dynamic Signaling and Metabolic Transitions during Human Spermatogonial Stem Cell Development. Cell Stem Cell 2017, 21, 533–546.e6. [Google Scholar] [CrossRef] [PubMed]
- Hermann, B.P.; Cheng, K.; Singh, A.; Roa-De la Cruz, L.; Mutoji, K.N.; Chen, I.C.; Gildersleeve, H.; Lehle, J.D.; Mayo, M.; Westernstroer, B.; et al. The Mammalian Spermatogenesis Single-Cell Transcriptome, from Spermatogonial Stem Cells to Spermatids. Cell Rep. 2018, 25, 1650–1667.e8. [Google Scholar] [CrossRef]
- Guo, J.; Nie, X.; Giebler, M.; Mlcochova, H.; Wang, Y.; Grow, E.J.; Donor, C.; Kim, R.; Tharmalingam, M.; Matilionyte, G.; et al. The Dynamic Transcriptional Cell Atlas of Testis Development during Human Puberty. Cell Stem Cell 2020, 26, 262–276.e4. [Google Scholar] [CrossRef]
- Caldeira-Brant, A.L.; Martinelli, L.M.; Marques, M.M.; Reis, A.B.; Martello, R.; Almeida, F.R.C.L.; Chiarini-Garcia, H. A subpopulation of human Adark spermatogonia behaves as the reserve stem cell. Reproduction 2020, 159, 437–451. [Google Scholar] [CrossRef]
- Tan, K.; Song, H.-W.; Thompson, M.; Munyoki, S.; Sukhwani, M.; Hsieh, T.-C.; Orwig, K.E.; Wilkinson, M.F. Transcriptome profiling reveals signaling conditions dictating human spermatogonia fate in vitro. Proc. Natl. Acad. Sci. USA 2020, 117, 17832–17841. [Google Scholar] [CrossRef]
- von Kopylow, K.; Kirchhoff, C.; Jezek, D.; Schulze, W.; Feig, C.; Primig, M.; Steinkraus, V.; Spiess, A.-N. Screening for biomarkers of spermatogonia within the human testis: A whole genome approach. Hum. Reprod. 2010, 25, 1104–1112. [Google Scholar] [CrossRef]
- Song, W.; Shi, X.; Xia, Q.; Yuan, M.; Liu, J.; Hao, K.; Qian, Y.; Zhao, X.; Zou, K. PLZF suppresses differentiation of mouse spermatogonial progenitor cells via binding of differentiation associated genes. J. Cell. Physiol. 2020, 235, 3033–3042. [Google Scholar] [CrossRef]
- Qiu, Q.; Yu, X.; Yao, C.; Hao, Y.; Fan, L.; Li, C.; Xu, P.; An, G.; Li, Z.; He, Z. FOXP3 pathogenic variants cause male infertility through affecting the proliferation and apoptosis of human spermatogonial stem cells. Aging 2019, 11, 12581–12599. [Google Scholar] [CrossRef]
- Fu, H.; Zhang, W.; Yuan, Q.; Niu, M.; Zhou, F.; Qiu, Q.; Mao, G.; Wang, H.; Wen, L.; Sun, M.; et al. PAK1 Promotes the Proliferation and Inhibits Apoptosis of Human Spermatogonial Stem Cells via PDK1/KDR/ZNF367 and ERK1/2 and AKT Pathways. Mol. Ther. -Nucleic Acids 2018, 12, 769–786. [Google Scholar] [CrossRef]
- Fu, H.; Zhou, F.; Yuan, Q.; Zhang, W.; Qiu, Q.; Yu, X.; He, Z. miRNA-31–5p Mediates the Proliferation and Apoptosis of Human Spermatogonial Stem Cells via Targeting JAZF1 and Cyclin A2. Mol. Ther. Nucleic Acids 2019, 14, 90–100. [Google Scholar] [CrossRef]
- Chen, W.; Cui, Y.; Liu, B.; Li, C.; Du, L.; Tang, R.; Qin, L.; Jiang, Y.; Li, J.; Yu, X.; et al. Hsa-miR-1908–3p Mediates the Self-Renewal and Apoptosis of Human Spermatogonial Stem Cells via Targeting KLF2. Mol. Ther. Nucleic Acids 2020, 20, 788–800. [Google Scholar] [CrossRef]
- Zhou, F.; Chen, W.; Cui, Y.; Liu, B.; Yuan, Q.; Li, Z.; He, Z. miRNA-122–5p stimulates the proliferation and DNA synthesis and inhibits the early apoptosis of human spermatogonial stem cells by targeting CBL and competing with lncRNA CASC7. Aging 2020, 12, 25528–25546. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Yuan, Q.; Zhang, W.; Niu, M.; Fu, H.; Qiu, Q.; Mao, G.; Wang, H.; Wen, L.; Wang, H.; et al. MiR-663a Stimulates Proliferation and Suppresses Early Apoptosis of Human Spermatogonial Stem Cells by Targeting NFIX and Regulating Cell Cycle. Mol. Ther. Nucleic Acids 2018, 12, 319–336. [Google Scholar] [CrossRef] [PubMed]
- Chan, F.; Oatley, M.J.; Kaucher, A.V.; Yang, Q.-E.; Bieberich, C.J.; Shashikant, C.S.; Oatley, J.M. Functional and molecular features of the Id4(+) germline stem cell population in mouse testes. Genes Dev. 2014, 28, 1351–1362. [Google Scholar] [CrossRef] [PubMed]
- Oatley, M.J.; Kaucher, A.V.; Racicot, K.E.; Oatley, J.M. Inhibitor of DNA Binding 4 Is Expressed Selectively by Single Spermatogonia in the Male Germline and Regulates the Self-Renewal of Spermatogonial Stem Cells in Mice. Biol. Reprod. 2011, 85, 347–356. [Google Scholar] [CrossRef]
- Sada, A.; Suzuki, A.; Suzuki, H.; Saga, Y. The RNA-Binding Protein NANOS2 Is Required to Maintain Murine Spermatogonial Stem Cells. Science 2009, 325, 1394–1398. [Google Scholar] [CrossRef]
- Zhou, Z.; Kawabe, H.; Suzuki, A.; Shinmyozu, K.; Saga, Y. NEDD4 controls spermatogonial stem cell homeostasis and stress response by regulating messenger ribonucleoprotein complexes. Nat. Commun. 2017, 8, 15662. [Google Scholar] [CrossRef]
- Green, C.D.; Ma, Q.; Manske, G.L.; Shami, A.N.; Zheng, X.; Marini, S.; Moritz, L.; Sultan, C.; Gurczynski, S.J.; Moore, B.B.; et al. A Comprehensive Roadmap of Murine Spermatogenesis Defined by Single-Cell RNA-Seq. Dev. Cell 2018, 46, 651–667.e610. [Google Scholar] [CrossRef]
- Grive, K.J.; Hu, Y.; Shu, E.; Grimson, A.; Elemento, O.; Grenier, J.K.; Cohen, P.E. Dynamic transcriptome profiles within spermatogonial and spermatocyte populations during postnatal testis maturation revealed by single-cell sequencing. PLoS Genet. 2019, 15, e1007810. [Google Scholar] [CrossRef]
- Chen, Y.; Zheng, Y.; Gao, Y.; Lin, Z.; Yang, S.; Wang, T.; Wang, Q.; Xie, N.; Hua, R.; Liu, M.; et al. Single-cell RNA-seq uncovers dynamic processes and critical regulators in mouse spermatogenesis. Cell Res. 2018, 28, 879–896. [Google Scholar] [CrossRef]
- Liu, W.; Li, N.; Zhang, M.; Liu, Y.; Sun, J.; Zhang, S.; Peng, S.; Hua, J. Eif2s3y regulates the proliferation of spermatogonial stem cells via Wnt6/<beta> -catenin signaling pathway. Biochim. Biophys. Acta Mol. Cell Res. 2020, 1867. [Google Scholar] [CrossRef]
- Tokue, M.; Ikami, K.; Mizuno, S.; Takagi, C.; Miyagi, A.; Takada, R.; Noda, C.; Kitadate, Y.; Hara, K.; Mizuguchi, H.; et al. SHISA6 Confers Resistance to Differentiation-Promoting Wnt/β-Catenin Signaling in Mouse Spermatogenic Stem Cells. Stem Cell Rep. 2017, 8, 561–575. [Google Scholar] [CrossRef]
- Niu, Z.; Mu, H.; Zhu, H.; Wu, J.; Hua, J. p38 MAPK pathway is essential for self-renewal of mouse male germline stem cells (mGSCs). Cell Prolif. 2017, 50. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Whelan, E.; Guan, X.; Deng, B.; Wang, S.; Sun, J.; Avarbock, M.; Wu, X.; Brinster, R. FGF9 promotes mouse spermatogonial stem cell proliferation mediated by p38 MAPK signalling. Cell Prolif. 2020, 54, e12933. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Zhu, Z.; Yao, C.; Huang, Y.; Zhi, E.; Chen, H.; Tian, R.; Li, P.; Yuan, Q.; Xue, Y.; et al. VEGFC/VEGFR3 Signaling Regulates Mouse Spermatogonial Cell Proliferation via the Activation of AKT/MAPK and Cyclin D1 Pathway and Mediates the Apoptosis by affecting Caspase 3/9 and Bcl-2. Cell Cycle 2018, 17, 225–239. [Google Scholar] [CrossRef]
- Goertz, M.J.; Wu, Z.; Gallardo, T.D.; Hamra, F.K.; Castrillon, D.H. Foxo1 is required in mouse spermatogonial stem cells for their maintenance and the initiation of spermatogenesis. J. Clin. Investig. 2011, 121, 3456–3466. [Google Scholar] [CrossRef]
- Kang, H.; Chen, L.; Lichti-Kaiser, K.; Liao, G.; Gerrish, K.; Bortner, C.; Yao, H.; Eddy, E.; Jetten, A. Transcription Factor GLIS3: A New and Critical Regulator of Postnatal Stages of Mouse Spermatogenesis. Stem Cells 2016, 34, 2772–2783. [Google Scholar] [CrossRef]
- Tan, J.; Wollmann, H.; van Pelt, A.; Kaldis, P.; Messerschmidt, D. Infertility-Causing Haploinsufficiency Reveals TRIM28/KAP1 Requirement in Spermatogonia. Stem Cell Rep. 2020, 14, 818–827. [Google Scholar] [CrossRef]
- Du, G.; Wang, X.; Luo, M.; Xu, W.; Zhou, T.; Wang, M.; Yu, L.; Li, L.; Cai, L.; Wang, P.; et al. mRBPome capture identifies the RNA-binding protein TRIM71, an essential regulator of spermatogonial differentiation. Development 2020, 147. [Google Scholar] [CrossRef]
- Mikedis, M.M.; Fan, Y.; Nicholls, P.K.; Endo, T.; Jackson, E.K.; Cobb, S.A.; de Rooij, D.G.; Page, D.C. DAZL mediates a broad translational program regulating expansion and differentiation of spermatogonial progenitors. eLife 2020, 9. [Google Scholar] [CrossRef]
- Wang, Z.; Xu, X.; Li, J.L.; Palmer, C.; Maric, D.; Dean, J. Sertoli cell-only phenotype and scRNA-seq define PRAMEF12 as a factor essential for spermatogenesis in mice. Nat. Commun. 2019, 10, 5196. [Google Scholar] [CrossRef]
- Lee, J.; Kanatsu-Shinohara, M.; Morimoto, H.; Kazuki, Y.; Takashima, S.; Oshimura, M.; Toyokuni, S.; Shinohara, T. Genetic Reconstruction of Mouse Spermatogonial Stem Cell Self-Renewal In Vitro by Ras-Cyclin D2 Activation. Cell Stem Cell 2009, 5, 76–86. [Google Scholar] [CrossRef]
- Aloisio, G.M.; Nakada, Y.; Saatcioglu, H.D.; Pena, C.G.; Baker, M.D.; Tarnawa, E.D.; Mukherjee, J.; Manjunath, H.; Bugde, A.; Sengupta, A.L.; et al. PAX7 expression defines germline stem cells in the adult testis. J. Clin. Investig. 2014, 124, 3929–3944. [Google Scholar] [CrossRef]
- Song, H.; Bettegowda, A.; Lake, B.; Zhao, A.; Skarbrevik, D.; Babajanian, E.; Sukhwani, M.; Shum, E.; Phan, M.; Plank, T.; et al. The Homeobox Transcription Factor RHOX10 Drives Mouse Spermatogonial Stem Cell Establishment. Cell Rep. 2016, 17, 149–164. [Google Scholar] [CrossRef]
- Wu, X.; Oatley, J.M.; Oatley, M.J.; Kaucher, A.V.; Avarbock, M.R.; Brinster, R.L. The POU Domain Transcription Factor POU3F1 Is an Important Intrinsic Regulator of GDNF-Induced Survival and Self-Renewal of Mouse Spermatogonial Stem Cells. Biol. Reprod. 2010, 82, 1103–1111. [Google Scholar] [CrossRef]
- Martinot, E.; Sèdes, L.; Baptissart, M.; Holota, H.; Rouaisnel, B.; Damon-Soubeyrand, C.; De Haze, A.; Saru, J.; Thibault-Carpentier, C.; Keime, C.; et al. The Bile Acid Nuclear Receptor FXRα Is a Critical Regulator of Mouse Germ Cell Fate. Stem Cell Rep. 2017, 9, 315–328. [Google Scholar] [CrossRef]
- He, Z.; Jiang, J.; Kokkinaki, M.; Tang, L.; Zeng, W.; Gallicano, I.; Dobrinski, I.; Dym, M. MiRNA-20 and mirna-106a regulate spermatogonial stem cell renewal at the post-transcriptional level via targeting STAT3 and Ccnd1. Stem Cells 2013, 31, 2205–2217. [Google Scholar] [CrossRef]
- Yamaji, M.; Jishage, M.; Meyer, C.; Suryawanshi, H.; Der, E.; Yamaji, M.; Garzia, A.; Morozov, P.; Manickavel, S.; McFarland, H.; et al. DND1 maintains germline stem cells via recruitment of the CCR4-NOT complex to target mRNAs. Nature 2017, 543, 568–572. [Google Scholar] [CrossRef]
- Matson, C.K.; Murphy, M.W.; Griswold, M.D.; Yoshida, S.; Bardwell, V.J.; Zarkower, D. The Mammalian Doublesex Homolog DMRT1 Is a Transcriptional Gatekeeper that Controls the Mitosis versus Meiosis Decision in Male Germ Cells. Dev. Cell 2010, 19, 612–624. [Google Scholar] [CrossRef]
- Zhu, Z.; Li, C.; Yang, S.; Tian, R.; Wang, J.; Yuan, Q.; Dong, H.; He, Z.; Wang, S.; Li, Z. Dynamics of the Transcriptome during Human Spermatogenesis: Predicting the Potential Key Genes Regulating Male Gametes Generation. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef]
- Hua, R.; Wei, H.; Liu, C.; Zhang, Y.; Liu, S.; Guo, Y.; Cui, Y.; Zhang, X.; Guo, X.; Li, W.; et al. FBXO47 regulates telomere-inner nuclear envelope integration by stabilizing TRF2 during meiosis. Nucleic Acids Res. 2019, 47, 11755–11770. [Google Scholar] [CrossRef]
- Legrand, J.M.D.; Chan, A.-L.; La, H.M.; Rossello, F.J.; Anko, M.-L.; Fuller-Pace, F.V.; Hobbs, R.M. DDX5 plays essential transcriptional and post-transcriptional roles in the maintenance and function of spermatogonia. Nat. Commun. 2019, 10. [Google Scholar] [CrossRef]
- Gao, X.; Chen, H.; Liu, J.; Shen, S.; Wang, Q.; Clement, T.; Deskin, B.; Chen, C.; Zhao, D.; Wang, L.; et al. The REGγ-Proteasome Regulates Spermatogenesis Partially by P53-PLZF Signaling. Stem Cell Rep. 2019, 13, 559–571. [Google Scholar] [CrossRef]
- Meng, C.; Liao, J.; Zhao, D.; Huang, H.; Qin, J.; Lee, T.-L.; Chen, D.; Chan, W.-Y.; Xia, Y. L3MBTL2 regulates chromatin remodeling during spermatogenesis. Cell Death Differ. 2019, 26, 2194–2207. [Google Scholar] [CrossRef]
- Hu, X.; Shen, B.; Liao, S.; Ning, Y.; Ma, L.; Chen, J.; Lin, X.; Zhang, D.; Li, Z.; Zheng, C.; et al. Gene knockout of Zmym3 in mice arrests spermatogenesis at meiotic metaphase with defects in spindle assembly checkpoint. Cell Death Dis. 2017, 8, e2910. [Google Scholar] [CrossRef]
- Che, L.; Alavattam, K.G.; Stambrook, P.J.; Namekawa, S.H.; Du, C. BRUCE preserves genomic stability in the male germline of mice. Cell Death Differ. 2020, 27, 2402–2416. [Google Scholar] [CrossRef] [PubMed]
- Lei, W.-L.; Han, F.; Hu, M.-W.; Liang, Q.-X.; Meng, T.-G.; Zhou, Q.; Ouyang, Y.-C.; Hou, Y.; Schatten, H.; Wang, Z.-B.; et al. Protein phosphatase 6 is a key factor regulating spermatogenesis. Cell Death Differ. 2020, 27, 1952–1964. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Tan-Tai, W.; Li, X.; Liu, M.; Shi, H.; Martin-DeLeon, P.; O, W.; Chen, H. PHB regulates meiotic recombination via JAK2-mediated histone modifications in spermatogenesis. Nucleic Acids Res. 2020, 48, 4780–4796. [Google Scholar] [CrossRef] [PubMed]
- Guan, Y.; Leu, N.; Ma, J.; Chmátal, L.; Ruthel, G.; Bloom, J.; Lampson, M.; Schimenti, J.; Luo, M.; Wang, P. SKP1 drives the prophase I to metaphase I transition during male meiosis. Sci. Adv. 2020, 6, eaaz2129. [Google Scholar] [CrossRef] [PubMed]
- Chihara, M.; Ikebuchi, R.; Otsuka, S.; Ichii, O.; Hashimoto, Y.; Suzuki, A.; Saga, Y.; Kon, Y. Mice Stage-Specific Claudin 3 Expression Regulates Progression of Meiosis in Early Stage Spermatocytes. Biol. Reprod. 2013, 89. [Google Scholar] [CrossRef]
- Naro, C.; Pellegrini, L.; Jolly, A.; Farini, D.; Cesari, E.; Bielli, P.; de la Grange, P.; Sette, C. Functional Interaction between U1snRNP and Sam68 Insures Proper 3 End Pre-mRNA Processing during Germ Cell Differentiation. Cell Rep. 2019, 26, 2929–2941.e5. [Google Scholar] [CrossRef]
- Paronetto, M.P.; Messina, V.; Bianchi, E.; Barchi, M.; Vogel, G.; Moretti, C.; Palombi, F.; Stefanini, M.; Geremia, R.; Richard, S.; et al. Sam68 regulates translation of target mRNAs in male germ cells, necessary for mouse spermatogenesis. J. Cell Biol. 2009, 185, 235–249. [Google Scholar] [CrossRef]
- Vasileva, A.; Tiedau, D.; Firooznia, A.; Mueller-Reichert, T.; Jessberger, R. Tdrd6 Is Required for Spermiogenesis, Chromatoid Body Architecture, and Regulation of miRNA Expression. Curr. Biol. 2009, 19, 630–639. [Google Scholar] [CrossRef]
- Tanaka, T.; Hosokawa, M.; Vagin, V.V.; Reuter, M.; Hayashi, E.; Mochizuki, A.L.; Kitamura, K.; Yamanaka, H.; Kondoh, G.; Okawa, K.; et al. Tudor domain containing 7 (Tdrd7) is essential for dynamic ribonucleoprotein (RNP) remodeling of chromatoid bodies during spermatogenesis. Proc. Natl. Acad. Sci. USA 2011, 108, 10579–10584. [Google Scholar] [CrossRef]
- Yabuta, Y.; Ohta, H.; Abe, T.; Kurimoto, K.; Chuma, S.; Saitou, M. TDRD5 is required for retrotransposon silencing, chromatoid body assembly, and spermiogenesis in mice. J. Cell Biol. 2011, 192, 781–795. [Google Scholar] [CrossRef]
- Huang, Q.; Liu, Y.; Zhang, S.; Yap, Y.T.; Li, W.; Zhang, D.; Gardner, A.; Zhang, L.; Song, S.; Hess, R.A.; et al. Autophagy core protein ATG5 is required for elongating spermatid development, sperm individualization and normal fertility in male mice. Autophagy 2020, 17, 1–15. [Google Scholar] [CrossRef]
- Wang, H.; Wan, H.; Li, X.; Liu, W.; Chen, Q.; Wang, Y.; Yang, L.; Tang, H.; Zhang, X.; Duan, E.; et al. Atg7 is required for acrosome biogenesis during spermatogenesis in mice. Cell Res. 2014, 24, 852–869. [Google Scholar] [CrossRef]
- Yuan, S.; Stratton, C.J.; Bao, J.; Zheng, H.; Bhetwal, B.P.; Yanagimachi, R.; Yan, W. Spata6 is required for normal assembly of the sperm connecting piece and tight head-tail conjunction. Proc. Natl. Acad. Sci. USA 2015, 112, E430–E439. [Google Scholar] [CrossRef]
- Crapster, J.A.; Rack, P.G.; Hellmann, Z.J.; Le, A.D.; Adams, C.M.; Leib, R.D.; Elias, J.E.; Perrino, J.; Behr, B.; Li, Y.; et al. HIPK4 is essential for murine spermiogenesis. eLife 2020, 9. [Google Scholar] [CrossRef]
- Liu, S.; Yu, H.; Liu, Y.; Liu, X.; Zhang, Y.; Bu, C.; Yuan, S.; Chen, Z.; Xie, G.; Li, W.; et al. Chromodomain Protein CDYL Acts as a Crotonyl-CoA Hydratase to Regulate Histone Crotonylation and Spermatogenesis. Mol. Cell 2017, 67, 853–866.e5. [Google Scholar] [CrossRef]
- Inoue, S.; Tomasini, R.; Rufini, A.; Elia, A.; Agostini, M.; Amelio, I.; Cescon, D.; Dinsdale, D.; Zhou, L.; Harris, I.; et al. TAp73 is required for spermatogenesis and the maintenance of male fertility. Proc. Natl. Acad. Sci. USA 2014, 111, 1843–1848. [Google Scholar] [CrossRef]
- Xu, K.; Yang, Y.; Feng, G.-H.; Sun, B.-F.; Chen, J.-Q.; Li, Y.-F.; Chen, Y.-S.; Zhang, X.-X.; Wang, C.-X.; Jiang, L.-Y.; et al. Mettl3-mediated m(6)A regulates spermatogonial differentiation and meiosis initiation. Cell Res. 2017, 27, 1100–1114. [Google Scholar] [CrossRef]
- Lin, Z.; Hsu, P.J.; Xing, X.; Fang, J.; Lu, Z.; Zou, Q.; Zhang, K.-J.; Zhang, X.; Zhou, Y.; Zhang, T.; et al. Mettl3-/Mettl14-mediated mRNA N-6-methyladenosine modulates murine spermatogenesis. Cell Res. 2017, 27, 1216–1230. [Google Scholar] [CrossRef]
- Hsu, P.J.; Zhu, Y.; Ma, H.; Guo, Y.; Shi, X.; Liu, Y.; Qi, M.; Lu, Z.; Shi, H.; Wang, J.; et al. Ythdc2 is an N-6-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 2017, 27, 1115–1127. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.; Klukovich, R.; Peng, H.; Wang, Z.; Yu, T.; Zhang, Y.; Zheng, H.; Klungland, A.; Yan, W. ALKBH5-dependent m6A demethylation controls splicing and stability of long 3-UTR mRNAs in male germ cells. Proc. Natl. Acad. Sci. USA 2018, 115, E325–E333. [Google Scholar] [CrossRef]
- Wang, H.; Yuan, Q.; Sun, M.; Niu, M.; Wen, L.; Fu, H.; Zhou, F.; Chen, Z.; Yao, C.; Hou, J.; et al. BMP6 Regulates Proliferation and Apoptosis of Human Sertoli Cells Via Smad2/3 and Cyclin D1 Pathway and DACH1 and TFAP2A Activation. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Hai, Y.; Sun, M.; Niu, M.; Yuan, Q.; Guo, Y.; Li, Z.; He, Z. BMP4 promotes human Sertoli cell proliferation via Smad1/5 and ID2/3 pathway and its abnormality is associated with azoospermia. Discov. Med. 2015, 19, 311–325. [Google Scholar] [PubMed]
- Yao, C.; Sun, M.; Yuan, Q.; Niu, M.; Chen, Z.; Hou, J.; Wang, H.; Wen, L.; Liu, Y.; Li, Z.; et al. MiRNA-133b promotes the proliferation of human Sertoli cells through targeting GLI3. Oncotarget 2016, 7, 2201–2219. [Google Scholar] [CrossRef] [PubMed]
- Fouchécourt, S.; Livera, G.; Messiaen, S.; Fumel, B.; Parent, A.; Marine, J.; Monget, P. Apoptosis of Sertoli cells after conditional ablation of murine double minute 2 (Mdm2) gene is p53-dependent and results in male sterility. Cell Death Differ. 2016, 23, 521–530. [Google Scholar] [CrossRef] [PubMed]
- Pitetti, J.-L.; Calvel, P.; Zimmermann, C.; Conne, B.; Papaioannou, M.D.; Aubry, F.; Cederroth, C.R.; Urner, F.; Fumel, B.; Crausaz, M.; et al. An Essential Role for Insulin and IGF1 Receptors in Regulating Sertoli Cell Proliferation, Testis Size, and FSH Action in Mice. Mol. Endocrinol. 2013, 27, 814–827. [Google Scholar] [CrossRef]
- Hilbold, E.; Distl, O.; Hoedemaker, M.; Wilkening, S.; Behr, R.; Rajkovic, A.; Langeheine, M.; Rode, K.; Jung, K.; Metzger, J.; et al. Loss of Cx43 in Murine Sertoli Cells Leads to Altered Prepubertal Sertoli Cell Maturation and Impairment of the Mitosis-Meiosis Switch. Cells 2020, 9, 676. [Google Scholar] [CrossRef] [PubMed]
- Giese, S.; Hossain, H.; Markmann, M.; Chakraborty, T.; Tchatalbachev, S.; Guillou, F.; Bergmann, M.; Failing, K.; Weider, K.; Brehm, R. Sertoli-cell-specific knockout of connexin 43 leads to multiple alterations in testicular gene expression in prepubertal mice. Dis. Models Mech. 2012, 5, 895–913. [Google Scholar] [CrossRef]
- Gerber, J.; Rode, K.; Hambruch, N.; Langeheine, M.; Schnepel, N.; Brehm, R. Establishment and functional characterization of a murine primary Sertoli cell line deficient of connexin43. Cell Tissue Res. 2020, 381, 309–326. [Google Scholar] [CrossRef]
- Hastie, N.; Wang, X.N.; Li, Z.S.; Ren, Y.; Jiang, T.; Wang, Y.Q.; Chen, M.; Zhang, J.; Hao, J.X.; Wang, Y.B.; et al. The Wilms Tumor Gene, Wt1, Is Critical for Mouse Spermatogenesis via Regulation of Sertoli Cell Polarity and Is Associated with Non-Obstructive Azoospermia in Humans. PLoS Genet. 2013, 9, e1003645. [Google Scholar] [CrossRef]
- Tanwar, P.S.; Kaneko-Tarui, T.; Zhang, L.; Rani, P.; Taketo, M.M.; Teixeira, J. Constitutive WNT/Beta-Catenin Signaling in Murine Sertoli Cells Disrupts Their Differentiation and Ability to Support Spermatogenesis. Biol. Reprod. 2010, 82, 422–432. [Google Scholar] [CrossRef]
- Thomas, P.A.; Schafler, E.D.; Ruff, S.E.; Voisin, M.; Ha, S.; Logan, S.K. UXT in Sertoli Cells is Required for Blood-Testis Barrier Integrity. Biol. Reprod. 2020, 103, 880–891. [Google Scholar] [CrossRef] [PubMed]
- Pradhan, B.S.; Bhattacharya, I.; Sarkar, R.; Majumdar, S.S. Pubertal down regulation of Tetraspanin 8 in testicular Sertoli cells is crucial for male fertility. Mol. Hum. Reprod. 2020, 26, 760–772. [Google Scholar] [CrossRef] [PubMed]
- Mendis, S.H.S.; Meachem, S.J.; Sarraj, M.A.; Loveland, K.L. Activin A Balances Sertoli and Germ Cell Proliferation in the Fetal Mouse Testis. Biol. Reprod. 2011, 84, 379–391. [Google Scholar] [CrossRef]
- Kyroenlahti, A.; Euler, R.; Bielinska, M.; Schoeller, E.L.; Moley, K.H.; Toppari, J.; Heikinheimo, M.; Wilson, D.B. GATA4 regulates Sertoli cell function and fertility in adult male mice. Mol. Cell. Endocrinol. 2011, 333, 85–95. [Google Scholar] [CrossRef]
- Mazaud-Guittot, S.; Meugnier, E.; Pesenti, S.; Wu, X.; Vidal, H.; Gow, A.; Le Magueresse-Battistoni, B. Claudin 11 Deficiency in Mice Results in Loss of the Sertoli Cell Epithelial Phenotype in the Testis. Biol. Reprod. 2010, 82, 202–213. [Google Scholar] [CrossRef]
- Willems, A.; Batlouni, S.R.; Esnal, A.; Swinnen, J.V.; Saunders, P.T.K.; Sharpe, R.M.; Franca, L.R.; De Gendt, K.; Verhoeven, G. Selective Ablation of the Androgen Receptor in Mouse Sertoli Cells Affects Sertoli Cell Maturation, Barrier Formation and Cytoskeletal Development. PLoS ONE 2010, 5, e14168. [Google Scholar] [CrossRef]
- Chen, L.; Willis, W.; Eddy, E. Targeting the Gdnf Gene in peritubular myoid cells disrupts undifferentiated spermatogonial cell development. Proc. Natl. Acad. Sci. USA 2016, 113, 1829–1834. [Google Scholar] [CrossRef]
- Hannigan, M.M.; Zagore, L.L.; Licatalosi, D.D. Ptbp2 Controls an Alternative Splicing Network Required for Cell Communication during Spermatogenesis. Cell Rep. 2017, 19, 2598–2612. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Kanatsu-Shinohara, M.; Lei, Z.; Rao, C.; Shinohara, T. The Luteinizing Hormone-Testosterone Pathway Regulates Mouse Spermatogonial Stem Cell Self-Renewal by Suppressing WNT5A Expression in Sertoli Cells. Stem Cell Rep. 2016, 7, 279–291. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, C.; Stevant, I.; Borel, C.; Conne, B.; Pitetti, J.-L.; Calvel, P.; Kaessmann, H.; Jegou, B.; Chalmel, F.; Nef, S. Research Resource: The Dynamic Transcriptional Profile of Sertoli Cells During the Progression of Spermatogenesis. Mol. Endocrinol. 2015, 29, 627–642. [Google Scholar] [CrossRef]
- Lee, S.; Park, J.; Lee, D.; Otsu, K.; Kim, P.; Mizuno, S.; Lee, M.; Kim, H.; Harada, H.; Takahashi, S.; et al. Mast4 knockout shows the regulation of spermatogonial stem cell self-renewal via the FGF2/ERM pathway. Cell Death Differ. 2020. [Google Scholar] [CrossRef] [PubMed]
- Winters, B.R.; Walsh, T.J. The epidemiology of male infertility. Urol. Clin. N. Am. 2014, 41, 195–204. [Google Scholar] [CrossRef]
- Sun, H.; Jiang, Y.T.; Zhang, S.; Zhao, Y.H.; Wu, Q.J. Global, regional, and national prevalence and disability-adjusted life-years for infertility in 195 countries and territories, 1990–2017: Results from a global burden of disease study, 2017. Aging 2019, 11, 10952–10991. [Google Scholar] [CrossRef]
- Hu, Z.; Xia, Y.; Guo, X.; Dai, J.; Li, H.; Hu, H.; Jiang, Y.; Lu, F.; Wu, Y.; Yang, X.; et al. A genome-wide association study in Chinese men identifies three risk loci for non-obstructive azoospermia. Nat. Genet. 2012, 44, 183–186. [Google Scholar] [CrossRef]
- Kasak, L.; Punab, M.; Nagirnaja, L.; Grigorova, M.; Minajeva, A.; Lopes, A.M.; Punab, A.M.; Aston, K.I.; Carvalho, F.; Laasik, E.; et al. Bi-allelic Recessive Loss-of-Function Variants in FANCM Cause Non-obstructive Azoospermia. Am. J. Hum. Genet. 2018, 103, 200–212. [Google Scholar] [CrossRef]
- Wu, X.; Luo, C.; Hu, L.; Chen, X.; Chen, Y.; Fan, J.; Cheng, C.Y.; Sun, F. Unraveling epigenomic abnormality in azoospermic human males by WGBS, RNA-Seq, and transcriptome profiling analyses. J. Assist. Reprod. Genet. 2020, 37, 789–802. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.-S.; Liang, A.; Dai, Y.-B.; Wu, X.-L.; Sun, F. Expression and localization of retinoid receptors in the testis of normal and infertile men. Mol. Reprod. Dev. 2020, 87, 978–985. [Google Scholar] [CrossRef] [PubMed]
- Arafat, M.; Zeadna, A.; Levitas, E.; Har Vardi, I.; Samueli, B.; Shaco-Levy, R.; Dabsan, S.; Lunenfeld, E.; Huleihel, M.; Parvari, R. Novel mutation in USP26 associated with azoospermia in a Sertoli cell-only syndrome patient. Mol. Genet. Genom. Med. 2020, 8. [Google Scholar] [CrossRef]
- Yang, C.; Yao, C.; Tian, R.; Zhu, Z.; Zhao, L.; Li, P.; Chen, H.; Huang, Y.; Zhi, E.; Gong, Y.; et al. miR-202–3p Regulates Sertoli Cell Proliferation, Synthesis Function, and Apoptosis by Targeting LRP6 and Cyclin D1 of Wnt/beta-Catenin Signaling. Mol. Ther. Nucleic Acids 2019, 14, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Stahl, P.J.; Mielnik, A.N.; Barbieri, C.E.; Schlegel, P.N.; Paduch, D.A. Deletion or underexpression of the Y-chromosome genes CDY2 and HSFY is associated with maturation arrest in American men with nonobstructive azoospermia. Asian J. 2012, 14, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, T.; Iijima, M.; Shin, T.; Minase, G.; Ueda, H.; Saijo, Y.; Okada, H.; Sengoku, K. An association study of the single-nucleotide polymorphism c190C>T (Arg64Cys) in the human testis-specific histone variant, H3t, of Japanese patients with Sertoli cell-only syndrome. Asian J. 2018, 20, 527–528. [Google Scholar] [CrossRef]
- Miyamoto, T.; Bando, Y.; Koh, E.; Tsujimura, A.; Miyagawa, Y.; Iijima, M.; Namiki, M.; Shiina, M.; Ogata, K.; Matsumoto, N.; et al. A PLK4 mutation causing azoospermia in a man with Sertoli cell-only syndrome. Andrology 2016, 4, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Cui, W.; He, X.; Zhai, X.; Zhang, H.; Zhang, Y.; Jin, F.; Song, X.; Wu, D.; Shi, Q.; Li, L. CARF promotes spermatogonial self-renewal and proliferation through Wnt signaling pathway. Cell Discov. 2020, 6, 85. [Google Scholar] [CrossRef]
- Chung, C.L.; Lu, C.W.; Cheng, Y.S.; Lin, C.Y.; Sun, H.S.; Lin, Y.M. Association of aberrant expression of sex-determining gene fibroblast growth factor 9 with Sertoli cell-only syndrome. Fertil. Steril. 2013, 100, 1547–1554.e4. [Google Scholar] [CrossRef]
- Zhang, H.; Zhou, D.; Zhu, F.; Chen, F.; Zhu, Y.; Yu, R.; Fan, L. Disordered APC/C-mediated cell cycle progression and IGF1/PI3K/AKT signalling are the potential basis of Sertoli cell-only syndrome. Andrologia 2019, 51, e13288. [Google Scholar] [CrossRef]
- O’Bryan, M.K.; Grealy, A.; Stahl, P.J.; Schlegel, P.N.; McLachlan, R.I.; Jamsai, D. Genetic variants in the ETV5 gene in fertile and infertile men with nonobstructive azoospermia associated with Sertoli cell-only syndrome. Fertil. Steril. 2012, 98, 827–835.e3. [Google Scholar] [CrossRef]
- Li, J.; Guo, W.; Li, F.; He, J.; Yu, Q.; Wu, X.; Li, J.; Mao, X. HnRNPL as a key factor in spermatogenesis: Lesson from functional proteomic studies of azoospermia patients with sertoli cell only syndrome. J. Proteom. 2012, 75, 2879–2891. [Google Scholar] [CrossRef]
- Lei, B.; Wan, B.; Peng, J.; Yang, Y.; Lv, D.; Zhou, X.; Shu, F.; Li, F.; Zhong, L.; Wu, H.; et al. PRPS2 Expression Correlates with Sertoli-Cell Only Syndrome and Inhibits the Apoptosis of TM4 Sertoli Cells. J. Urol. 2015, 194, 1491–1497. [Google Scholar] [CrossRef]
- Tan, Y.Q.; Tu, C.; Meng, L.; Yuan, S.; Sjaarda, C.; Luo, A.; Du, J.; Li, W.; Gong, F.; Zhong, C.; et al. Loss-of-function mutations in TDRD7 lead to a rare novel syndrome combining congenital cataract and nonobstructive azoospermia in humans. Genet. Med. 2019, 21, 1209–1217. [Google Scholar] [CrossRef]
- Arafat, M.; Har-Vardi, I.; Harlev, A.; Levitas, E.; Zeadna, A.; Abofoul-Azab, M.; Dyomin, V.; Sheffield, V.C.; Lunenfeld, E.; Huleihel, M.; et al. Mutation in TDRD9 causes non-obstructive azoospermia in infertile men. J. Med. Genet. 2017, 54, 633–639. [Google Scholar] [CrossRef]
- Babakhanzadeh, E.; Khodadadian, A.; Rostami, S.; Alipourfard, I.; Aghaei, M.; Nazari, M.; Hosseinnia, M.; Mehrjardi, M.Y.V.; Jamshidi, Y.; Ghasemi, N. Testicular expression of TDRD1, TDRD5, TDRD9 and TDRD12 in azoospermia. BMC Med. Genet. 2020, 21, 33. [Google Scholar] [CrossRef]
- Colombo, R.; Pontoglio, A.; Bini, M. Two Novel TEX15 Mutations in a Family with Nonobstructive Azoospermia. Gynecol. Obs. Investig. 2017, 82, 283–286. [Google Scholar] [CrossRef]
- Boroujeni, P.B.; Sabbaghian, M.; Totonchi, M.; Sodeifi, N.; Sarkardeh, H.; Samadian, A.; Sadighi-Gilani, M.A.; Gourabi, H. Expression analysis of genes encoding TEX11, TEX12, TEX14 and TEX15 in testis tissues of men with non-obstructive azoospermia. JBRA Assist. Reprod. 2018, 22, 185–192. [Google Scholar] [CrossRef] [PubMed]
- He, W.B.; Tu, C.F.; Liu, Q.; Meng, L.L.; Yuan, S.M.; Luo, A.X.; He, F.S.; Shen, J.; Li, W.; Du, J.; et al. DMC1 mutation that causes human non-obstructive azoospermia and premature ovarian insufficiency identified by whole-exome sequencing. J. Med. Genet. 2018, 55, 198–204. [Google Scholar] [CrossRef] [PubMed]
- Babakhanzadeh, E.; Khodadadian, A.; Nazari, M.; Dehghan Tezerjani, M.; Aghaei, S.M.; Ghasemifar, S.; Hosseinnia, M.; Mazaheri, M. Deficient Expression of DGCR8 in Human Testis is Related to Spermatogenesis Dysfunction, Especially in Meiosis I. Int. J. Gen. Med. 2020, 13, 185–192. [Google Scholar] [CrossRef]
- Ramasamy, R.; Ridgeway, A.; Lipshultz, L.I.; Lamb, D.J. Integrative DNA methylation and gene expression analysis identifies discoidin domain receptor 1 association with idiopathic nonobstructive azoospermia. Fertil. Steril. 2014, 102, 968–973.e963. [Google Scholar] [CrossRef]
- Li, L.J.; Zhang, F.B.; Liu, S.Y.; Tian, Y.H.; Le, F.; Lou, H.Y.; Huang, H.F.; Jin, F. Decreased expression of SAM68 in human testes with spermatogenic defects. Fertil. Steril. 2014, 102, 61–67.e63. [Google Scholar] [CrossRef]
- Tang, W.; Zhu, Y.; Qin, W.; Zhang, H.; Zhang, H.; Lin, H.; Zhen, X.; Zhuang, X.; Tang, Y.; Jiang, H. Ran-binding protein 3 is associated with human spermatogenesis and male infertility. Andrologia 2020, 52, e13446. [Google Scholar] [CrossRef] [PubMed]
- Riera-Escamilla, A.; Enguita-Marruedo, A.; Moreno-Mendoza, D.; Chianese, C.; Sleddens-Linkels, E.; Contini, E.; Benelli, M.; Natali, A.; Colpi, G.M.; Ruiz-Castane, E.; et al. Sequencing of a ‘mouse azoospermia’ gene panel in azoospermic men: Identification of RNF212 and STAG3 mutations as novel genetic causes of meiotic arrest. Hum. Reprod. 2019, 34, 978–988. [Google Scholar] [CrossRef] [PubMed]
- Ramasamy, R.; Cengiz, C.; Karaca, E.; Scovell, J.; Jhangiani, S.N.; Akdemir, Z.C.; Bainbridge, M.; Yu, Y.; Huff, C.; Gibbs, R.A.; et al. Whole-exome sequencing identifies novel homozygous mutation in NPAS2 in family with nonobstructive azoospermia. Fertil. Steril. 2015, 104, 286–291. [Google Scholar] [CrossRef]
- Bashamboo, A.; Ferraz-de-Souza, B.; Lourenco, D.; Lin, L.; Sebire, N.J.; Montjean, D.; Bignon-Topalovic, J.; Mandelbaum, J.; Siffroi, J.-P.; Christin-Maitre, S.; et al. Human Male Infertility Associated with Mutations in NR5A1 Encoding Steroidogenic Factor 1. Am. J. Hum. Genet. 2010, 87, 505–512. [Google Scholar] [CrossRef] [PubMed]
- Ferlin, A.; Rocca, M.; Vinanzi, C.; Ghezzi, M.; Di Nisio, A.; Foresta, C. Mutational screening of NR5A1 gene encoding steroidogenic factor 1 in cryptorchidism and male factor infertility and functional analysis of seven undescribed mutations. Fertil. Steril. 2015, 104, 163–169.e161. [Google Scholar] [CrossRef]
- Schilit, S.L.P.; Menon, S.; Friedrich, C.; Kammin, T.; Wilch, E.; Hanscom, C.; Jiang, S.; Kliesch, S.; Talkowski, M.E.; Tuettelmann, F.; et al. SYCP2 Translocation-Mediated Dysregulation and Frameshift Variants Cause Human Male Infertility. Am. J. Hum. Genet. 2020, 106, 41–57. [Google Scholar] [CrossRef]
- Wyrwoll, M.J.; Temel, S.G.; Nagirnaja, L.; Oud, M.S.; Lopes, A.M.; van der Heijden, G.W.; Heald, J.S.; Rotte, N.; Wistuba, J.; Woeste, M.; et al. Bi-allelic Mutations in M1AP Are a Frequent Cause of Meiotic Arrest and Severely Impaired Spermatogenesis Leading to Male Infertility. Am. J. Hum. Genet. 2020, 107, 342–351. [Google Scholar] [CrossRef]
- Wormser, O.; Levy, Y.; Bakhrat, A.; Bonaccorsi, S.; Graziadio, L.; Gatti, M.; AbuMadighem, A.; McKenney, R.J.; Okada, K.; El Riati, S.; et al. Absence of SCAPER causes male infertility in humans and Drosophila by modulating microtubule dynamics during meiosis. J. Med. Genet. 2020. [Google Scholar] [CrossRef]
- Wu, X.-L.; Yun, D.-M.; Gao, S.; Liang, A.J.; Duan, Z.-Z.; Wang, H.-S.; Wang, G.-S.; Sun, F. The testis-specific gene 1700102P08Rik is essential for male fertility. Mol. Reprod. Dev. 2020, 87, 231–240. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.; Wang, X.; Li, W.; Yang, X.; Li, Z.; Liu, W.; Li, C.; Zhu, Z.; Wang, L.; Wang, J.; et al. Biallelic Mutations in CFAP43 and CFAP44 Cause Male Infertility with Multiple Morphological Abnormalities of the Sperm Flagella. Am. J. Hum. Genet. 2017, 100, 854–864. [Google Scholar] [CrossRef] [PubMed]
- Coutton, C.; Vargas, A.S.; Amiri-Yekta, A.; Kherraf, Z.-E.; Ben Mustapha, S.F.; Le Tanno, P.; Wambergue-Legrand, C.; Karaouzene, T.; Martinez, G.; Crouzy, S.; et al. Mutations in CFAP43 and CFAP44 cause male infertility and flagellum defects in Trypanosoma and human. Nat. Commun. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Liu, C.; Yang, X.; Lv, M.; Ni, X.; Li, Q.; Cheng, H.; Liu, W.; Tian, S.; Wu, H.; et al. Bi-allelic Loss-of-function Variants in CFAP58 Cause Flagellar Axoneme and Mitochondrial Sheath Defects and Asthenoteratozoospermia in Humans and Mice. Am. J. Hum. Genet. 2020, 107, 514–526. [Google Scholar] [CrossRef]
- Dong, F.; Amiri-Yekta, A.; Martinez, G.; Saut, A.; Tek, J.; Stouvenel, L.; Lorès, P.; Karaouzène, T.; Thierry-Mieg, N.; Satre, V.; et al. Absence of CFAP69 Causes Male Infertility due to Multiple Morphological Abnormalities of the Flagella in Human and Mouse. Am. J. Hum. Genet. 2018, 102, 636–648. [Google Scholar] [CrossRef]
- Kherraf, Z.; Amiri-Yekta, A.; Dacheux, D.; Karaouzène, T.; Coutton, C.; Christou-Kent, M.; Martinez, G.; Landrein, N.; Le Tanno, P.; Fourati Ben Mustapha, S.; et al. A Homozygous Ancestral SVA-Insertion-Mediated Deletion in WDR66 Induces Multiple Morphological Abnormalities of the Sperm Flagellum and Male Infertility. Am. J. Hum. Genet. 2018, 103, 400–412. [Google Scholar] [CrossRef]
- Liu, C.; Miyata, H.; Gao, Y.; Sha, Y.; Tang, S.; Xu, Z.; Whitfield, M.; Patrat, C.; Wu, H.; Dulioust, E.; et al. Bi-allelic DNAH8 Variants Lead to Multiple Morphological Abnormalities of the Sperm Flagella and Primary Male Infertility. Am. J. Hum. Genet. 2020, 107, 330–341. [Google Scholar] [CrossRef]
- Coutton, C.; Martinez, G.; Kherraf, Z.; Amiri-Yekta, A.; Boguenet, M.; Saut, A.; He, X.; Zhang, F.; Cristou-Kent, M.; Escoffier, J.; et al. Bi-allelic Mutations in ARMC2 Lead to Severe Astheno-Teratozoospermia Due to Sperm Flagellum Malformations in Humans and Mice. Am. J. Hum. Genet. 2019, 104, 331–340. [Google Scholar] [CrossRef]
- Liu, W.; He, X.; Yang, S.; Zouari, R.; Wang, J.; Wu, H.; Kherraf, Z.; Liu, C.; Coutton, C.; Zhao, R.; et al. Bi-allelic Mutations in TTC21A Induce Asthenoteratospermia in Humans and Mice. Am. J. Hum. Genet. 2019, 104, 738–748. [Google Scholar] [CrossRef]
- Shen, Y.; Zhang, F.; Li, F.; Jiang, X.; Yang, Y.; Li, X.; Li, W.; Wang, X.; Cheng, J.; Liu, M.; et al. Loss-of-function mutations in QRICH2 cause male infertility with multiple morphological abnormalities of the sperm flagella. Nat. Commun. 2019, 10, 433. [Google Scholar] [CrossRef]
- Zhu, F.; Liu, C.; Wang, F.; Yang, X.; Zhang, J.; Wu, H.; Zhang, Z.; He, X.; Zhang, Z.; Zhou, P.; et al. Mutations in PMFBP1 Cause Acephalic Spermatozoa Syndrome. Am. J. Hum. Genet. 2018, 103, 188–199. [Google Scholar] [CrossRef]
- Zhu, F.; Wang, F.; Yang, X.; Zhang, J.; Wu, H.; Zhang, Z.; Zhang, Z.; He, X.; Zhou, P.; Wei, Z.; et al. Biallelic SUN5 Mutations Cause Autosomal-Recessive Acephalic Spermatozoa Syndrome. Am. J. Hum. Genet. 2016, 99, 942–949. [Google Scholar] [CrossRef] [PubMed]
- Whitfield, M.; Thomas, L.; Bequignon, E.; Schmitt, A.; Stouvenel, L.; Montantin, G.; Tissier, S.; Duquesnoy, P.; Copin, B.; Chantot, S.; et al. Mutations in DNAH17, Encoding a Sperm-Specific Axonemal Outer Dynein Arm Heavy Chain, Cause Isolated Male Infertility Due to Asthenozoospermia. Am. J. Hum. Genet. 2019, 105, 198–212. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Ma, H.; Khan, T.; Ma, A.; Li, T.; Zhang, H.; Gao, J.; Zhou, J.; Li, Y.; Yu, C.; et al. A DNAH17 missense variant causes flagella destabilization and asthenozoospermia. J. Exp. Med. 2020, 217. [Google Scholar] [CrossRef] [PubMed]
- El Khouri, E.; Thomas, L.; Jeanson, L.; Bequignon, E.; Vallette, B.; Duquesnoy, P.; Montantin, G.; Copin, B.; Dastot-Le Moal, F.; Blanchon, S.; et al. Mutations in DNAJB13, Encoding an HSP40 Family Member, Cause Primary Ciliary Dyskinesia and Male Infertility. Am. J. Hum. Genet. 2016, 99, 489–500. [Google Scholar] [CrossRef]
- Rahbari, R.; Wuster, A.; Lindsay, S.J.; Hardwick, R.J.; Alexandrov, L.B.; Al Turki, S.; Dominiczak, A.; Morris, A.; Porteous, D.; Smith, B.; et al. Timing, rates and spectra of human germline mutation. Nat. Genet. 2016, 48, 126–133. [Google Scholar] [CrossRef]
- Bell, A.; Mello, C.; Nemesh, J.; Brumbaugh, S.; Wysoker, A.; McCarroll, S. Insights into variation in meiosis from 31,228 human sperm genomes. Nature 2020, 583, 259–264. [Google Scholar] [CrossRef]
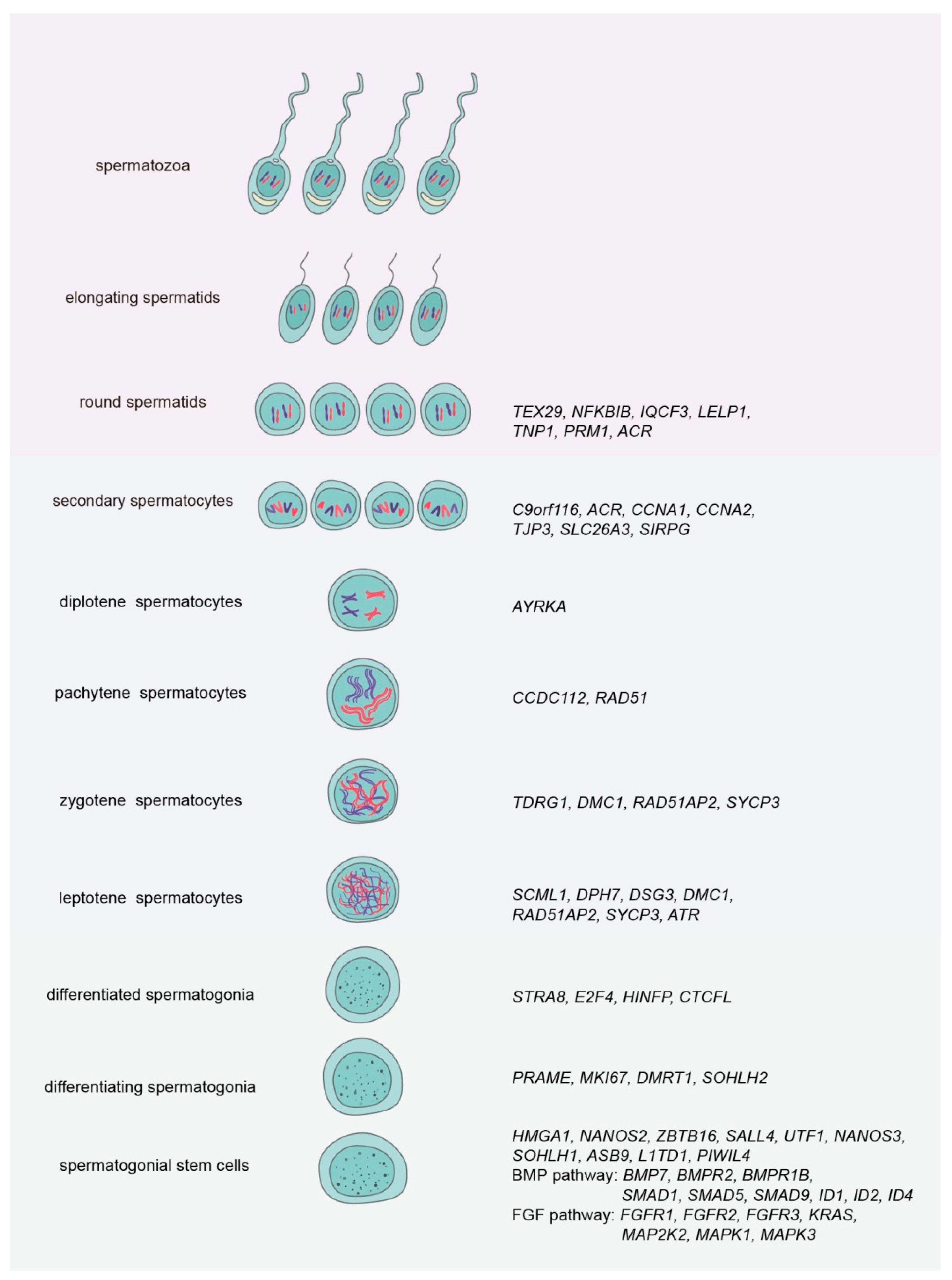
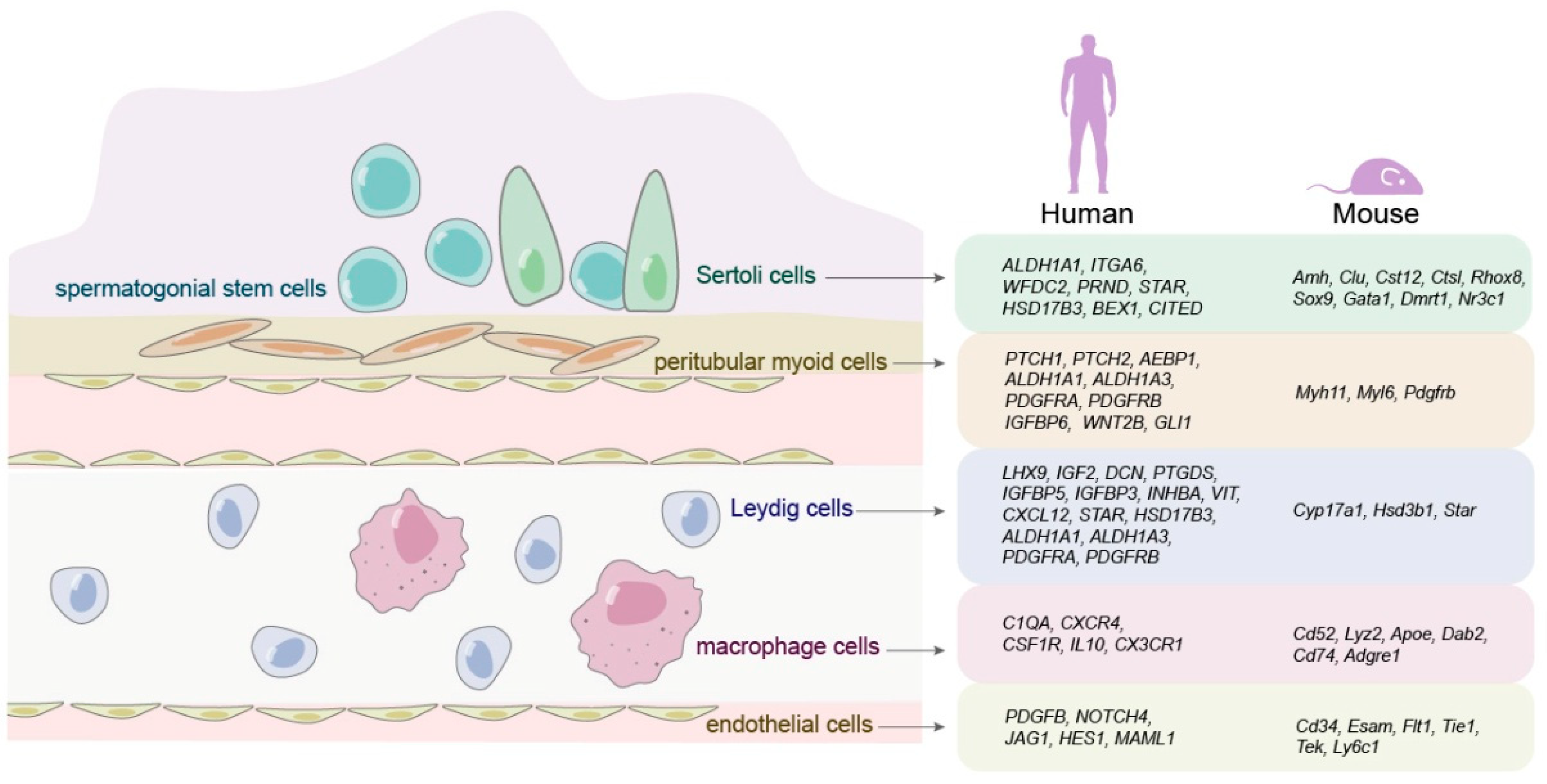
| Cells | Markers | Stages | Novel Genes | References |
|---|---|---|---|---|
| Germ cells | Ddx4 | Undifferentiated spermatogonia | Nanos2, Nanos3, Eomes, Pax7, Rhox10, Tspan8, Sall4, Sdc4, Bcl6, Taf4b, Lhx1, Dusp6, Epha2, Ptpn13, Pvr, Tcl1 | [12,28] |
| Differentiatingspermatogonia | Uchl1, Tcea3, Crabp1, Dmrtb1, Tex101, Hspa5, Stra8, Sycp3, Prdm9, Hormad1, Hormad2, Sycp1, Tex15 | [12,28] | ||
| Early spermatocytes | Meioc, Prdm3, Top2a, Smc3 | [29] | ||
| Spermatocytes | Piwil1, Pttg1, Insl6, Spag6, Tbpl1, Sycp1, Sycp2, Sycp3, Hzafx | [12,28,29] | ||
| Round spermatids | Acrv1, Tssk1, Spaca1, Tsga8, Pgk2, Cd37, Cd63, Cd96, Cd177, Ranbp9, Morf4l1, Catsper3, Cstsper4, Spata25, Izumo1 | [28,29,30] | ||
| Elongated spermatids | Prm1, Prm2, Prm3, Tnp1, Tnp2, Hspa1l, Izumo3, Tssk6, Dnajb3 | [12,28] |
| Types | Novel Genes | References | |
|---|---|---|---|
| Maturation arrest | CDY2, HSFY | [105] | |
| NOA | SCOS | Pramef12, H3t, PLK4, CARF, FGF9, IGF1, ETV5, HnRNPL, PRPS2 | [41,106,107,108,109,110,111,112,113] |
| Unclassified NOA | TDRD7, TDRD9, TEX15, DMC1, DGCR8, FANCM, DDR1, SAM68, RanBP3, RNF212, STAG3, NPAS2 | [100,114,115,116,117,118,119,120,121,122,123,124,125] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Du, L.; Chen, W.; Cheng, Z.; Wu, S.; He, J.; Han, L.; He, Z.; Qin, W. Novel Gene Regulation in Normal and Abnormal Spermatogenesis. Cells 2021, 10, 666. https://doi.org/10.3390/cells10030666
Du L, Chen W, Cheng Z, Wu S, He J, Han L, He Z, Qin W. Novel Gene Regulation in Normal and Abnormal Spermatogenesis. Cells. 2021; 10(3):666. https://doi.org/10.3390/cells10030666
Chicago/Turabian StyleDu, Li, Wei Chen, Zixin Cheng, Si Wu, Jian He, Lu Han, Zuping He, and Weibing Qin. 2021. "Novel Gene Regulation in Normal and Abnormal Spermatogenesis" Cells 10, no. 3: 666. https://doi.org/10.3390/cells10030666
APA StyleDu, L., Chen, W., Cheng, Z., Wu, S., He, J., Han, L., He, Z., & Qin, W. (2021). Novel Gene Regulation in Normal and Abnormal Spermatogenesis. Cells, 10(3), 666. https://doi.org/10.3390/cells10030666





