Functional Expression of Choline Transporters in Human Neural Stem Cells and Its Link to Cell Proliferation, Cell Viability, and Neurite Outgrowth
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. RNA Extraction and the Quantitative Real-Time Polymerase Chain Reaction (qPCR)
2.3. Western Blotting
2.4. Immunocytochemistry
2.5. Extracellular [3H]choline Uptake Assay
2.6. Cell Proliferation and Cell Viability Assay
2.7. Caspase-3/7 Activity Assay
2.8. Neurite Outgrowth Assay
2.9. Statistics Analysis
3. Results
3.1. Expression of Choline Transporter mRNA and Protein in hNSCs
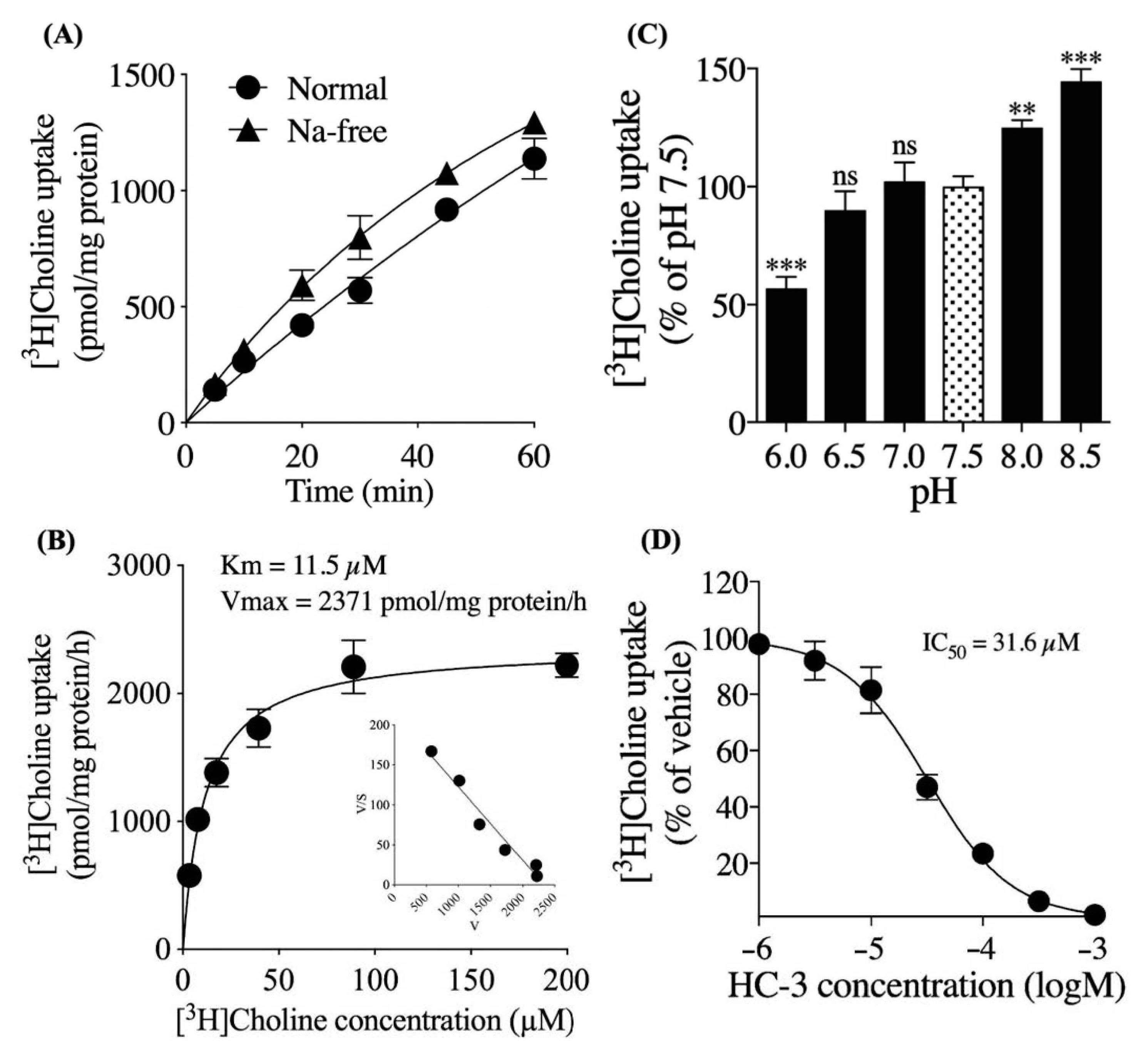
3.2. Characteristics of Extracellular [3H]choline Uptake in hNSCs
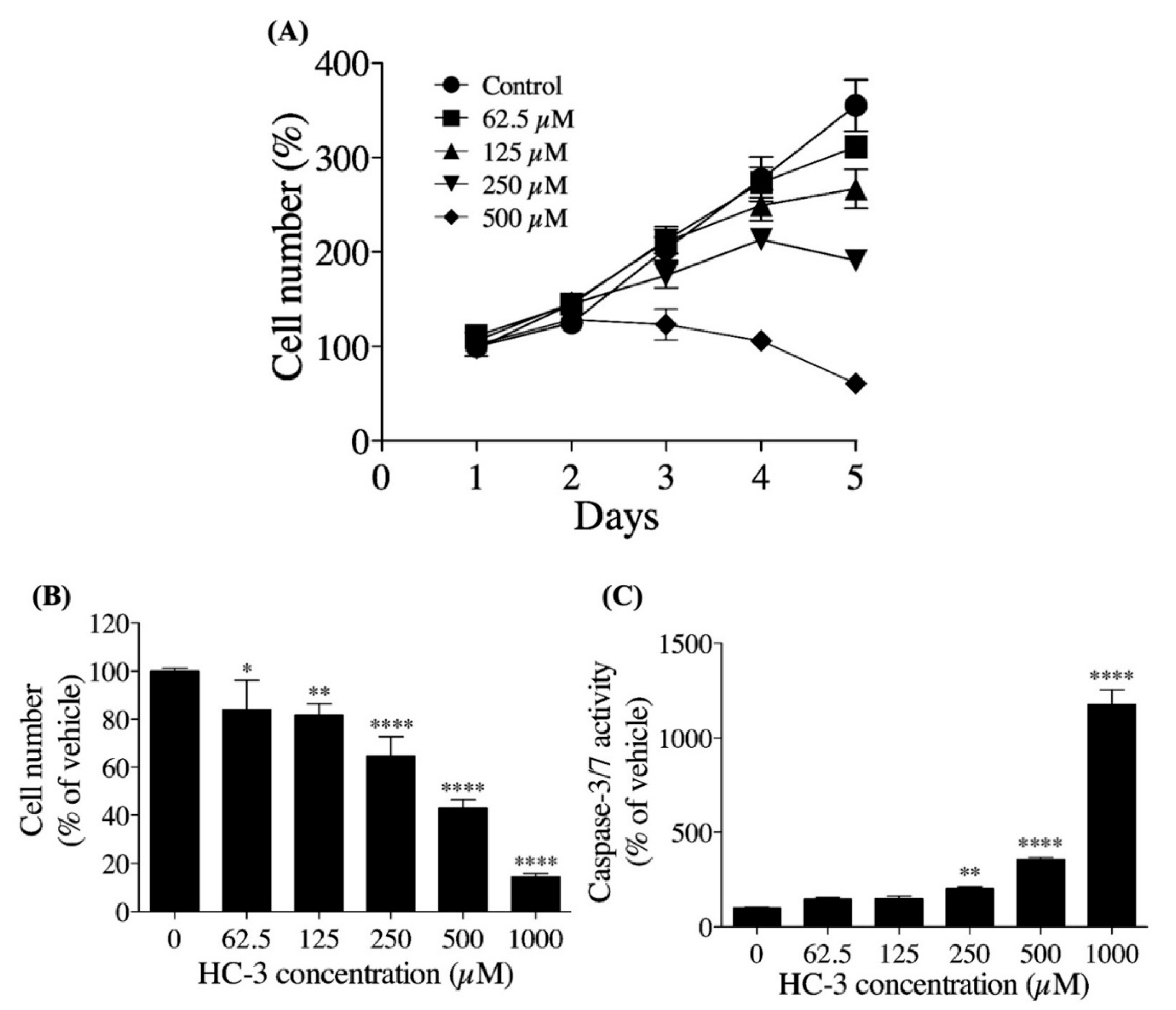
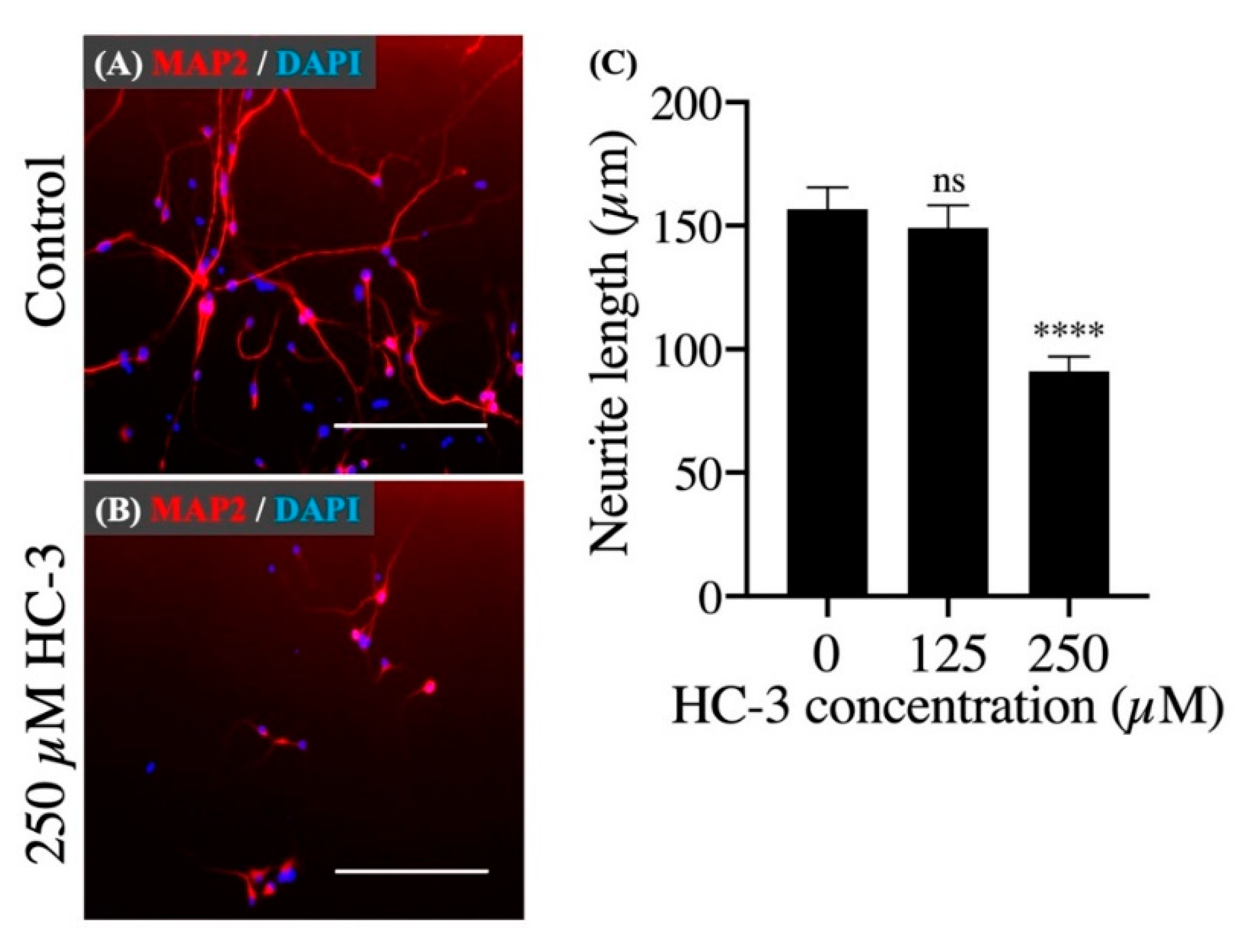
3.3. Extracellular Choline Uptake Inhibition on Cellular Activities in hNSCs
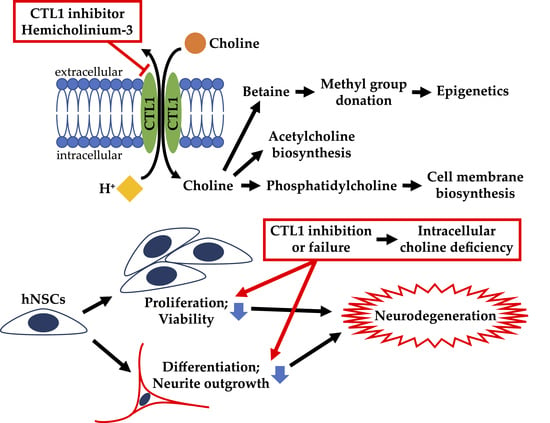
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Reynolds, B.A.; Weiss, S. Clonal and Population Analyses Demonstrate That an EGF-Responsive Mammalian Embryonic CNS Precursor Is a Stem Cell. Dev. Biol. 1996, 175, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Weiss, S.; Reynolds, B.A.; Vescovi, A.L.; Morshead, C.; Craig, C.G.; van der Kooy, D. Is There a Neural Stem Cell in the Mammalian Forebrain? Trends Neurosci. 1996, 19, 387–393. [Google Scholar] [CrossRef]
- Song, H.; Stevens, C.F.; Gage, F.H. Astroglia Induce Neurogenesis from Adult Neural Stem Cells. Nature 2002, 417, 39–44. [Google Scholar] [CrossRef]
- Nagoshi, N.; Tsuji, O.; Nakamura, M.; Okano, H. Cell Therapy for Spinal Cord Injury Using Induced Pluripotent Stem Cells. Regen. Ther. 2019, 11, 75–80. [Google Scholar] [CrossRef]
- Zeisel, S.H. Choline: Critical Role during Fetal Development and Dietary Requirements in Adults. Annu. Rev. Nutr. 2006, 26, 229–250. [Google Scholar] [CrossRef]
- Sarter, M.; Parikh, V. Choline Transporters, Cholinergic Transmission and Cognition. Nat. Rev. Neurosci. 2005, 6, 48–56. [Google Scholar] [CrossRef]
- Kempson, S.A.; Montrose, M.H. Osmotic Regulation of Renal Betaine Transport: Transcription and Beyond. Pflug. Arch. 2004, 449, 227–234. [Google Scholar] [CrossRef]
- Krzysztof Blusztajn, J.; J Mellott, T. Choline Nutrition Programs Brain Development via DNA and Histone Methylation. Cent. Nerv. Syst. Agents Med. Chem. 2012, 12, 82–94. [Google Scholar] [CrossRef] [PubMed]
- Michel, V.; Yuan, Z.; Ramsubir, S.; Bakovic, M. Choline Transport for Phospholipid Synthesis. Exp. Biol. Med. 2006, 231, 490–504. [Google Scholar] [CrossRef]
- Misawa, H.; Nakata, K.; Matsuura, J.; Nagao, M.; Okuda, T.; Haga, T. Distribution of the High-Affinity Choline Transporter in the Central Nervous System of the Rat. Neuroscience 2001, 105, 87–98. [Google Scholar] [CrossRef]
- Traiffort, E.; Ruat, M.; O’Regan, S.; Meunier, F.M. Molecular Characterization of the Family of Choline Transporter-like Proteins and Their Splice Variants: The Choline Transporter-like Proteins. J. Neurochem. 2005, 92, 1116–1125. [Google Scholar] [CrossRef]
- Inazu, M. Choline Transporter-like Proteins CTLs/SLC44 Family as a Novel Molecular Target for Cancer Therapy. Biopharm. Drug Dispos. 2014, 35, 431–449. [Google Scholar] [CrossRef] [PubMed]
- Inazu, M. Functional Expression of Choline Transporters in the Blood-Brain Barrier. Nutrients 2019, 11, 2265. [Google Scholar] [CrossRef]
- Yara, M.; Iwao, B.; Hara, N.; Yamanaka, T.; Uchino, H.; Inazu, M. Molecular and Functional Characterization of Choline Transporter in the Human Trophoblastic Cell Line JEG-3 Cells. Placenta 2015, 36, 631–637. [Google Scholar] [CrossRef] [PubMed]
- Bennett, J.A.; Mastrangelo, M.A.; Ture, S.K.; Smith, C.O.; Loelius, S.G.; Berg, R.A.; Shi, X.; Burke, R.M.; Spinelli, S.L.; Cameron, S.J.; et al. The Choline Transporter Slc44a2 Controls Platelet Activation and Thrombosis by Regulating Mitochondrial Function. Nat. Commun. 2020, 11, 3479. [Google Scholar] [CrossRef] [PubMed]
- Yabuki, M.; Inazu, M.; Yamada, T.; Tajima, H.; Matsumiya, T. Molecular and Functional Characterization of Choline Transporter in Rat Renal Tubule Epithelial NRK-52E Cells. Arch. Biochem. Biophys. 2009, 485, 88–96. [Google Scholar] [CrossRef] [PubMed]
- Nishiyama, R.; Nagashima, F.; Iwao, B.; Kawai, Y.; Inoue, K.; Midori, A.; Yamanaka, T.; Uchino, H.; Inazu, M. Identification and Functional Analysis of Choline Transporter in Tongue Cancer: A Novel Molecular Target for Tongue Cancer Therapy. J. Pharmacol. Sci. 2016, 131, 101–109. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nagashima, F.; Nishiyama, R.; Iwao, B.; Kawai, Y.; Ishii, C.; Yamanaka, T.; Uchino, H.; Inazu, M. Molecular and Functional Characterization of Choline Transporter-like Proteins in Esophageal Cancer Cells and Potential Therapeutic Targets. Biomol. Ther. 2018, 26, 399–408. [Google Scholar] [CrossRef]
- Koepsell, H.; Lips, K.; Volk, C. Polyspecific Organic Cation Transporters: Structure, Function, Physiological Roles, and Biopharmaceutical Implications. Pharm. Res. 2007, 24, 1227–1251. [Google Scholar] [CrossRef]
- Craciunescu, C.N.; Albright, C.D.; Mar, M.H.; Song, J.; Zeisel, S.H. Choline Availability during Embryonic Development Alters Progenitor Cell Mitosis in Developing Mouse Hippocampus. J. Nutr. 2003, 133, 3614–3618. [Google Scholar] [CrossRef] [PubMed]
- Glenn, M.J.; Gibson, E.M.; Kirby, E.D.; Mellott, T.J.; Blusztajn, J.K.; Williams, C.L. Prenatal Choline Availability Modulates Hippocampal Neurogenesis and Neurogenic Responses to Enriching Experiences in Adult Female Rats: Prenatal Choline and Adult Neurogenesis. Eur. J. Neurosci. 2007, 25, 2473–2482. [Google Scholar] [CrossRef] [PubMed]
- Albright, C.D.; Friedrich, C.B.; Brown, E.C.; Mar, M.H.; Zeisel, S.H. Maternal Dietary Choline Availability Alters Mitosis, Apoptosis and the Localization of TOAD-64 Protein in the Developing Fetal Rat Septum. Brain Res. Dev. Brain Res. 1999, 115, 123–129. [Google Scholar] [CrossRef]
- Albright, C.D.; Tsai, A.Y.; Friedrich, C.B.; Mar, M.H.; Zeisel, S.H. Choline Availability Alters Embryonic Development of the Hippocampus and Septum in the Rat. Brain Res. Dev. Brain Res. 1999, 113, 13–20. [Google Scholar] [CrossRef]
- Williams, C.L.; Meck, W.H.; Heyer, D.D.; Loy, R. Hypertrophy of Basal Forebrain Neurons and Enhanced Visuospatial Memory in Perinatally Choline-Supplemented Rats. Brain Res. 1998, 794, 225–238. [Google Scholar] [CrossRef]
- Meck, W.H.; Williams, C.L.; Cermak, J.M.; Blusztajn, J.K. Developmental Periods of Choline Sensitivity Provide an Ontogenetic Mechanism for Regulating Memory Capacity and Age-Related Dementia. Front. Integr. Neurosci. 2007, 1, 7. [Google Scholar] [CrossRef] [PubMed]
- Cermak, J.M.; Holler, T.; Jackson, D.A.; Blusztajn, J.K. Prenatal Availability of Choline Modifies Development of the Hippocampal Cholinergic System. FASEB J. 1998, 12, 349–357. [Google Scholar] [CrossRef]
- Van der Veen, J.N.; Kennelly, J.P.; Wan, S.; Vance, J.E.; Vance, D.E.; Jacobs, R.L. The Critical Role of Phosphatidylcholine and Phosphatidylethanolamine Metabolism in Health and Disease. Biochim. Biophys. Acta 2017, 1859, 1558–1572. [Google Scholar] [CrossRef] [PubMed]
- Mellott, T.J.; Follettie, M.T.; Diesl, V.; Hill, A.A.; Lopez-Coviella, I.; Blusztajn, J.K. Prenatal Choline Availability Modulates Hippocampal and Cerebral Cortical Gene Expression. FASEB J. 2007, 21, 1311–1323. [Google Scholar] [CrossRef] [PubMed]
- Honda, M.; Nakashima, K.; Katada, S. Epigenetic Regulation of Human Neural Stem Cell Differentiation. Results Probl. Cell Differ. 2018, 66, 125–136. [Google Scholar] [CrossRef]
- Yung-Chi, C.; Prusoff, W.H. Relationship between the Inhibition Constant (KI) and the Concentration of Inhibitor Which Causes 50 per Cent Inhibition (I50) of an Enzymatic Reaction. Biochem. Pharmacol. 1973, 22, 3099–3108. [Google Scholar] [CrossRef]
- Brentnall, M.; Rodriguez-Menocal, L.; De Guevara, R.L.; Cepero, E.; Boise, L.H. Caspase-9, Caspase-3 and Caspase-7 Have Distinct Roles during Intrinsic Apoptosis. BMC Cell Biol. 2013, 14, 32. [Google Scholar] [CrossRef]
- Kim, J.W.; Hahn, K.R.; Yoo, D.Y.; Jung, H.Y.; Hwang, I.K.; Seong, J.K.; Yoon, Y.S. Methionine-Choline Deprivation Impairs Adult Hippocampal Neurogenesis in C57BL/6 Mice. J. Med. Food 2019, 22, 344–354. [Google Scholar] [CrossRef] [PubMed]
- Hedtke, V.; Bakovic, M. Choline Transport for Phospholipid Synthesis: An Emerging Role of Choline Transporter-like Protein 1. Exp. Biol. Med. 2019, 244, 655–662. [Google Scholar] [CrossRef]
- Nair, T.S.; Kommareddi, P.K.; Galano, M.M.; Miller, D.M.; Kakaraparthi, B.N.; Telian, S.A.; Arts, H.A.; El-Kashlan, H.; Kilijanczyk, A.; Lassig, A.A.D.; et al. SLC44A2 Single Nucleotide Polymorphisms, Isoforms, and Expression: Association with Severity of Meniere’s Disease? Genomics 2016, 108, 201–208. [Google Scholar] [CrossRef]
- Haga, T. Molecular Properties of the High-Affinity Choline Transporter CHT1. J. Biochem. 2014, 156, 181–194. [Google Scholar] [CrossRef]
- Zeisel, S.H. Choline: Needed for Normal Development of Memory. J. Am. Coll. Nutr. 2000, 19, 528S–531S. [Google Scholar] [CrossRef] [PubMed]
- O’Donoghue, N.; Sweeney, T.; Donagh, R.; Clarke, K.J.; Porter, R.K. Control of Choline Oxidation in Rat Kidney Mitochondria. Biochim. Biophys. Acta 2009, 1787, 1135–1139. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, T.; Suwanai, H.; Shikuma, J.; Suzuki, R.; Yamanaka, T.; Odawara, M.; Inazu, M. Protein Kinase C Promotes Choline Transporter-like Protein 1 Function via Improved Cell Surface Expression in Immortalized Human Hepatic Cells. Mol. Med. Rep. 2020, 21, 777–785. [Google Scholar] [CrossRef]
- Ridgway, N.D. The Role of Phosphatidylcholine and Choline Metabolites to Cell Proliferation and Survival. Crit. Rev. Biochem. Mol. Biol. 2013, 48, 20–38. [Google Scholar] [CrossRef] [PubMed]
- Yen, C.L.; Mar, M.H.; Meeker, R.B.; Fernandes, A.; Zeisel, S.H. Choline Deficiency Induces Apoptosis in Primary Cultures of Fetal Neurons. FASEB J. 2001, 15, 1704–1710. [Google Scholar] [CrossRef] [PubMed]
- Niculescu, M.D.; Craciunescu, C.N.; Zeisel, S.H. Dietary Choline Deficiency Alters Global and Gene-Specific DNA Methylation in the Developing Hippocampus of Mouse Fetal Brains. FASEB J. 2006, 20, 43–49. [Google Scholar] [CrossRef]
- Huang, M.; Huang, N.N.; Li, Y.Y.; Wu, K.X.; Guo, M.Z.; Zhu, C.D. Effects of paraquat on the proliferation of human neural stem cells via DNA methylation. Chin. J. Ind. Hyg. Occup. Dis. 2019, 37, 161–168. [Google Scholar] [CrossRef]
- Paoletti, L.; Elena, C.; Domizi, P.; Banchio, C. Role of Phosphatidylcholine during Neuronal Differentiation. IUBMB Life 2011, 63, 714–720. [Google Scholar] [CrossRef]
- Ryu, J.R.; Hong, C.J.; Kim, J.Y.; Kim, E.K.; Sun, W.; Yu, S.W. Control of Adult Neurogenesis by Programmed Cell Death in the Mammalian Brain. Mol. Brain 2016, 9, 43. [Google Scholar] [CrossRef]
- Doe, C.Q. Neural Stem Cells: Balancing Self-Renewal with Differentiation. Development 2008, 135, 1575–1587. [Google Scholar] [CrossRef]
- Greene, N.D.E.; Copp, A.J. Neural Tube Defects. Annu. Rev. Neurosci. 2014, 37, 221–242. [Google Scholar] [CrossRef] [PubMed]
- Shaw, G.M.; Finnell, R.H.; Blom, H.J.; Carmichael, S.L.; Vollset, S.E.; Yang, W.; Ueland, P.M. Choline and Risk of Neural Tube Defects in a Folate-Fortified Population. Epidemiology 2009, 20, 714–719. [Google Scholar] [CrossRef] [PubMed]
- Okae, H.; Iwakura, Y. Neural Tube Defects and Impaired Neural Progenitor Cell Proliferation in Gbeta1-Deficient Mice. Dev. Dyn. 2010, 239, 1089–1101. [Google Scholar] [CrossRef] [PubMed]
- Khoramian, T.S.; Babaei, A.S.; Amini, M.; Khodagholi, F. Modulation of H2O2-Induced Neurite Outgrowth Impairment and Apoptosis in PC12 Cells by a 1,2,4-Triazine Derivative. Basic Clin. Neurosci. 2012, 3, 49–60. [Google Scholar]
- Hou, S.T.; Jiang, S.X.; Smith, R.A. Permissive and Repulsive Cues and Signalling Pathways of Axonal Outgrowth and Regeneration. Int. Rev. Cell Mol. Biol. 2008, 267, 125–181. [Google Scholar] [CrossRef] [PubMed]
- McKeon, R.J.; Schreiber, R.C.; Rudge, J.S.; Silver, J. Reduction of Neurite Outgrowth in a Model of Glial Scarring Following CNS Injury Is Correlated with the Expression of Inhibitory Molecules on Reactive Astrocytes. J. Neurosci. 1991, 11, 3398–3411. [Google Scholar] [CrossRef] [PubMed]
- Fagerberg, C.R.; Taylor, A.; Distelmaier, F.; Schrøder, H.D.; Kibæk, M.; Wieczorek, D.; Tarnopolsky, M.; Brady, L.; Larsen, M.J.; Jamra, R.A.; et al. Choline Transporter-like 1 Deficiency Causes a New Type of Childhood-Onset Neurodegeneration. Brain 2020, 143, 94–111. [Google Scholar] [CrossRef] [PubMed]


| Target Gene | Accession Number | Assay ID |
|---|---|---|
| CTL1 (SCL44A1) | NM_080546 | Hs_00223114m1 |
| CTL2 (SCL44A2) | NM_020428 | Hs_01105936m1 |
| CTL3 (SCL44A3) | NM_001114106 | Hs_00537043m1 |
| CTL4 (SCL44A4) | NM_001178044 | Hs_00228901m1 |
| CTL5 (SCL44A5) | NM_152697 | Hs_01120485m1 |
| CHT1 (SCL5A7) | NM_021815 | Hs_00222367m1 |
| OCT1 (SLC22A1) | NM_003057 | Hs_00427554m1 |
| OCT2 (SLC22A2) | NM_003058 | Hs_01010723m1 |
| OCT3 (SLC22A3) | NM_021977 | Hs_01009568m1 |
| GAPDH | NM_002046 | Hs_99999905m1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fujita, Y.; Nagakura, T.; Uchino, H.; Inazu, M.; Yamanaka, T. Functional Expression of Choline Transporters in Human Neural Stem Cells and Its Link to Cell Proliferation, Cell Viability, and Neurite Outgrowth. Cells 2021, 10, 453. https://doi.org/10.3390/cells10020453
Fujita Y, Nagakura T, Uchino H, Inazu M, Yamanaka T. Functional Expression of Choline Transporters in Human Neural Stem Cells and Its Link to Cell Proliferation, Cell Viability, and Neurite Outgrowth. Cells. 2021; 10(2):453. https://doi.org/10.3390/cells10020453
Chicago/Turabian StyleFujita, Yosuke, Tomoki Nagakura, Hiroyuki Uchino, Masato Inazu, and Tsuyoshi Yamanaka. 2021. "Functional Expression of Choline Transporters in Human Neural Stem Cells and Its Link to Cell Proliferation, Cell Viability, and Neurite Outgrowth" Cells 10, no. 2: 453. https://doi.org/10.3390/cells10020453
APA StyleFujita, Y., Nagakura, T., Uchino, H., Inazu, M., & Yamanaka, T. (2021). Functional Expression of Choline Transporters in Human Neural Stem Cells and Its Link to Cell Proliferation, Cell Viability, and Neurite Outgrowth. Cells, 10(2), 453. https://doi.org/10.3390/cells10020453