Abstract
Some genetic abnormalities of multiple myeloma (MM) detected more than two decades ago remain major prognostic factors. In recent years, the introduction of cutting-edge genomic methodologies has enabled the extensive deciphering of genomic events in MM. Although none of the alterations newly discovered have significantly improved the stratification of the outcome of patients with MM, some of them, point mutations in particular, are promising targets for the development of personalized medicine. This review summarizes the main genetic abnormalities described in MM together with their prognostic impact, and the therapeutic approaches potentially aimed at abrogating the undesirable pathogenic effect of each alteration.
1. Introduction
Multiple myeloma (MM) is a genetically complex and heterogeneous neoplasm in which the concurrency of multiple genomic events leads to tumor development and progression (Figure 1). Moreover, genetic abnormalities are major prognostic factors in MM. Molecular cytogenetic methodologies such as G-band karyotyping, fluorescence in situ hybridization (FISH), and comparative genomic hybridization (CGH) combined with more advanced genetic techniques, encompassing single nucleotide polymorphism (SNP) arrays and, more recently, next-generation sequencing (NGS) [1], have made it possible to identify numerous recurrent chromosomal and genetic alterations in MM that can be categorized into three types: chromosomal translocations, copy number abnormalities (CNAs), and point mutations [2]. Many of these abnormalities have been tested in several powerful studies involving a large number of patients. The new revised international staging system (R-ISS), developed by the International Myeloma Working Group (IMWG), requires the analysis of translocations t(4;14) and t(14;16), and 17p deletion to stratify risk in MM patients [3].
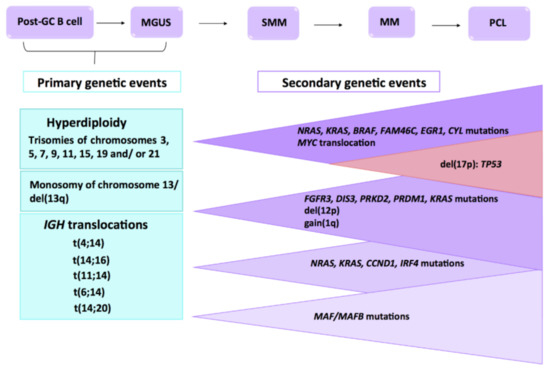
Figure 1.
Multistep molecular pathogenesis of multiple myeloma (MM) Primary and secondary genetic events involved in the MM transformation and progression from the precursor disease entities, such as monoclonal gammopathy of undetermined significance (MGUS) and smoldering multiple myeloma (SMM) to MM, and eventually to extramedullary myeloma/plasma cell leukemia (PCL). Adapted from Chesi M et al. and Manier S et al. [4,5].
Most of the abnormalities present in MM patients are not druggable, but progress in biological, clinical, and therapeutic research has helped identify drugs and combinations thereof that are more effective for treating particular abnormalities, as well as making several specific target agents available.
2. Genetic Abnormalities
2.1. Chromosomal Translocations
The most common translocations observed in MM patients are those involving the IGH gene (14q32), which is translocated to diverse oncogenes whose expression is upregulated under the influence of the powerful IGH enhancer. Higher levels of expression of certain oncogenes may confer a selective advantage on those subclones that carry the translocations [4]. IGH translocations are considered initiating events and are therefore named primary translocations. They are present in up to 50% of patients, and mainly involve five chromosomal loci, 11q13, 6p21, 4p16, 16q23, and 20q11, that contain the CCND1, CCND3, FGFR3/NSD2, MAF, and MAFB oncogenes, respectively. These translocations lead to the overexpression of the oncogene juxtaposed to the 3’ intronic IGH enhancer, and, in the particular case of t(4;14), provoke the simultaneous dysregulation of two oncogenes [5].
Rearrangements of the MYC oncogene, which cause it to be overexpressed, occur frequently in MM, and are considered secondary cytogenetic events. MYC translocations are observed in approximately 15–20% of patients newly diagnosed with MM [1,6].
2.1.1. Translocations t(11;14) and t(6;14)
Translocations t(11;14) and t(6;14) juxtapose the IGH enhancer with CCND1 (15–20%) and CCND3 (1–4%), respectively. Most of them occur at the switch region breakpoints, although a subset of CCND1 translocations has been described as originating from errors in V(D)J recombination [7]. The cyclin D dysregulation induced by both translocations inactivates RB1 (retinoblastoma), allowing cell-cycle progression [8].
Prognosis
Traditionally, newly diagnosed MM (NDMM) patients with t(11;14) have been categorized as standard risk [9,10] (Table 1). However, some recent studies indicate that t(11;14) is associated with shorter survival in the era of novel agents, acting as a marker of intermediate risk [11,12,13,14]. These data also suggest that high-dose chemotherapy benefits patients with t(11;14) [13,15]. Although the low prevalence of t(6;14) precludes the estimation of survival, this translocation is included in the standard-risk category [10]. Patients with one of these translocations are more likely to have bone disease [16].

Table 1.
Prognosis for each cytogenetic abnormality according to the risk stratification models for multiple myeloma.
Therapeutic Implications
In vitro experiments with MM cell lines and primary samples with t(11;14) treated with venetoclax revealed a high degree of sensitivity to this BCL2 inhibitor. These preclinical results were confirmed when patients with relapsed or refractory multiple myeloma (RRMM) bearing t(11;14) were treated with venetoclax in monotherapy, yielding an overall response rate (ORR) and median progression-free survival (PFS) of 86% and 6.6 months, respectively. Responses to venetoclax were associated with higher ratios of BCL2/BCL2L1 or BCL2/MCL1 [1,17]. One of the mechanisms of resistance to venetoclax is the overexpression of Mcl-1. Bortezomib has been reported to overcome this resistance by upregulating Noxa and subsequently neutralizing Mcl-1 [18]. On this basis, venetoclax has been tested in combination with dexamethasone and bortezomib in a phase Ib study, which yielded promising results in terms of response (ORR 67%) and survival (median PFS 9.5 months) [19]. Recently, a phase 3 trial testing the addition of venetoclax to bortezomib and dexamethasone showed significantly increased response rates (ORR 85% vs. 70%) and PFS (not reached vs. 9.9 months, p < 0.0001) without increasing mortality in patients with high BCL2 levels or t(11;14). However, the mortality in the experimental arm was higher in patients without t(11;14) due to increased infection rate [20,21]. According to these data, two phase 2 trials with venetoclax in combination with carfilzomib, and with daratumumab plus bortezomib, are now recruiting, and a phase 3 trial involving a combination with pomalidomide and dexamethasone is progress. Several clinical trials with Mcl-1 inhibitors, such as ABC294640 (sphingosine kinase 2 inhibitor), and with AZD5991 are underway [22,23,24,25]. AZD5991 is a small Mcl-1 inhibitor that activates the Bak-dependent mitochondrial apoptotic pathway [24], and its combination with venetoclax has shown synergism. Furthermore, dexamethasone enhances the expression of Bcl-2, which sensitizes MM cells to these kinds of agents [25].
2.1.2. Translocation t(4;14)
This translocation is observed in approximately 15% of patients, its incidence decreasing with age [5,26]. Translocation t(4;14) leads to simultaneous overexpression of two genes, FGFR3 (fibroblast growth factor receptor 3) and NSD2 (nuclear receptor binding SET domain protein 2), the latter also known as MMSET (multiple myeloma SET domain protein) and WHSC1 (Wolf–Hirschhorn syndrome candidate 1) [8]. Whereas MMSET is overexpressed in all cases with t(4;14), overexpression of FGFR3 is observed in 70% of patients with t(4;14) due to the deletion of the der(14) chromosome in one-third of cases. This suggests that MMSET activation may play a more critical role than FGFR3 in the pathogenesis of MM with t(4;14), contributing to increased proliferation, a change in cellular adhesion, and high tumorigenicity [27]. MMSET encodes histone 3 lysine 36 (H3K36) methyltransferase, and its overactivation in MM has been shown to affect the expression of several genes, including some related to cancer [8,28]. High levels of expression of CCND2 have been observed in myeloma cells with t(4;14), although the underlying mechanisms are not completely understood [29,30]. FGFR3 encodes a receptor tyrosine kinase for fibroblast growth factors and plays an essential role in regulating cell proliferation, differentiation, and apoptosis. The cytotoxic effect of targeting FGFR3 in MM cells with t(4;14) is also evidence of the contribution of FGFR3 in myelomagenesis.
Prognosis
The t(4;14) translocation was associated with unfavorable prognosis in several large studies [5] and patients bearing it were classified in the high-risk category [9,31,32] (Table 1). Several studies demonstrated that bortezomib overcomes the negative prognostic impact of t(4;14) (Table 2). Induction therapy based on bortezomib-dexamethasone along with high-dose melphalan improved event-free survival and overall survival (OS) of NDMM patients with t(4;14) [33]. Bortezomib in combination with thalidomide and dexamethasone (VTD) followed by double autologous hematopoietic stem-cell transplantation (ASCT) could overcome the bad prognosis associated with t(4;14) [34,35], though this drug applied on maintenance hardly had any influence on prognosis compared with thalidomide maintenance [36,37].

Table 2.
Impact of cytogenetic abnormalities on survival outcomes in MM patients according to different treatment regimens.
In RRMM patients with t(4;14), the combination of lenalidomide and dexamethasone has been reported to achieve an OS similar to patients without this cytogenetic abnormality, suggesting that this regimen could also overcome its poor prognosis [38]. On the other hand, a retrospective study showed lower ORRs and shorter median PFS and OS in RRMM patients with t(4;14) using the same scheme [39]. The difference between the two studies could be related to differences in age, in the median number of previous therapies, and in the proportion of patients with earlier thalidomide or bortezomib therapy [39]. Strikingly, pomalidomide, despite also being an immunomodulatory drug (IMID), did not seem to benefit this group of patients [40]. Combining elotuzumab with lenalidomide and dexamethasone produced interesting results such as a PFS of about 20 months for patients with t(4;14), similar to the survival observed in standard-risk patients [41]. Moreover, the proteasome inhibitor (PI) carfilzomib used as monotherapy was able to improve the ORR and OS by up to 12 months in patients with RRMM and t(4;14) [42] (Table 2).
Therapeutic Implications
Targeted therapy has been employed to treat patients with MM bearing t(4;14) that uses monoclonal antibodies (mAbs) that block ligand binding sites to FGFR3. Most of these mAbs are designed to recognize the mutant residue serine249 (S249) of FGFR3 since it results in ligand-independent activation of FGFR3. Mouse studies using R3Mab and MFGR1877A mAbs revealed that both of them induce antibody-dependent cell-mediated cytotoxicity [43,44]. Additionally, PRO-001 mAb was demonstrated to bind to FGFR3 and inhibit FGFR3 autophosphorylation and downstream signaling [45]. Moreover, the activity of dovitinib, a small molecule inhibitor of FGFR3, was tested in vitro and in xenograft animal models carrying t(4;14) [46]. However, phase II trials were only able to demonstrate the stabilization of the disease [47,48]. Further studies with FGFR3 mAbs in combination with other treatments, such as chemotherapy, bortezomib, or lenalidominde, are needed to optimize anti-FGFR3 antibody-based treatment. MMSET is involved in pathways related to DNA repair, modulating the response to chemotherapy [49,50]. However, at present, inhibitory molecules against this gene are, at best, at the developmental stage.
2.1.3. Translocations t(14;16) and t(14;20)
Translocations t(14;16), observed in 5% of MM, and t(14;20), seen in fewer than 2% of patients, deregulate the MAF and MAFB genes, respectively. Both genes belong to the MAF family and are leucine zipper-containing transcription factors. Increased levels of MAF induce upregulation of cyclin D2 through its transactivation function, resulting in an accelerated rate of division and DNA synthesis [8,29,51]. Overexpression of MAFB in MM induces proliferation and protects cells from drug-induced apoptosis, conferring resistance. Myeloma cells with t(14;16) or t(14;20) present genetic instability, similar to that observed for t(4;14) [52].
Prognosis
Translocations t(14;16) and t(14;20) are considered high-risk cytogenetic factors [53] (Table 1). The MAF molecular subgroup has also been associated with high levels of free light chain in serum and therefore a higher incidence of acute renal failure [54]. Although data from the Mayo Clinic associated t(14;16) with poor prognosis [55], these results have not been corroborated by the IFM group [56]. Nevertheless, this discrepancy could be related to the fact that the patients from the first study were treated with conventional chemotherapy, whereas more than 50% of the patients from the second study received a double transplant.
Therapeutic Implications
MAF transcription may be regulated by the MEK–ERK pathway. Thus, the extracellular signal-regulated kinase (ERK) has been demonstrated to activate the transcription factor FOS (a subunit of AP1), which is bound to the MAF promoter, inducing its transcription. Some studies reported that MEK inhibition induces apoptosis of MAF-expressing myelomas and blocks survival signals provided by the microenvironment [57,58,59]. The blockade of FOS activity is also toxic for MM cells harboring t(4;14), which is related to the MAF upregulation detected in myeloma cells with MMSET translocation.
AZD6244 is a novel oral and highly specific MEK inhibitor that targets not only MM cells, but also the microenvironment. Its combination with the HSP90 inhibitor TAS-116 is known to enhance the anti-myeloma effect [59]. Trametinib (GSK1120212) is a reversible MEK inhibitor that could improve ORRs when combined with AKT inhibitors [60]. Some studies show that the presence of t(14;20) in MM cell lines confers resistance to PIs, such as bortezomib [61,62]. Bortezomib leads to the stabilization and accumulation of phosphorylated MAF, which could partially explain the low efficacy of this therapy. Consequently, the inhibition of the serine/threonine protein kinase GSK3, which is responsible for the MAF and MAFB phosphorylation, could block proliferation and colony formation of MAF-expressing cells [63].
2.1.4. Translocation of MYC Oncogene
Most of the MYC rearrangements bring about the juxtaposition of a super-enhancer adjacent to MYC [64], which leads to increased expression levels of MYC mRNA [65]. MYC overexpression results in an increased DNA replication rate. This causes DNA damage and an increase in reactive oxygen species (ROS). MYC lesions are also one of the most important events promoting disease progression [66,67,68] and have recently been associated with a high tumoral burden [69].
Prognosis
Over time, MYC deregulation has come to be regarded as a central event of MM biology. A MYC activation signature was identified in the transition from premalignant conditions of plasma cell dyscrasias to symptomatic myeloma [66,70,71]. The impact of MYC rearrangements in the prognosis of MM patients is controversial. A large study by the IFM group did not find any association between MYC translocations assessed by FISH and MM prognosis [72]. Conversely, other groups had inferior survival in patients with MYC rearrangements compared to patients without these abnormalities [5,6,73,74]. Recently, another study highlighted the crucial role of MYC rearrangements as an independent prognostic factor in NDMM [69].
Therapeutic Implications
The direct inhibition of transcription factors remains a great therapeutic challenge. Different indirect ways of reducing MYC activity have been explored in MM. Thus, the amplification of MYC generates high levels of replicative stress and ROS, with the consequence that myeloma cells harboring this translocation could be attacked with combinations of agents that block the stress response. ATR inhibitors combined with drugs that increase ROS or bortezomib can give rise to synergistic cytotoxicity [75]. Bromodomain and extra-terminal (BET) protein belong to a protein family that interacts with acetylated histones. These interactions allow the recruitment of chromatin regulators to precise chromatin sites, which results in the expression of specific genes. Different in vitro and in vivo preclinical studies using BET inhibitors demonstrated the downregulation of MYC expression [76,77]. Based on those preclinical studies, the BET inhibitor CPI-0610 progressed to a phase I clinical trial. Primary targets of CPI-0610 are MYC, IKZF1, and IRF4 [78]. Recently, the therapeutic potential of the proteolysis targeting chimera (PROTAC) molecule ARV-825 was proven in MM. This molecule induced degradation of BET proteins and, subsequently, downregulation of MYC expression [79].
Additionally, leflunomide, an agent approved for the treatment of the rheumatoid arthritis, also exhibits anti-myeloma activity through the downregulation of MYC protein in preclinical studies [80]. Leflunomide blocks the PIM family, which stabilizes MYC, and on the other hand, its active metabolite teriflunomide induces the proteasome degradation of MYC. Moreover, leflunomide in combination with lenalidomide inhibits cell growth [80]. A phase 2 trial with leflunomide in combination with pomalidomide and dexamethasone is currently underway.
Recently, MYC has been associated with higher levels of PARP-1, which has been implicated in DNA repair and indicates poor prognosis. Accordingly, PARP inhibitors could be a treatment option for these patients, although clinical trials are needed to confirm this effect [81].
2.2. Copy Number Abnormalities
2.2.1. Hyperdiploid/Hypodiploid
Most MM cases are aneuploid, in which there are frequent gains and losses of complete chromosomes or chromosome arms. According to the ploidy status, MM is usually categorized in hyperdiploid and non-hyperdiploid MM. The hyperdiploid (H-MM) group, which accounts for 50–60% of all MM cases, is characterized by the presence of trisomies that typically affect the odd chromosomes [82]. Hyperdiploidy seems to be an early event in MM evolution since it has been described in monoclonal gammopathy of undetermined significance (MGUS) [83]. The non-hyperdiploid MM (NH-MM) group includes hypodiploid (up to 44/45 chromosomes), pseudodiploid (44/45 to 46/47), and near-tetraploid (more than 74) cases. NH-MM is frequently characterized by the loss of chromosomes 13, 14, 16, and 22. Hyperhaploid karyotypes, as a result of the loss of nearly a haploid set of chromosomes, have also been found in MM. This MM group is monosomic for many autosomes, excluding chromosomes 3, 5, 7, 9, 11, 15, 18, 19, and 21 found in disomy. The most frequent monosomies/deletions present in hyperhaploid cases affect 17p, 1p, 13q, and 16q [84,85]. In particular, monosomy 17 or del(17p) was identified in all hyperhaploid patients.
The high proportion of chromosomal imbalances observed in MM is a clear sign of underlying genomic instability. In this context, the phenomenon of chromothripsis, defined by tens to hundreds of random chromosomal rearrangements involving localized genomic regions, has been observed in a very low percentage of NDMMs [86].
Prognosis
Several studies have shown that hyperdiploid patients have better response rates to treatment and longer survival than patients with other aneuploidies [87]. This favorable prognosis was observed irrespective of the therapeutic context [88,89,90]. However, not all trisomies have the same impact on survival; in particular, trisomies of chromosomes 3 and 5 improve the prognosis in patients treated with chemotherapy or bortezomib followed by ASCT, and also in those treated with non-intensive protocols, even in patients with t(4;14) [91].
On the other hand, patients treated with chemotherapy belonging to the non-hyperdiploid group are characterized by more aggressive clinical features [92] and worse outcomes [88,93], especially those with hypodiploid karyotypes. The introduction of new drugs has not significantly changed this scenario. Hyperhaploid karyotypes are associated with an adverse prognosis, even worse than that of the hypodyploid group, with an estimated 20–25% survival after five years, despite intensive treatments based on PIs, IMIDs, and tandem ASCT [85].
Finally, the presence of chromothripsis seems to confer a higher risk of relapse and shorter survival [86,94].
Therapeutic Implications
The search for therapeutic targets associated with gains and losses of genetic material is difficult because unlike translocations and mutations, chromosomal imbalances can influence the expression of thousands of genes with very different and sometimes opposite functions. Nevertheless, some specific CNAs involve oncogenes or tumor suppressor genes of known or potential relevance in myeloma pathogenesis, as can be seen in the following sections.
2.2.2. Deletion of 1p
Deletions of 1p, del(1p), have been observed in up to 30% of patients with MM [92,95,96,97]. This incidence increases up to 60% in plasma cell leukemia [98], indicating that del(1p) may be related to clonal evolution [99]. The minimally deleted regions (MDRs) identified in the interstitial deletions are the 1p12, 1p21, 1p22.1, and 1p32.3 regions, were the FAM46C, CDC14A, MTF2, and CDKN2C genes are located, respectively [95,99,100]. The most frequent MDR is 1p22, which accounts for 15–22% of cases [95,101].
Prognosis
Del(1p) is associated with shorter survival in transplant-eligible patients, according to data from both the Myeloma IX trial and the IFM group [95,101]. Patients with a loss of the CDC14A gene, located at 1p21, also had a poor prognosis after chemotherapy treatment followed by ASCT consolidation [102]. This adverse prognosis was also confirmed in patients with RRMM treated with lenalidomide and dexamethasone [103]. FAM46C downregulation as a consequence of del(1p12), mutation, or both, has been reported to confer resistance to dexamethasone and lenalidomide in vitro [104], although only the FAM46C loss, but not the mutation, was associated with a worse outcome in patients treated with triplet induction therapies [105].
Therapeutic Implications
Loss of FAM46C is associated in MM cell lines with an increase of cell migration mediated by the activation of the PI3K/Rac1 pathway, so it is reasonable to speculate that patients with del(1p12) or FAM46C mutations could benefit from PI3K and Rac1 inhibitors [106].
2.2.3. Gain of 1q
The long arm of chromosome 1 is gained (three copies) or amplified (more than three copies) in nearly 50% of patients with NDMM [107,108,109]. These percentages increase as the disease progresses [110,111,112], reaching up to 68% in RRMM [107]. Overexpression of the CKS1B gene, located at 1q21, was initially linked to myeloma cell growth and survival, and consequently to drug resistance [113,114,115]. Other genes mapped at 1q, such as MUC1, MCL1, ANP32E, BCL9, PSMD4, and PDZK1, have been proposed as candidate participants in myelomagenesis [92,116,117,118,119,120].
Prognosis
1q gains have been associated with poor prognosis in patients treated with chemotherapy [108,113], PIs [107], and IMIDs [121], even in transplant-eligible patients [108,113,122,123]. Lenalidomide maintenance was able to improve PFS without any impact on OS in this set of patients [124] (Table 2).
The triple combination VRD can overcome the negative prognostic impact of 1q gains occurring in isolation, but not when they are in combination with other high-risk genetic abnormalities or in the case of 1q amplification [121]. The concept of “double-hit” myeloma includes those patients with 1q amplification together with international staging system ISS stage 3, which are considered to have an ultra-high risk disease in spite of novel therapeutic regimens [105,125].
Therapeutic Implications
Anti-CD46 antibody conjugated with monomethyl auristatin F was tested in preclinical studies with successful outcomes. This drug is especially appealing in patients with gain(1q), given that the CD46 gene is located on the long arm of chromosome 1 and the CD46 antigen is overexpressed in this group of patients [126]. In this regard, in vitro studies recently demonstrated higher Mcl1 expression and greater efficacy of Mcl1 inhibitors for MM samples harboring amp1q [127], although clinical trials are required to confirm these results. A phase 1 clinical trial is currently in progress.
2.2.4. Deletion of 13q
Deletion of 13q, del(13q), detected by FISH, is present in around 45% of patients, taking into account both monosomy 13, which is the most frequent alteration involving chromosome 13, and interstitial deletions observed in up to 15% of cases [128,129]. Its actual incidence was ascertained through the introduction of FISH with locus-specific probes [130,131]. The tumor suppressor RB1 is lost in these chromosome 13 aberrations.
Prognosis
Del(13q) was described in MM for the first time in 1995 and has consistently been related to adverse prognosis [132] in almost all large series of patients treated with both conventional and high-dose therapy [93,130,133]. However, increasing evidence suggests that this unfavorable prognosis arises from its close association with other high-risk genetic features, such as t(4;14), which appears in combination with del(13q) in 80% of cases. Therefore, RB1 loss on its own would not be a negative prognostic factor [134]. An intriguing opposite effect of monosomy 13 and del(13q) was recently described in NDMM patients treated with PIs and/or IMIDs, so del(13q) was shown to have an independent favorable impact on OS, whereas monosomy 13 was associated with shorter OS, regardless of the co-occurrence of high-risk cytogenetic abnormalities [135].
Therapeutic Implications
RB deletion seems to increase PDL1 expression, mediated by the NFkB pathway [136], so PDL1 inhibitors could be more active in this subset of patients.
2.2.5. Deletion of 17p
The prevalence of deletion of 17p13, del(17p), varies between 5 and 12% in patients with NDMM [11,108,137,138], and increases as the disease develops, reaching as much as 75% in secondary plasma cell, which is the most aggressive expression of plasma cell dyscrasias [139]. It should be borne in mind that these percentages may vary across different studies depending on the cut-off chosen, although 20% is the most broadly used [140,141,142] and is recommended by the European Myeloma Network (EMN) [143]. This deletion entails the loss of the TP53 gene, which is a key suppressor gene that organizes multiple functions associated with cell cycle control and DNA damage response [144]. In spite of the infrequent presence of TP53 mutations in MM, the TP53 gene is mutated in about half of patients who harbor del(17p), giving rise to its biallelic inactivation [145].
Prognosis
Del(17p), although an infrequent genetic abnormality in NDMM, has consistently been associated with shorter survival (Table 1). Nevertheless, there are studies that only found a negative prognostic impact when del(17p) was present in a high proportion, namely between 50 and 60% [146]. Bortezomib-based induction and maintenance regimens seem to improve the prognosis [11,147], according to the HOVON-65/GMMG-HD4 [37] and Total Therapy 3 trials [148] (Table 2). However, response rates and survival are still lower than in patients who do not feature high-risk cytogenetic alterations. Given the results obtained with bortezomib, several studies were carried out with other PIs such as carfilzomib, which yielded better results than those with bortezomib [149], but failed to overcome the poor prognosis associated with del(17p) [149,150]. The trial performed with ixazomib in combination with lenalidomide and dexamethasone gave a better PFS in patients with del(17p) compared with lenalidomide and dexamethasone [151]. The combination of pomalidomide and dexamethasone showed promising results in RRMM patients harboring del(17p), whose PFS was similar to that of patients categorized as being at standard risk. This regimen is thought to be able to overcome the poor prognosis of patients with del(17p), despite the fact that PFS and TTP were less than 1 year (approximately 5 and 8 months, respectively) [40,152].
The introduction of drugs with different mechanisms of action, such as elotuzumab, which is a monoclonal anti-SLAM-F7 antibody, in combination with lenalidomide and dexamethasone, produced better outcomes for patients with del(17p) [41], although the results are difficult to extrapolate since del(17p) was considered positive when at least one cell presented the event, which is at odds with most of the other published studies, which used the threshold of 20% for numerical abnormalities recommended by the EMN [143]. Given this background, there are currently several active trials with elotuzumab in combination with other agents, and even with triplets (elotuzumab + VRD). The anti-CD38 monoclonal antibody isatuximab has also been studied in MM, whereby a phase 2 trial demonstrated better ORR in patients with del(17p) than in those with standard risk (40% vs. 17%) [153].
In essence, del(17p) confers a clear negative impact on survival and has always been considered one of the most important independent prognostic factors. Consequently, del(17p) is one of the high-risk cytogenetic abnormalities included in the R-ISS prognostic index [3,154]. More recently, the bi-allelic inactivation of TP53 due to the presence of mutations in one allele and deletion in the other has been considered a marker of ultra-high-risk disease in spite of the incorporation of new drugs. In fact, this genetic event is also included in the definition of “double-hit” myeloma along with the combination of 1q amplification and ISS stage 3 [105,155].
Therapeutic Implications
The POL2RA gene, mapped at 17p, can be lost along with TP53 in del(17p). This gene encodes an RNA polymerase II enzyme, which can be inhibited by amanitins [156,157]. This drug, conjugated with a targeted antibody against BCMA, has been tested in MM cell lines and mouse models with promising results [158]. Furthermore, the decrease of polymerase activity can be compensated by the overexpression of RBX1. Against this background, combinations of RBX1 inhibitors with amanitins have been proposed to treat patients with metastatic prostate cancer harboring del(17p) [159].
2.3. Mutations
Whole-genome and whole-exome sequencing (WGS/WES) by next-generation sequencing of thousands of MM samples has led to the detection of around 60 exonic mutations per patient, with an average 1.6 mutations per Mb [4]. This frequency of mutations is higher than that in acute leukemias, but much lower than that in solid tumors, which tend to have hundreds of mutations [166]. In contrast with other hematological malignancies, there is no universal, unique, and specific mutation in MM, although many recurrently mutated genes have been detected. In fact, about 250 mutated genes have been described in MM [166], about 60 of which are considered driver genes [167]. Most mutations are single nucleotide variants with consequences for the structure of the final protein. These can be present at both the clonal and sub-clonal levels, and evolve as the disease develops.
Various WGS and WES studies have been carried out to characterize the mutational landscape of MM at diagnosis [109] and relapse [168,169]. Some have even compared the landscapes on both occasions by using paired samples [155,170]. Even though there are many mutated genes, only a few of them are mutated in more than 5% of patients [171]: KRAS (20–25%), NRAS (20–25%), TP53 (8–15%), DIS3 (11%), FAM46C (11%), BRAF (6–15%), TRAF3 (3–6%), ROBO1 (2–5%), EGR1 (4–6%), SP140 (5–7%), and FAT3 (4–7%) [172].
2.3.1. Prognosis
Many of the genes that are mutated in MM can be grouped into different pathways whose deregulation may be essential for myeloma cells to survive and proliferate (Figure 2). The pathway most frequently affected by mutations is the mitogen-activated protein kinase (MAPK) pathway, which includes the KRAS, NRAS, BRAF, EGR1, and FGFR3 genes, whose mutations have been detected in almost 50% of MM patients. Although KRAS mutations remain neutral, NRAS mutations are associated with worse outcomes in patients treated with bortezomib in monotherapy [173], and those affecting BRAF are known to have a negative influence on survival [174]. Conversely, mutations of the EGR1 gene have a favorable effect on outcomes [109]. Mutations affecting the DNA repair pathway, such as those involving the TP53, ATR, ATM, and ZFHX4 genes, occur in approximately 15% of patients. All these genes, in conjunction with CHEK1/2, participate in this pathway, which is responsible for controlling the apoptotic process in response to DNA damage, among other functions. These mutations have been considered prognostic biomarkers associated with shorter survival [105,109]. TRAF3, a component of the NF-kB pathway, which regulates the inflammatory response and cell proliferation, is mutated in about 5% of MM patients. Other components of this pathway, such as NFKBIA, BIRC2/3, and CYLD, are also recurrently mutated, but at frequencies less than 5%. Overall, these mutations account for approximately 20% of the mutations observed in MM. A favorable impact on PFS and OS has been described for TRAF3 mutations [4,105,175]. DIS3 and FAM46C are two of the genes most frequently mutated after KRAS and NRAS. Both belong to the RNA processing pathway, which is responsible for regulating gene expression depending on specific environmental conditions. Even though FAM46C acts as a tumor-suppressor gene in MM [106,176], no prognostic impact has yet been demonstrated [105]. On the other hand, DIS3 mutations have recently been described as associated with a deleterious effect on the outcomes of patients treated with intensive therapy [174]. Mutations affecting genes involved in the cell-cycle control pathway, such as CCND1/2/3, CDK4/6, and RB1, are essential for the immortality that characterizes the oncogenic process. Accordingly, mutations in CCND1, CDKN2C, and RB1 genes are associated with an unfavorable prognosis [105,109]. Genes participating in the B cell differentiation pathway are also recurrently targeted by mutations. Notably, SP140 mutations affect about 5% of patients, the gene being implicated in the immune system response mediated by B cell receptors [169]. Another gene prone to mutation is PRDM1, whose role in promoting terminal differentiation of B cells is controlled by IRF4 [4]. Strikingly, IRF4 mutations seem to confer good prognosis [109]. Cereblon participates jointly with IRF4 in the mechanism of action of IMIDs, and its mutations have been associated with shorter PFS and OS in patients treated with lenalidomide-based regimens [177]. Lastly, deregulation of the JAK–STAT pathway by STAT3 mutations has a negative impact on PFS and OS [178].
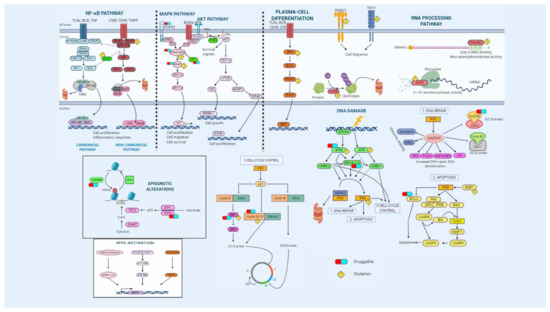
Figure 2.
Overview of the main signaling pathways dysregulated in multiple myeloma. Genetic alterations affect essential pathways and biological processes such as NF-κB and MAPK pathways, plasma-cell differentiation, RNA processing, cell cycle, apoptosis, and DNA-damage repair mechanisms. Some druggable targets are indicated.
Other genes that are affected by mutations but are not included in the aforementioned main pathways include ROBO1, FAT3, and ATP13A4, which are known to play a role in cell migration, adhesion, and regulation of neurons during development [172]. HUWE1, mutated in 5% of MM cases, encodes a ubiquitin ligase that blocks MYC-activated genes [179]. Finally, mutations of TGDS gene have been related to a negative prognosis, shortening both OS and PFS [105,179].
2.3.2. Therapeutic Implications
In recent years, our knowledge of the genetic bases of MM has improved considerably, although this has not yet been translated into more precise molecular classifications aimed at providing personalized medicine for treating MM. Current efforts focus on targeted therapy driven by molecular and cytogenetic abnormalities.
Therapeutic agents that inactivate different pathways have demonstrated efficacy in clinical trials at early stages (Table 3). In this context, several clinical trials in various types of cancer, including MM (TAPUR, MATCH, and CAPTURE trials) have begun. Another phase 1/2 clinical trial, MyDRUG (Myeloma-Developing Regimens Using Genomics), has been developed exclusively for RRMM.

Table 3.
Targeted therapy in MM.
CDK mutations, despite having no impact on survival, might be essential to oncogenesis. Two CDK inhibitors, dinaciclib and palbociclib, are effective in RRMM, alone or in combination with bortezomib and dexamethasone, respectively [180,181]. Two case reports of a combination of BRAF and MEK inhibitors have been published [182,183].
TP53 mutations have a clearly unfavorable impact on survival and its poor prognosis remains despite the broad range of treatments available. Consequently, different approaches have been used to increase the activity of the p53 protein [144]. One of them restores the proper protein tertiary structure of mutant p53 by zinc metallochaperone agents [184], PRIMA-1 [185], or APR246 [186], and the other impairs the binding of wild type p53 with its inhibitor, MDM2, by means of nutlin [187], RITA [188], benzodiazepines [189], and spiro-oxindole [190]. Additionally, cyclotherapy pauses the cell cycle and gives wild-type p53 cells an advantage over cells with mutant p53 [191]. Apart from that, CDK1/6, PLK1, AURKA, NEK2, and MYC genes, among others, are known to be synthetic lethal genes in patients with mutated TP53 [192], which suggests new therapeutic possibilities.
Furthermore, checkpoint inhibitors have been proposed to treat patients with a high rate of mutations, with patients with MAPK mutations having the highest rates [193].
3. Future Perspectives
The survival of patients with MM has been significantly extended in the last two decades thanks to the use of triple-drug combinations including PI, IMIDs, and glucocorticoids, and the approval of new agents with proven efficacy against MM. At the same time, we have witnessed a revolution in genomic methodologies that have revealed the enormous genetic complexity, in terms of the number of chromosomal and molecular alterations present in each patient, and of the clonal evolution that these abnormalities undergo in the course of the disease.
Nevertheless, our extensive knowledge of the molecular mechanisms that contribute to the development and progression of MM is yet to have any great impact on therapeutic decisions. In recent years, substantial progress has been made towards enabling more accurate prognostic stratification. However, considerable heterogeneity remains within each cytogenetic risk category, and this makes it challenging to recommend risk-adapted therapeutic solutions, except in the case of the benefit that PI-based regimens provide to high-risk MM patients. On the other hand, it will become increasingly necessary to reformulate new risk stratification models that incorporate not only the prognostic effect of a few cytogenetic abnormalities, but also the weighted impact of each of the numerous genetic lesions that can be detected in every patient with MM [201]. Nor can it be said that the promise of precision medicine will be a reality in the therapeutic approach of MM in the near future, given the difficulty of eradicating genetically highly heterogeneous cell populations whose sub-clonal content evolves over time.
Faced with this scenario, the scientific community will need to direct its efforts in several directions simultaneously. First, it should create platforms and repositories encompassing as much genomic information from standardized NGS panels as possible. Second, it should aim to generate prognostic and predictive biomarkers that allow the currently available therapeutic arsenal to be individualized in such a way as to avoid ineffective drug combinations and thereby minimize unnecessary toxicities. Finally, in parallel, it should promote clinical trials with innovative designs, particularly with respect to the modality of master protocols, in which genetic biomarkers guide the increasing number of therapeutic alternatives.
Author Contributions
Conceptualization, N.C.G.; writing—original draft preparation, I.J.C.-B. and C.d.R.; writing—review and editing, I.J.C.-B., C.d.R., and N.C.G.; supervision, N.C.G. All authors have read and agreed to the published version of the manuscript.
Funding
I.J.C.-B. was supported by a fellowship (contract PFIS-2020: FI20/00226) from the Instituto de Salud Carlos III; C.d.R. was supported by a fellowship from the Asociación Española Contra el Cancer (AECC). This study was funded by the Instituto de Salud Carlos III and co-financed by FEDER (PI16/01074 and PI19/00674); by the Asociación Española Contra el Cancer (AECC) (Proyectos Estratégicos: PROYE20047GUTI); and by the Gerencia Regional de Salud, Junta de Castilla y León grants (GRS1654/A/17, GRS1849/A/18, and GRS 2058/A/19).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Acknowledgments
The authors would like to thank Philip Mason for his help in reviewing the English language of the manuscript.
Conflicts of Interest
I.J.C.-B. and C.d.R. declare no conflict of interest. N.C.G.: honoraria from Janssen.
References
- Pawlyn, C.; Davies, F.E. Toward personalized treatment in multiple myeloma based on molecular characteristics. Blood 2019, 133, 660–675. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, N.C.; García-Sanz, R.; San Miguel, J.F. Molecular biology of myeloma. Clin. Transl. Oncol. Off. Publ. Fed. Span. Oncol. Soc. Natl. Cancer Inst. Mex. 2007, 9, 618–624. [Google Scholar] [CrossRef]
- Palumbo, A.; Avet-Loiseau, H.; Oliva, S.; Lokhorst, H.M.; Goldschmidt, H.; Rosinol, L.; Richardson, P.; Caltagirone, S.; Lahuerta, J.J.; Facon, T.; et al. Revised international staging system for multiple myeloma: A report from International Myeloma Working Group. J. Clin. Oncol. 2015, 33, 2863–2869. [Google Scholar] [CrossRef] [PubMed]
- Manier, S.; Salem, K.Z.; Park, J.; Landau, D.A.; Getz, G.; Ghobrial, I.M. Genomic complexity of multiple myeloma and its clinical implications. Nat. Rev. Clin. Oncol. 2017, 14, 100–113. [Google Scholar] [CrossRef]
- Chesi, M.; Bergsagel, P.L. Advances in the pathogenesis and diagnosis of multiple myeloma. Int. J. Lab. Hematol. 2015, 37 (Suppl. 1), 108–114. [Google Scholar] [CrossRef]
- Walker, B.A.; Wardell, C.P.; Murison, A.; Boyle, E.M.; Begum, D.B.; Dahir, N.M.; Proszek, P.Z.; Melchor, L.; Pawlyn, C.; Kaiser, M.F.; et al. APOBEC family mutational signatures are associated with poor prognosis translocations in multiple myeloma. Nat. Commun. 2015, 6, 6997. [Google Scholar] [CrossRef] [PubMed]
- Morgan, G.; Walker, B.; Wardell, C.; Johnson, D.; Kaiser, M.; Begum, D.; Dahir, N.; Ross, F.; Davies, F.; Gonzalez, D. Characterization of IgH breakpoints in multiple myeloma indicates a subset of translocations appear to occur in pre-germinal center B Cells. Blood 2013, 121, 3413–3420. [Google Scholar] [CrossRef]
- Barwick, B.G.; Gupta, V.A.; Vertino, P.M.; Boise, L.H. Cell of origin and genetic alterations in the pathogenesis of multiple myeloma. Front Immunol. 2019, 10, 1121. [Google Scholar] [CrossRef]
- Rajkumar, S.V.; Dimopoulos, M.A.; Palumbo, A.; Blade, J.; Merlini, G.; Mateos, M.-V.; Kumar, S.; Hillengass, J.; Kastritis, E.; Richardson, P.; et al. International Myeloma Working Group updated criteria for the diagnosis of multiple myeloma. Lancet Oncol. 2014, 15, e538–e548. [Google Scholar] [CrossRef]
- Mikhael, J.R.; Dingli, D.; Roy, V.; Reeder, C.B.; Buadi, F.K.; Hayman, S.R.; Dispenzieri, A.; Fonseca, R.; Sher, T.; Kyle, R.A.; et al. Management of newly diagnosed symptomatic multiple myeloma: Updated Mayo stratification of myeloma and risk-adapted therapy (mSMART) consensus guidelines 2013. Mayo Clin. Proc. 2013, 88, 360–376. [Google Scholar] [CrossRef]
- Kaufman, G.P.; Gertz, M.A.; Dispenzieri, A.; Lacy, M.Q.; Buadi, F.K.; Dingli, D.; Hayman, S.R.; Kapoor, P.; Lust, J.A.; Russell, S.; et al. Impact of cytogenetic classification on outcomes following early high-dose therapy in multiple myeloma. Leukemia 2016, 30, 633–639. [Google Scholar] [CrossRef]
- Lakshman, A.; Alhaj Moustafa, M.; Rajkumar, S.V.; Dispenzieri, A.; Gertz, M.A.; Buadi, F.K.; Lacy, M.Q.; Dingli, D.; Fonder, A.L.; Hayman, S.R.; et al. Natural history of t(11;14) multiple myeloma. Leukemia 2018, 32, 131–138. [Google Scholar] [CrossRef]
- Gran, C.; Uttervall, K.; Borg Bruchfeld, J.; Wallblom, A.; Alici, E.; Gahrton, G.; Nahi, H. Translocation (11;14) in newly diagnosed multiple myeloma, time to reclassify this standard risk chromosomal aberration? Eur. J. Haematol. 2019, ejh.13325. [Google Scholar] [CrossRef]
- Paner, A.; Patel, P.; Dhakal, B. The evolving role of translocation t(11;14) in the biology, prognosis, and management of multiple myeloma. Blood Rev. 2020, 41, 100643. [Google Scholar] [CrossRef] [PubMed]
- Soverini, S.; Cavo, M.; Cellini, C.; Terragna, C.; Zamagni, E.; Ruggeri, D.; Testoni, N.; Tosi, P.; De Vivo, A.; Amabile, M.; et al. Cyclin D1 overexpression is a favorable prognostic variable for newly diagnosed multiple myeloma patients treated with high-dose chemotherapy and single or double autologous transplantation. Blood 2003, 102, 1588–1594. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, A.J.; Rajkumar, S.V.; Therneau, T.M.; Singh, P.P.; Dispenzieri, A.; Kumar, S.K. Relationship between initial clinical presentation and the molecular cytogenetic classification of myeloma. Leukemia 2014, 28, 398–403. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Kaufman, J.L.; Gasparetto, C.; Mikhael, J.; Vij, R.; Pegourie, B.; Benboubker, L.; Facon, T.; Amiot, M.; Moreau, P.; et al. Efficacy of venetoclax as targeted therapy for relapsed/refractory t(11;14) multiple myeloma. Blood 2017, 130, 2401–2409. [Google Scholar] [CrossRef]
- Punnoose, E.A.; Leverson, J.D.; Peale, F.; Boghaert, E.R.; Belmont, L.D.; Tan, N.; Young, A.; Mitten, M.; Ingalla, E.; Darbonne, W.C.; et al. Expression profile of BCL-2, BCL-XL, and MCL-1 predicts pharmacological response to the BCL-2 selective antagonist venetoclax in multiple myeloma models. Mol. Cancer Ther. 2016, 15, 1132–1144. [Google Scholar] [CrossRef]
- Moreau, P.; Chanan-Khan, A.; Roberts, A.W.; Agarwal, A.B.; Facon, T.; Kumar, S.; Touzeau, C.; Punnoose, E.A.; Cordero, J.; Munasinghe, W.; et al. Promising efficacy and acceptable safety of venetoclax plus bortezomib and dexamethasone in relapsed/refractory MM. Blood 2017, 130, 2392–2400. [Google Scholar] [CrossRef]
- Ghobrial, I. BELLINI: A renaissance for an era of precision therapy in multiple myeloma. Lancet Oncol. 2020, 21, 1547–1549. [Google Scholar] [CrossRef]
- Kumar, S.K.; Harrison, S.J.; Cavo, M.; de la Rubia, J.; Popat, R.; Gasparetto, C.; Hungria, V.; Salwender, H.; Suzuki, K.; Kim, I.; et al. Venetoclax or placebo in combination with bortezomib and dexamethasone in patients with relapsed or refractory multiple myeloma (BELLINI): A randomised, double-blind, multicentre, phase 3 trial. Lancet Oncol. 2020, 21, 1630–1642. [Google Scholar] [CrossRef]
- Venkata, J.K.; An, N.; Stuart, R.; Costa, L.J.; Cai, H.; Coker, W.; Song, J.H.; Gibbs, K.; Matson, T.; Garrett-Mayer, E.; et al. Inhibition of sphingosine kinase 2 downregulates the expression of c-myc and Mcl-1 and induces apoptosis in multiple myeloma. Blood 2014, 124, 1915–1925. [Google Scholar] [CrossRef]
- Sundaramoorthy, P.; Gasparetto, C.; Kang, Y. The combination of a sphingosine kinase 2 inhibitor (ABC294640) and a Bcl-2 inhibitor (ABT-199) displays synergistic anti-myeloma effects in myeloma cells without a t(11;14) translocation. Cancer Med. 2018, 7, 3257–3268. [Google Scholar] [CrossRef] [PubMed]
- Tron, A.E.; Belmonte, M.A.; Adam, A.; Aquila, B.M.; Boise, L.H.; Chiarparin, E.; Cidado, J.; Embrey, K.J.; Gangl, E.; Gibbons, F.D.; et al. Discovery of Mcl-1-specific inhibitor AZD5991 and preclinical activity in multiple myeloma and acute myeloid leukemia. Nat. Commun. 2018, 9, 5341. [Google Scholar] [CrossRef] [PubMed]
- Siu, K.T.; Huang, C.; Panaroni, C.; Mukaihara, K.; Fulzele, K.; Soucy, R.; Thorburn, C.; Cidado, J.; Drew, L.; Chattopadhyay, S.; et al. BCL2 blockade overcomes MCL1 resistance in multiple myeloma. Leukemia 2019, 33, 2098–2102. [Google Scholar] [CrossRef]
- Avet-Loiseau, H.; Hulin, C.; Campion, L.; Rodon, P.; Marit, G.; Attal, M.; Royer, B.; Dib, M.; Voillat, L.; Bouscary, D.; et al. Chromosomal abnormalities are major prognostic factors in elderly patients with multiple myeloma: The Intergroupe Francophone Du Myélome experience. J. Clin. Oncol. 2013, 31, 2806–2809. [Google Scholar] [CrossRef]
- Pawlyn, C.; Morgan, G.J. Evolutionary biology of high-risk multiple myeloma. Nat. Rev. Cancer 2017, 17, 543–556. [Google Scholar] [CrossRef] [PubMed]
- Brito, J.L.R.; Walker, B.; Jenner, M.; Dickens, N.J.; Brown, N.J.M.; Ross, F.M.; Avramidou, A.; Irving, J.A.E.; Gonzalez, D.; Davies, F.E.; et al. MMSET deregulation affects cell cycle progression and adhesion regulons in t(4;14) myeloma plasma cells. Haematologica 2009, 94, 78–86. [Google Scholar] [CrossRef]
- Misiewicz-Krzeminska, I.; Sarasquete, M.E.; Vicente-Dueñas, C.; Krzeminski, P.; Wiktorska, K.; Corchete, L.A.; Quwaider, D.; Rojas, E.A.; Corral, R.; Martín, A.A.; et al. Post-transcriptional modifications contribute to the upregulation of cyclin D2 in multiple myeloma. Clin. Cancer Res. 2016, 22, 207–217. [Google Scholar] [CrossRef] [PubMed]
- Bergsagel, P.L.; Kuehl, W.M.; Zhan, F.; Sawyer, J.; Barlogie, B.; Shaughnessy, J., Jr. Cyclin D dysregulation: An early and unifying pathogenic event in multiple myeloma. Blood 2005, 106, 296–303. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, R.; Bergsagel, P.L.; Drach, J.; Shaughnessy, J.; Gutierrez, N.; Stewart, A.K.; Morgan, G.; Van Ness, B.; Chesi, M.; Minvielle, S.; et al. International Myeloma Working Group molecular classification of multiple myeloma: Spotlight review. Leukemia 2009, 23, 2210–2221. [Google Scholar] [CrossRef]
- Avet-Loiseau, H.; Durie, B.G.M.; Cavo, M.; Attal, M.; Gutierrez, N.; Haessler, J.; Goldschmidt, H.; Hajek, R.; Lee, J.H.; Sezer, O.; et al. Combining fluorescent in situ hybridization data with ISS staging improves risk assessment in myeloma: An International Myeloma Working Group collaborative project. Leukemia 2013, 27, 711–717. [Google Scholar] [CrossRef]
- Avet-Loiseau, H.; Leleu, X.; Roussel, M.; Moreau, P.; Guerin-Charbonnel, C.; Caillot, D.; Marit, G.; Benboubker, L.; Voillat, L.; Mathiot, C.; et al. Bortezomib plus dexamethasone induction improves outcome of patients with t(4;14) myeloma but not outcome of patients with del(17p). J. Clin. Oncol. 2010, 28, 4630–4634. [Google Scholar] [CrossRef] [PubMed]
- Tacchetti, P.; Pantani, L.; Patriarca, F.; Petrucci, M.T.; Zamagni, E.; Dozza, L.; Galli, M.; Di Raimondo, F.; Crippa, C.; Boccadoro, M.; et al. Bortezomib, thalidomide, and dexamethasone followed by double autologous haematopoietic stem-cell transplantation for newly diagnosed multiple myeloma (GIMEMA-MMY-3006): Long-term follow-up analysis of a randomised phase 3, open-label study. Lancet Haematol. 2020, 7, e861–e873. [Google Scholar] [CrossRef]
- Cavo, M.; Tacchetti, P.; Patriarca, F.; Petrucci, M.T.; Pantani, L.; Galli, M.; Di Raimondo, F.; Crippa, C.; Zamagni, E.; Palumbo, A.; et al. Bortezomib with thalidomide plus dexamethasone compared with thalidomide plus dexamethasone as induction therapy before, and consolidation therapy after, double autologous stem-cell transplantation in newly diagnosed multiple myeloma: A randomised phase 3 study. Lancet Lond. Engl. 2010, 376, 2075–2085. [Google Scholar] [CrossRef]
- Sonneveld, P.; Schmidt-Wolf, I.G.H.; van der Holt, B.; El Jarari, L.; Bertsch, U.; Salwender, H.; Zweegman, S.; Vellenga, E.; Broyl, A.; Blau, I.W.; et al. Bortezomib induction and maintenance treatment in patients with newly diagnosed multiple myeloma: Results of the randomized phase III HOVON-65/GMMG-HD4 trial. J. Clin. Oncol. 2012, 30, 2946–2955. [Google Scholar] [CrossRef]
- Neben, K.; Lokhorst, H.M.; Jauch, A.; Bertsch, U.; Hielscher, T.; van der Holt, B.; Salwender, H.; Blau, I.W.; Weisel, K.; Pfreundschuh, M.; et al. Administration of bortezomib before and after autologous stem cell transplantation improves outcome in multiple myeloma patients with deletion 17p. Blood 2012, 119, 940–948. [Google Scholar] [CrossRef] [PubMed]
- Reece, D.; Song, K.W.; Fu, T.; Roland, B.; Chang, H.; Horsman, D.E.; Mansoor, A.; Chen, C.; Masih-Khan, E.; Trieu, Y.; et al. Influence of cytogenetics in patients with relapsed or refractory multiple myeloma treated with lenalidomide plus dexamethasone: Adverse effect of deletion 17p13. Blood 2009, 114, 522–525. [Google Scholar] [CrossRef] [PubMed]
- Avet-Loiseau, H.; Soulier, J.; Fermand, J.-P.; Yakoub-Agha, I.; Attal, M.; Hulin, C.; Garderet, L.; Belhadj, K.; Dorvaux, V.; Minvielle, S.; et al. Impact of high-risk cytogenetics and prior therapy on outcomes in patients with advanced relapsed or refractory multiple myeloma treated with lenalidomide plus dexaméthasone. Leukemia 2010, 24, 623–628. [Google Scholar] [CrossRef]
- Dimopoulos, M.A.; Weisel, K.C.; Song, K.W.; Delforge, M.; Karlin, L.; Goldschmidt, H.; Moreau, P.; Banos, A.; Oriol, A.; Garderet, L.; et al. Cytogenetics and long-term survival of patients with refractory or relapsed and refractory multiple myeloma treated with pomalidomide and low-dose dexamethasone. Haematologica 2015, 100, 1327–1333. [Google Scholar] [CrossRef]
- Lonial, S.; Dimopoulos, M.; Palumbo, A.; White, D.; Grosicki, S.; Spicka, I.; Walter-Croneck, A.; Moreau, P.; Mateos, M.-V.; Magen, H.; et al. Elotuzumab therapy for relapsed or refractory multiple myeloma. N. Engl. J. Med. 2015, 373, 621–631. [Google Scholar] [CrossRef] [PubMed]
- Jakubowiak, A.J.; Siegel, D.S.; Martin, T.; Wang, M.; Vij, R.; Lonial, S.; Trudel, S.; Kukreti, V.; Bahlis, N.; Alsina, M.; et al. Treatment outcomes in patients with relapsed and refractory multiple myeloma and high-risk cytogenetics receiving single-agent carfilzomib in the PX-171-003-A1 study. Leukemia 2013, 27, 2351–2356. [Google Scholar] [CrossRef]
- Hadari, Y.; Schlessinger, J. FGFR3-targeted MAb therapy for bladder cancer and multiple myeloma. J. Clin. Invest. 2009, 119, 1077–1079. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kamath, A.V.; Lu, D.; Gupta, P.; Jin, D.; Xin, Y.; Brady, A.; Stephan, J.P.; Li, H.; Tien, J.; Qing, J.; et al. Preclinical pharmacokinetics of MFGR1877A, a human monoclonal antibody to FGFR3, and prediction of its efficacious clinical dose for the treatment of t(4;14)-positive multiple myeloma. Cancer Chemother. Pharmacol. 2012, 69, 1071–1078. [Google Scholar] [CrossRef] [PubMed]
- Trudel, S.; Stewart, A.K.; Rom, E.; Wei, E.; Zhi, H.L.; Kotzer, S.; Chumakov, I.; Singer, Y.; Chang, H.; Liang, S.B.; et al. The inhibitory anti-FGFR3 antibody, PRO-001, is cytotoxic to t(4;14) multiple myeloma Cells. Blood 2006, 107, 4039–4046. [Google Scholar] [CrossRef]
- Kalff, A.; Spencer, A. The t(4;14) translocation and FGFR3 overexpression in multiple myeloma: Prognostic implications and current clinical strategies. Blood Cancer J. 2012, 2, e89-8. [Google Scholar] [CrossRef]
- Scheid, C.; Reece, D.; Beksac, M.; Spencer, A.; Callander, N.; Sonneveld, P.; Kalimi, G.; Cai, C.; Shi, M.; Scott, J.W.; et al. Phase 2 study of dovitinib in patients with relapsed or refractory multiple myeloma with or without t(4;14) translocation. Eur. J. Haematol. 2015, 95, 316–324. [Google Scholar] [CrossRef]
- Majumder, M.M.; Silvennoinen, R.; Anttila, P.; Tamborero, D.; Eldfors, S.; Yadav, B.; Karjalainen, R.; Kuusanmäki, H.; Lievonen, J.; Parsons, A.; et al. Identification of precision treatment strategies for relapsed/refractory multiple myeloma by functional drug sensitivity testing. Oncotarget 2017, 8, 56338–56350. [Google Scholar] [CrossRef]
- Xie, Z.; Bi, C.; Chooi, J.Y.; Chan, Z.L.; Mustafa, N.; Chng, W.J. MMSET regulates expression of IRF4 in t(4;14) myeloma and its silencing potentiates the effect of bortezomib. Leukemia 2015, 29, 2347–2354. [Google Scholar] [CrossRef]
- Shah, M.Y.; Martinez-Garcia, E.; Phillip, J.M.; Chambliss, A.B.; Popovic, R.; Ezponda, T.; Small, E.C.; Will, C.; Phillip, M.P.; Neri, P.; et al. MMSET/WHSC1 enhances DNA damage repair leading to an increase in resistance to chemotherapeutic agents. Oncogene 2016, 35, 5905–5915. [Google Scholar] [CrossRef]
- Tonon, G.; Anderson, K.C. Multiple myeloma. Mol. Basis Cancer 2015, 455–466.e4. [Google Scholar] [CrossRef]
- Lauring, J.; Abukhdeir, A.M.; Konishi, H.; Garay, J.P.; Gustin, J.P.; Wang, Q.; Arceci, R.J.; Matsui, W.; Park, B.H. The multiple myeloma associated MMSET gene contributes to cellular adhesion, clonogenic growth, and tumorigenicity. Blood 2008, 111, 856–864. [Google Scholar] [CrossRef] [PubMed]
- International myeloma Working Group. Criteria for the classification of monoclonal gammopathies, multiple myeloma and related disorders: A report of the International Myeloma Working Group. Br. J. Haematol. 2003, 121, 749–757. [Google Scholar] [CrossRef]
- Rajkumar, S.V. Multiple myeloma: 2018 update on diagnosis, risk-stratification, and management. Am. J. Hematol. 2018, 93, 1091–1110. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, R.; Blood, E.; Rue, M.; Harrington, D.; Oken, M.M.; Kyle, R.A.; Dewald, G.W.; Van Ness, B.; Van Wier, S.A.; Henderson, K.J.; et al. Clinical and biologic implications of recurrent genomic aberrations in myeloma. Blood 2003, 101, 4569–4575. [Google Scholar] [CrossRef] [PubMed]
- Avet-Loiseau, H.; Malard, F.; Campion, L.; Magrangeas, F.; Sebban, C.; Lioure, B.; Decaux, O.; Lamy, T.; Legros, L.; Fuzibet, J.G.; et al. Translocation t(14;16) and multiple myeloma: Is it really an independent prognostic factor? Blood 2011, 117, 2009–2011. [Google Scholar] [CrossRef]
- Tai, Y.T.; Fulciniti, M.; Hideshima, T.; Song, W.; Leiba, M.; Li, X.F.; Rumizen, M.; Burger, P.; Morrison, A.; Podar, K.; et al. Targeting MEK induces myeloma-cell cytotoxicity and inhibits osteoclastogenesis. Blood 2007, 110, 1656–1663. [Google Scholar] [CrossRef]
- Annunziata, C.M.; Hernandez, L.; Davis, R.E.; Zingone, A.; Lamy, L.; Lam, L.T.; Hurt, E.M.; Shaffer, A.L.; Kuehl, W.M.; Staudt, L.M. A mechanistic rationale for MEK inhibitor therapy in myeloma based on blockade of MAF oncogene expression. Blood 2011, 117, 2396–2404. [Google Scholar] [CrossRef]
- Suzuki, R.; Kikuchi, S.; Harada, T.; Mimura, N.; Minami, J.; Ohguchi, H.; Yoshida, Y.; Sagawa, M.; Gorgun, G.; Cirstea, D.; et al. Combination of a selective HSP90α/β inhibitor and a RAS-RAF-MEK-ERK signaling pathway inhibitor triggers synergistic cytotoxicity in multiple myeloma cells. PLoS ONE 2015, 10, 1–20. [Google Scholar] [CrossRef]
- Ragon, B.K.; Odenike, O.; Baer, M.R.; Stock, W.; Borthakur, G.; Patel, K.; Han, L.; Chen, H.; Ma, H.; Joseph, L.; et al. Oral MEK 1/2 inhibitor trametinib in combination with AKT inhibitor GSK2141795 in patients with acute myeloid leukemia with RAS mutations: A phase II study. Clin. Lymphoma Myeloma Leuk. 2019, 19, 431–440.e13. [Google Scholar] [CrossRef]
- Qiang, Y.-W.; Ye, S.; Chen, Y.; Buros, A.F.; Edmonson, R.; van Rhee, F.; Barlogie, B.; Epstein, J.; Morgan, G.J.; Davies, F.E. MAF protein mediates innate resistance to proteasome inhibition therapy in multiple myeloma. Blood 2016, 128, 2919–2930. [Google Scholar] [CrossRef]
- Qiang, Y.-W.; Ye, S.; Huang, Y.; Chen, Y.; Van Rhee, F.; Epstein, J.; Walker, B.A.; Morgan, G.J.; Davies, F.E. MAFb protein confers intrinsic resistance to proteasome inhibitors in multiple myeloma. BMC Cancer 2018, 18, 724. [Google Scholar] [CrossRef]
- Herath, N.I.; Rocques, N.; Garancher, A.; Eychène, A.; Pouponnot, C. GSK3-mediated MAF phosphorylation in multiple myeloma as a potential therapeutic target. Blood Cancer J. 2014, 4, e175. [Google Scholar] [CrossRef] [PubMed]
- Misund, K.; Keane, N.; Stein, C.K.; Asmann, Y.W.; Day, G.; Welsh, S.; Van Wier, S.A.; Riggs, D.L.; Ahmann, G.; Chesi, M.; et al. MYC dysregulation in the progression of multiple myeloma. Leukemia 2019, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Affer, M.; Chesi, M.; Chen, W.-D.G.; Keats, J.J.; Demchenko, Y.N.; Roschke, A.V.; Van Wier, S.; Fonseca, R.; Bergsagel, P.L.; Kuehl, W.M. Promiscuous MYC locus rearrangements hijack enhancers but mostly super-enhancers to dysregulate MYC expression in multiple myeloma. Leukemia 2014, 28, 1725–1735. [Google Scholar] [CrossRef] [PubMed]
- Kuehl, W.M.; Bergsagel, P.L. MYC addiction: A potential therapeutic target in MM. Blood 2012, 120, 2351–2352. [Google Scholar] [CrossRef]
- Cottini, F.; Hideshima, T.; Suzuki, R.; Tai, Y.-T.; Bianchini, G.; Richardson, P.G.; Anderson, K.C.; Tonon, G. Synthetic lethal approaches exploiting DNA damage in aggressive myeloma. Cancer Discov. 2015, 5, 972–987. [Google Scholar] [CrossRef] [PubMed]
- Jovanović, K.K.; Roche-Lestienne, C.; Ghobrial, I.M.; Facon, T.; Quesnel, B.; Manier, S. Targeting MYC in multiple myeloma. Leukemia 2018, 32, 1295–1306. [Google Scholar] [CrossRef]
- Abdallah, N.; Baughn, L.B.; Rajkumar, S.V.; Kapoor, P.; Gertz, M.A.; Dispenzieri, A.; Lacy, M.Q.; Hayman, S.R.; Buadi, F.K.; Dingli, D.; et al. Implications of MYC rearrangements in newly diagnosed multiple myeloma. Clin. Cancer Res. 2020. [Google Scholar] [CrossRef]
- Chng, W.J.; Huang, G.F.; Chung, T.H.; Ng, S.B.; Gonzalez-Paz, N.; Troska-Price, T.; Mulligan, G.; Chesi, M.; Bergsagel, P.L.; Fonseca, R. Clinical and biological implications of MYC activation: A common difference between MGUS and newly diagnosed multiple myeloma. Leukemia 2011, 25, 1026–1035. [Google Scholar] [CrossRef]
- Brioli, A.; Melchor, L.; Walker, B.A.; Davies, F.E.; Morgan, G.J. Biology and treatment of myeloma. Clin. Lymphoma Myeloma Leuk. 2014, 14, S65–S70. [Google Scholar] [CrossRef]
- Avet-Loiseau, H.; Attal, M.; Moreau, P.; Charbonnel, C.; Garban, F.; Hulin, C.; Leyvraz, S.; Michallet, M.; Yakoub-Agha, I.; Garderet, L.; et al. Genetic abnormalities and survival in multiple myeloma: The experience of the Intergroupe Francophone Du Myélome. Blood 2007, 109, 3489–3495. [Google Scholar] [CrossRef]
- Walker, B.A.; Wardell, C.P.; Brioli, A.; Boyle, E.; Kaiser, M.F.; Begum, D.B.; Dahir, N.B.; Johnson, D.C.; Ross, F.M.; Davies, F.E.; et al. Translocations at 8q24 juxtapose MYC with genes that harbor superenhancers resulting in overexpression and poor prognosis in myeloma patients. Blood Cancer J. 2014, 4, e191-7. [Google Scholar] [CrossRef]
- Glitza, I.C.; Lu, G.; Shah, R.; Bashir, Q.; Shah, N.; Champlin, R.E.; Shah, J.; Orlowski, R.Z.; Qazilbash, M.H. Chromosome 8q24.1/c-MYC abnormality: A marker for high-risk myeloma. Leuk. Lymphoma 2015, 56, 602–607. [Google Scholar] [CrossRef] [PubMed]
- Kenneth, C. Anderson Progress and paradigms in multiple myeloma. Clin. Cancer Res. 2017, 176, 139–148. [Google Scholar] [CrossRef]
- Delmore, J.E.; Issa, G.C.; Lemieux, M.E.; Rahl, P.B.; Shi, J.; Jacobs, H.M.; Kastritis, E.; Gilpatrick, T.; Paranal, R.M.; Qi, J.; et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 2011, 146, 904–917. [Google Scholar] [CrossRef]
- Stubbs, M.C.; Burn, T.C.; Sparks, R.; Maduskuie, T.; Diamond, S.; Rupar, M.; Wen, X.; Volgina, A.; Zolotarjova, N.; Waeltz, P.; et al. The novel bromodomain and extraterminal domain inhibitor INCB054329 induces vulnerabilities in myeloma cells that inform rational combination strategies. Clin. Cancer Res. 2019, 25, 300–311. [Google Scholar] [CrossRef] [PubMed]
- Siu, K.T.; Ramachandran, J.; Yee, A.J.; Eda, H.; Santo, L.; Panaroni, C.; Mertz, J.A.; Sims Iii, R.J.; Cooper, M.R.; Raje, N. Preclinical activity of CPI-0610, a novel small-molecule bromodomain and extra-terminal protein inhibitor in the therapy of multiple myeloma. Leukemia 2017, 31, 1760–1769. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.L.; Damnernsawad, A.; Shyamsunder, P.; Chng, W.J.; Han, B.C.; Xu, L.; Pan, J.; Pravin, D.P.; Alkan, S.; Tyner, J.W.; et al. Proteolysis targeting chimeric molecules as therapy for multiple myeloma: Efficacy, biomarker and drug combinations. Haematologica 2019, 104, 1209–1220. [Google Scholar] [CrossRef] [PubMed]
- Buettner, R.; Morales, C.; Caserta, E.; Troadec, E.; Gunes, E.G.; Viola, D.; Khalife, J.; Li, H.; Keats, J.J.; Christofferson, A.; et al. Leflunomide regulates c-Myc expression in myeloma cells through PIM targeting. Blood Adv. 2019, 3, 1027–1032. [Google Scholar] [CrossRef]
- Caracciolo, D.; Scionti, F.; Juli, G.; Altomare, E.; Golino, G.; Todoerti, K.; Grillone, K.; Riillo, C.; Arbitrio, M.; Iannone, M.; et al. Exploiting MYC-induced PARPness to target genomic instability in multiple myeloma. Haematologica 2020. [Google Scholar] [CrossRef] [PubMed]
- Chng, W.J.; Ketterling, R.P.; Fonseca, R. Analysis of genetic abnormalities provides insights into genetic evolution of hyperdiploid myeloma. Genes Chromosomes Cancer 2006, 45, 1111–1120. [Google Scholar] [CrossRef] [PubMed]
- Chng, W.J.; Van Wier, S.A.; Ahmann, G.J.; Winkler, J.M.; Jalal, S.M.; Bergsagel, P.L.; Chesi, M.; Trendle, M.C.; Oken, M.M.; Blood, E.; et al. A validated FISH trisomy index demonstrates the hyperdiploid and nonhyperdiploid dichotomy in MGUS. Blood 2005, 106, 2156–2161. [Google Scholar] [CrossRef]
- Hoctor, V.T.; Campbell, L.J. Hyperhaploid plasma cell myeloma. Cancer Genet 2012, 205, 414–418. [Google Scholar] [CrossRef]
- Sawyer, J.R.; Tian, E.; Shaughnessy, J.D.; Epstein, J.; Swanson, C.M.; Stangeby, C.; Hale, C.L.; Parr, L.; Lynn, M.; Sammartino, G.; et al. Hyperhaploidy is a novel high-risk cytogenetic subgroup in multiple myeloma. Leukemia 2017, 31, 637–644. [Google Scholar] [CrossRef]
- Magrangeas, F.; Avet-Loiseau, H.; Munshi, N.C.; Minvielle, S. Chromothripsis identifies a rare and aggressive entity among newly diagnosed multiple myeloma patients. Blood 2011, 118, 675–678. [Google Scholar] [CrossRef] [PubMed]
- Chng, W.J.; Santana-Dávila, R.; Van Wier, S.A.; Ahmann, G.J.; Jalal, S.M.; Bergsagel, P.L.; Chesi, M.; Trendle, M.C.; Jacobus, S.; Blood, E.; et al. Prognostic factors for hyperdiploid-myeloma: Effects of chromosome 13 deletions and IgH translocations. Leukemia 2006, 20, 807–813. [Google Scholar] [CrossRef] [PubMed]
- Smadja, N.V.; Bastard, C.; Brigaudeau, C.; Leroux, D.; Fruchart, C. Hypodiploidy is a major prognostic factor in multiple myeloma. Blood 2001, 98. [Google Scholar] [CrossRef] [PubMed]
- Pandey, S.; Rajkumar, S.V.; Kapoor, P.; Ketterling, R.P.; Lacy, M.Q.; Gertz, M.A.; Buadi, F.; Dingli, D.; Hayman, S.R.; Dispenzieri, A.; et al. Impact of FISH abnormalities on response to lenalidomide in patients with multiple myeloma. Blood 2013, 122, 3210. [Google Scholar] [CrossRef]
- Mei, J.; Zhai, Y.; Li, H.; Li, F.; Zhou, X.; Song, P.; Zhao, Q.; Yu, Y.; An, Z.; Wang, L. Prognostic impact of hyperdiploidy in multiple myeloma patients with high-risk cytogenetics: A pilot study in China. J. Cancer Res. Clin. Oncol. 2018, 144, 2263–2273. [Google Scholar] [CrossRef]
- Chretien, M.-L.; Corre, J.; Lauwers-Cances, V.; Magrangeas, F.; Cleynen, A.; Yon, E.; Hulin, C.; Leleu, X.; Orsini-Piocelle, F.; Blade, J.-S.; et al. Understanding the role of hyperdiploidy in myeloma prognosis: Which trisomies really matter? Blood 2015, 126, 2713–2719. [Google Scholar] [CrossRef]
- Walker, B.A.; Leone, P.E.; Chiecchio, L.; Dickens, N.J.; Jenner, M.W.; Boyd, K.D.; Johnson, D.C.; Gonzalez, D.; Dagrada, G.P.; Protheroe, R.K.M.; et al. A compendium of myeloma-associated chromosomal copy number abnormalities and their prognostic value. Blood 2010, 116, e56–e65. [Google Scholar] [CrossRef]
- Fassas, A.B.-T.; Spencer, T.; Sawyer, J.; Zangari, M.; Lee, C.-K.; Anaissie, E.; Muwalla, F.; Morris, C.; Barlogie, B.; Tricot, G. Both hypodiploidy and deletion of chromosome 13 independently confer poor prognosis in multiple myeloma. Br. J. Haematol. 2002, 118, 1041–1047. [Google Scholar] [CrossRef]
- Kaur, G.; Gupta, R.; Mathur, N.; Rani, L.; Kumar, L.; Sharma, A.; Singh, V.; Gupta, A.; Sharma, O.D. Clinical impact of chromothriptic complex chromosomal rearrangements in newly diagnosed multiple myeloma. Leuk. Res. 2019, 76, 58–64. [Google Scholar] [CrossRef]
- Boyd, K.D.; Ross, F.M.; Walker, B.A.; Wardell, C.P.; Tapper, W.J.; Chiecchio, L.; Dagrada, G.; Konn, Z.J.; Gregory, W.M.; Jackson, G.H.; et al. Mapping of chromosome 1p deletions in myeloma identifies FAM46C at 1p12 and CDKN2C at 1p32.3 as being genes in regions associated with adverse survival. Clin. Cancer Res. 2011, 17, 7776–7784. [Google Scholar] [CrossRef]
- Qazilbash, M.H.; Saliba, R.M.; Ahmed, B.; Parikh, G.; Mendoza, F.; Ashraf, N.; Hosing, C.; Flosser, T.; Weber, D.M.; Wang, M.; et al. Deletion of the short arm of chromosome 1 (Del 1p) is a strong predictor of poor outcome in myeloma patients undergoing an autotransplant. Biol. Blood Marrow Transpl. 2007, 13, 1066–1072. [Google Scholar] [CrossRef] [PubMed][Green Version]
- López-Corral, L.; Sarasquete, M.E.; Beà, S.; García-Sanz, R.; Mateos, M.V.; Corchete, L.A.; Sayagués, J.M.; García, E.M.; Bladé, J.; Oriol, A.; et al. SNP-based mapping arrays reveal high genomic complexity in monoclonal gammopathies, from MGUS to myeloma status. Leukemia 2012, 26, 2521–2529. [Google Scholar] [CrossRef]
- Li, F.; Hu, L.; Xu, Y.; Li, Z.; Yi, S.; Gu, Z.; Li, C.; Hao, M.; Ru, K.; Zhan, F.; et al. Identification of characteristic and prognostic values of chromosome 1p abnormality by multi-gene fluorescence in situ hybridization in multiple myeloma. Leukemia 2016, 30, 1197–1201. [Google Scholar] [CrossRef] [PubMed]
- Leone, P.E.; Walker, B.A.; Jenner, M.W.; Chiecchio, L.; Dagrada, G.; Protheroe, R.K.M.; Johnson, D.C.; Dickens, N.J.; Brito, J.L.; Else, M.; et al. Deletions of CDKN2C in multiple myeloma: Biological and clinical implications. Clin. Cancer Res. 2008, 14, 6033–6041. [Google Scholar] [CrossRef] [PubMed]
- Wong, A.K.; Chen, Y.; Lian, L.; Ha, P.C.; Petersen, K.; Laity, K.; Carillo, A.; Emerson, M.; Heichman, K.; Gupte, J.; et al. Genomic structure, chromosomal location, and mutation analysis of the human CDC14A gene. Genomics 1999, 59, 248–251. [Google Scholar] [CrossRef] [PubMed]
- Hebraud, B.; Leleu, X.; Lauwers-Cances, V.; Roussel, M.; Caillot, D.; Marit, G.; Karlin, L.; Hulin, C.; Gentil, C.; Guilhot, F.; et al. Deletion of the 1p32 region is a major independent prognostic factor in young patients with myeloma: The IFM Experience on 1195 patients. Leukemia 2014, 28, 675–679. [Google Scholar] [CrossRef]
- Chang, H.; Ning, Y.; Qi, X.; Yeung, J.; Xu, W. Chromosome 1p21 deletion is a novel prognostic marker in patients with multiple myeloma. Br. J. Haematol. 2007, 139, 51–54. [Google Scholar] [CrossRef]
- Chang, H.; Jiang, A.; Qi, C.; Trieu, Y.; Chen, C.; Reece, D. Impact of genomic aberrations including chromosome 1 abnormalities on the outcome of patients with relapsed or refractory multiple myeloma treated with lenalidomide and dexamethasone. Leuk. Lymphoma 2010, 51, 2084–2091. [Google Scholar] [CrossRef]
- Zhu, Y.X.; Shi, C.-X.; Bruins, L.A.; Jedlowski, P.; Wang, X.; Kortüm, K.M.; Luo, M.; Ahmann, J.M.; Braggio, E.; Stewart, A.K. Loss of FAM46C promotes cell survival in myeloma. Cancer Res. 2017, 77, 4317–4327. [Google Scholar] [CrossRef]
- Walker, B.A.; Mavrommatis, K.; Wardell, C.P.; Ashby, T.C.; Bauer, M.; Davies, F.; Rosenthal, A.; Wang, H.; Qu, P.; Hoering, A.; et al. A high-risk, double-hit, group of newly diagnosed myeloma identified by genomic analysis. Leukemia 2019, 33, 159–170. [Google Scholar] [CrossRef]
- Herrero, A.B.; Quwaider, D.; Corchete, L.A.; Mateos, M.V.; García-Sanz, R.; Gutiérrez, N.C. FAM46C controls antibody production by the polyadenylation of immunoglobulin mRNAs and inhibits cell migration in multiple myeloma. J. Cell Mol. Med. 2020, 24, 4171–4182. [Google Scholar] [CrossRef] [PubMed]
- An, G.; Xu, Y.; Shi, L.; Shizhen, Z.; Deng, S.; Xie, Z.; Sui, W.; Zhan, F.; Qiu, L. Chromosome 1q21 gains confer inferior outcomes in multiple myeloma treated with bortezomib but copy number variation and percentage of plasma cells involved have no additional prognostic value. Haematologica 2014, 99, 353–359. [Google Scholar] [CrossRef]
- Avet-Loiseau, H.; Attal, M.; Campion, L.; Caillot, D.; Hulin, C.; Marit, G.; Stoppa, A.-M.; Voillat, L.; Wetterwald, M.; Pegourie, B.; et al. Long-term analysis of the IFM 99 trials for myeloma: Cytogenetic abnormalities [t(4;14), del(17p), 1q gains] play a major role in defining long-term survival. J. Clin. Oncol. 2012, 30, 1949–1952. [Google Scholar] [CrossRef] [PubMed]
- Walker, B.A.; Boyle, E.M.; Wardell, C.P.; Murison, A.; Begum, D.B.; Dahir, N.M.; Proszek, P.Z.; Johnson, D.C.; Kaiser, M.F.; Melchor, L.; et al. Mutational spectrum, copy number changes, and outcome: Results of a sequencing study of patients with newly diagnosed myeloma. J. Clin. Oncol. 2015, 33, 3911–3920. [Google Scholar] [CrossRef] [PubMed]
- Neben, K.; Jauch, A.; Hielscher, T.; Hillengass, J.; Lehners, N.; Seckinger, A.; Granzow, M.; Raab, M.S.; Ho, A.D.; Goldschmidt, H.; et al. Progression in smoldering myeloma is independently determined by the chromosomal abnormalities del(17p), t(4;14), gain 1q, hyperdiploidy, and tumor load. J. Clin. Oncol. 2013, 31, 4325–4332. [Google Scholar] [CrossRef]
- Chang, H.; Yeung, J.; Xu, W.; Ning, Y.; Patterson, B. Significant increase of CKS1B amplification from monoclonal gammopathy of undetermined significance to multiple myeloma and plasma cell leukaemia as demonstrated by interphase fluorescence in situ hybridisation. Br. J. Haematol. 2006, 134, 613–615. [Google Scholar] [CrossRef]
- Hanamura, I.; Stewart, J.P.; Huang, Y.; Zhan, F.; Santra, M.; Sawyer, J.R.; Hollmig, K.; Zangarri, M.; Pineda-Roman, M.; van Rhee, F.; et al. Frequent gain of chromosome band 1q21 in plasma-cell dyscrasias detected by fluorescence in situ hybridization: Incidence increases from MGUS to relapsed myeloma and is related to prognosis and disease progression following tandem stem-cell transplantat. Blood 2006, 108, 1724–1732. [Google Scholar] [CrossRef]
- Fonseca, R.; Van Wier, S.A.; Chng, W.J.; Ketterling, R.; Lacy, M.Q.; Dispenzieri, A.; Bergsagel, P.L.; Rajkumar, S.V.; Greipp, P.R.; Litzow, M.R.; et al. Prognostic value of chromosome 1q21 gain by fluorescent in situ hybridization and increase CKS1B expression in myeloma. Leukemia 2006, 20, 2034–2040. [Google Scholar] [CrossRef]
- Zhan, F.; Colla, S.; Wu, X.; Chen, B.; Stewart, J.P.; Kuehl, W.M.; Barlogie, B.; Shaughnessy, J.D. CKS1B, Overexpressed in aggressive disease, regulates multiple myeloma growth and survival through SKP2- and p27Kip1-dependent and -independent mechanisms. Blood 2007, 109, 4995–5001. [Google Scholar] [CrossRef]
- Shi, L.; Wang, S.; Zangari, M.; Xu, H.; Cao, T.M.; Xu, C.; Wu, Y.; Xiao, F.; Liu, Y.; Yang, Y.; et al. Over-expression of CKS1B activates both MEK/ERK and JAK/STAT3 signaling pathways and promotes myeloma cell drug-resistance. Oncotarget 2010, 1, 22–33. [Google Scholar] [CrossRef] [PubMed]
- Treon, S.P.; Maimonis, P.; Bua, D.; Young, G.; Raje, N.; Mollick, J.; Chauhan, D.; Tai, Y.T.; Hideshima, T.; Shima, Y.; et al. Elevated soluble MUC1 levels and decreased anti-MUC1 antibody levels in patients with multiple myeloma. Blood 2000, 96, 3147–3153. [Google Scholar] [CrossRef]
- Legartova, S.; Krejci, J.; Harnicarova, A.; Hajek, R.; Kozubek, S.; Bartova, E. Nuclear topography of the 1q21 genomic region and Mcl-1 protein levels associated with pathophysiology of multiple myeloma. Neoplasma 2009, 56, 404–413. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sawyer, J.R.; Tricot, G.; Lukacs, J.L.; Binz, R.L.; Tian, E.; Barlogie, B.; Shaughnessy, J. Genomic instability in multiple myeloma: Evidence for jumping segmental duplications of chromosome arm 1q. Genes Chromosomes Cancer 2005, 42, 95–106. [Google Scholar] [CrossRef] [PubMed]
- Shaughnessy, J.D.; Qu, P.; Usmani, S.; Heuck, C.J.; Zhang, Q.; Zhou, Y.; Tian, E.; Hanamura, I.; van Rhee, F.; Anaissie, E.; et al. Pharmacogenomics of bortezomib test-dosing identifies hyperexpression of proteasome genes, especially PSMD4, as novel high-risk feature in myeloma treated with total therapy 3. Blood 2011, 118, 3512–3524. [Google Scholar] [CrossRef] [PubMed]
- Inoue, J.; Otsuki, T.; Hirasawa, A.; Imoto, I.; Matsuo, Y.; Shimizu, S.; Taniwaki, M.; Inazawa, J. Overexpression of PDZK1 within the 1q12-Q22 amplicon is likely to be associated with drug-resistance phenotype in multiple myeloma. Am. J. Pathol. 2004, 165, 71–81. [Google Scholar] [CrossRef]
- Schmidt, T.M.; Barwick, B.G.; Joseph, N.; Heffner, L.T.; Hofmeister, C.C.; Bernal, L.; Dhodapkar, M.V.; Gupta, V.A.; Jaye, D.L.; Wu, J.; et al. Gain of chromosome 1q is associated with early progression in multiple myeloma patients treated with lenalidomide, bortezomib, and dexamethasone. Blood Cancer J. 2019, 9, 94. [Google Scholar] [CrossRef] [PubMed]
- Shah, G.L.; Landau, H.; Londono, D.; Devlin, S.M.; Kosuri, S.; Lesokhin, A.M.; Lendvai, N.; Hassoun, H.; Chung, D.J.; Koehne, G.; et al. Gain of chromosome 1q portends worse prognosis in multiple myeloma despite novel agent-based induction regimens and autologous transplantation. Leuk. Lymphoma 2017, 58, 1823–1831. [Google Scholar] [CrossRef] [PubMed]
- Bock, F.; Lu, G.; Srour, S.A.; Gaballa, S.; Lin, H.Y.; Baladandayuthapani, V.; Honhar, M.; Stich, M.; Das Shah, N.; Bashir, Q.; et al. Outcome of patients with multiple myeloma and CKS1B gene amplification after autologous hematopoietic stem cell transplantation. Biol. Blood Marrow Transpl. 2016, 22, 2159–2164. [Google Scholar] [CrossRef]
- Jackson, G.H.; Davies, F.E.; Pawlyn, C.; Cairns, D.A.; Striha, A.; Collett, C.; Hockaday, A.; Jones, J.R.; Kishore, B.; Garg, M.; et al. Lenalidomide maintenance versus observation for patients with newly diagnosed multiple myeloma (myeloma XI): A multicentre, open-label, randomised, phase 3 trial. Lancet Oncol. 2019, 20, 57–73. [Google Scholar] [CrossRef]
- Walker, B.A.; Morgan, G.J. The genomic features associated with high-risk multiple myeloma. Oncotarget 2018, 9, 35478–35479. [Google Scholar] [CrossRef]
- Sherbenou, D.W.; Aftab, B.T.; Su, Y.; Behrens, C.R.; Wiita, A.; Logan, A.C.; Acosta-Alvear, D.; Hann, B.C.; Walter, P.; Shuman, M.A.; et al. Antibody-drug conjugate targeting CD46 eliminates multiple myeloma cells. J. Clin. Invest. 2016, 126, 4640–4653. [Google Scholar] [CrossRef]
- Slomp, A.; Moesbergen, L.M.; Gong, J.-N.; Cuenca, M.; von dem Borne, P.A.; Sonneveld, P.; Huang, D.C.S.; Minnema, M.C.; Peperzak, V. Multiple myeloma with 1q21 amplification is highly sensitive to MCL-1 targeting. Blood Adv. 2019, 3, 4202–4214. [Google Scholar] [CrossRef]
- Avet-Louseau, H.; Daviet, A.; Sauner, S.; Bataille, R.; Intergroupe Francophone du Myélome. Chromosome 13 abnormalities in multiple myeloma are mostly monosomy 13. Br. J. Haematol. 2000, 111, 1116–1117. [Google Scholar]
- Fonseca, R.; Oken, M.M.; Harrington, D.; Bailey, R.J.; Van Wier, S.A.; Henderson, K.J.; Kay, N.E.; Van Ness, B.; Greipp, P.R.; Dewald, G.W. Deletions of chromosome 13 in multiple myeloma identified by interphase FISH usually denote large deletions of the q arm or monosomy. Leukemia 2001, 15, 981–986. [Google Scholar] [CrossRef] [PubMed]
- Zojer, N.; Königsberg, R.; Ackermann, J.; Fritz, E.; Dallinger, S.; Krömer, E.; Kaufmann, H.; Riedl, L.; Gisslinger, H.; Schreiber, S.; et al. Deletion of 13q14 remains an independent adverse prognostic variable in multiple myeloma despite its frequent detection by interphase fluorescence in situ hybridization. Blood 2000, 95, 1925–1930. [Google Scholar] [CrossRef] [PubMed]
- Shaughnessy, J.; Tian, E.; Sawyer, J.; Bumm, K.; Landes, R.; Badros, A.; Morris, C.; Tricot, G.; Epstein, J.; Barlogie, B. High incidence of chromosome 13 deletion in multiple myeloma detected by multiprobe interphase FISH. Blood 2000, 96, 1505–1511. [Google Scholar] [CrossRef]
- Tricot, G.; Barlogie, B.; Jagannath, S.; Bracy, D.; Mattox, S.; Vesole, D.H.; Naucke, S.; Sawyer, J.R. Poor prognosis in multiple myeloma is associated only with partial or complete deletions of chromosome 13 or abnormalities involving 11q and not with other karyotype abnormalities. Blood 1995, 86, 4250–4256. [Google Scholar] [CrossRef] [PubMed]
- Kiyota, M.; Kobayashi, T.; Fuchida, S.; Yamamoto-Sugitani, M.; Ohshiro, M.; Shimura, Y.; Mizutani, S.; Nagoshi, H.; Sasaki, N.; Nakayama, R.; et al. Monosomy 13 in metaphase spreads is a predictor of poor long-term outcome after bortezomib plus dexamethasone treatment for relapsed/refractory multiple myeloma. Int. J. Hematol. 2012, 95, 516–526. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, N.C.; Castellanos, M.V.; Martín, M.L.; Mateos, M.V.; Hernández, J.M.; Fernández, M.; Carrera, D.; Rosiñol, L.; Ribera, J.M.; Ojanguren, J.M.; et al. Prognostic and biological implications of genetic abnormalities in multiple myeloma undergoing autologous stem cell transplantation: t(4;14) is the most relevant adverse prognostic factor, whereas RB deletion as a unique abnormality is not associated with adverse prognosis. Leukemia 2007, 21, 143–150. [Google Scholar] [CrossRef]
- Binder, M.; Rajkumar, S.V.; Ketterling, R.P.; Greipp, P.T.; Dispenzieri, A.; Lacy, M.Q.; Gertz, M.A.; Buadi, F.K.; Hayman, S.R.; Hwa, Y.L.; et al. Prognostic implications of abnormalities of chromosome 13 and the presence of multiple cytogenetic high-risk abnormalities in newly diagnosed multiple myeloma. Blood Cancer J. 2017, 7, e600. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Ding, D.; Yan, Y.; Li, H.; Wang, B.; Ma, L.; Ye, Z.; Ma, T.; Wu, Q.; Rodrigues, D.N.; et al. Phosphorylated RB promotes cancer immunity by inhibiting NF-ΚB activation and PD-L1 expression. Mol. Cell 2019, 73, 22–35.e6. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.; Qi, C.; Yi, Q.-L.; Reece, D.; Stewart, A.K. P53 gene deletion detected by fluorescence in situ hybridization is an adverse prognostic factor for patients with multiple myeloma following autologous stem cell transplantation. Blood 2005, 105, 358–360. [Google Scholar] [CrossRef]
- Hu, B.; Thall, P.; Milton, D.R.; Sasaki, K.; Bashir, Q.; Shah, N.; Patel, K.; Popat, U.; Hosing, C.; Nieto, Y.; et al. High-risk myeloma and minimal residual disease postautologous-HSCT predict worse outcomes. Leuk. Lymphoma 2019, 60, 442–452. [Google Scholar] [CrossRef] [PubMed]
- Tiedemann, R.E.; Gonzalez-Paz, N.; Kyle, R.A.; Santana-Davila, R.; Price-Troska, T.; Van Wier, S.A.; Chng, W.J.; Ketterling, R.P.; Gertz, M.A.; Henderson, K.; et al. Genetic aberrations and survival in plasma cell leukemia. Leukemia 2008, 22, 1044–1052. [Google Scholar] [CrossRef]
- Merz, M.; Hielscher, T.; Seckinger, A.; Hose, D.; Mai, E.K.; Raab, M.S.; Goldschmidt, H.; Jauch, A.; Hillengass, J. Baseline Characteristics, chromosomal alterations, and treatment affecting prognosis of deletion 17p in newly diagnosed myeloma. Am. J. Hematol. 2016, 91, E473–E477. [Google Scholar] [CrossRef]
- An, G.; Li, Z.; Tai, Y.-T.; Acharya, C.; Li, Q.; Qin, X.; Yi, S.; Xu, Y.; Feng, X.; Li, C.; et al. The impact of clone size on the prognostic value of chromosome aberrations by fluorescence in situ hybridization in multiple myeloma. Clin. Cancer Res. 2015, 21, 2148–2156. [Google Scholar] [CrossRef]
- Lakshman, A.; Painuly, U.; Rajkumar, S.V.; Ketterling, R.P.; Kapoor, P.; Greipp, P.T.; Gertz, M.A.; Buadi, F.K.; Lacy, M.Q.; Dingli, D.; et al. Natural history of multiple myeloma with de novo del(17p). Blood Cancer J. 2019, 9, 32. [Google Scholar] [CrossRef]
- Ross, F.M.; Avet-Loiseau, H.; Ameye, G.; Gutiérrez, N.C.; Liebisch, P.; O’Connor, S.; Dalva, K.; Fabris, S.; Testi, A.M.; Jarosova, M.; et al. Report from the European Myeloma Network on interphase FISH in multiple myeloma and related disorders. Haematologica 2012, 97, 1272–1277. [Google Scholar] [CrossRef]
- Herrero, A.B.; Rojas, E.A.; Misiewicz-Krzeminska, I.; Krzeminski, P.; Gutiérrez, N.C. Molecular mechanisms of P53 deregulation in cancer: An overview in multiple myeloma. Int. J. Mol. Sci. 2016, 17, 2003. [Google Scholar] [CrossRef]
- Lodé, L.; Eveillard, M.; Trichet, V.; Soussi, T.; Wuillème, S.; Richebourg, S.; Magrangeas, F.; Ifrah, N.; Campion, L.; Traullé, C.; et al. Mutations in TP53 are exclusively associated with del(17p) in multiple myeloma. Haematologica 2010, 95, 1973–1976. [Google Scholar] [CrossRef]
- Thakurta, A.; Ortiz, M.; Blecua, P.; Towfic, F.; Corre, J.; Serbina, N.V.; Flynt, E.; Yu, Z.; Yang, Z.; Palumbo, A.; et al. High subclonal fraction of 17p deletion is associated with poor prognosis in multiple myeloma. Blood 2019, 133, 1217–1221. [Google Scholar] [CrossRef]
- Bergsagel, P.L.; Mateos, M.-V.; Gutierrez, N.C.; Rajkumar, S.V.; San Miguel, J.F. Improving overall survival and overcoming adverse prognosis in the treatment of cytogenetically high-risk multiple myeloma. Blood 2013, 121, 884–892. [Google Scholar] [CrossRef] [PubMed]
- Shaughnessy, J.D.; Zhou, Y.; Haessler, J.; van Rhee, F.; Anaissie, E.; Nair, B.; Waheed, S.; Alsayed, Y.; Epstein, J.; Crowley, J.; et al. TP53 deletion is not an adverse feature in multiple myeloma treated with total therapy 3. Br. J. Haematol. 2009, 147, 347–351. [Google Scholar] [CrossRef]
- Chng, W.-J.; Goldschmidt, H.; Dimopoulos, M.A.; Moreau, P.; Joshua, D.; Palumbo, A.; Facon, T.; Ludwig, H.; Pour, L.; Niesvizky, R.; et al. Carfilzomib-dexamethasone vs. bortezomib-dexamethasone in relapsed or refractory multiple myeloma by cytogenetic risk in the phase 3 study ENDEAVOR. Leukemia 2017, 31, 1368–1374. [Google Scholar] [CrossRef]
- Avet-Loiseau, H.; Fonseca, R.; Siegel, D.; Dimopoulos, M.A.; Špička, I.; Masszi, T.; Hájek, R.; Rosiñol, L.; Goranova-Marinova, V.; Mihaylov, G.; et al. Carfilzomib significantly improves the progression-free survival of high-risk patients in multiple myeloma. Blood 2016, 128, 1174–1180. [Google Scholar] [CrossRef]
- Moreau, P.; Masszi, T.; Grzasko, N.; Bahlis, N.J.; Hansson, M.; Pour, L.; Sandhu, I.; Ganly, P.; Baker, B.W.; Jackson, S.R.; et al. Oral ixazomib, lenalidomide, and dexamethasone for multiple myeloma. N. Engl. J. Med. 2016, 374, 1621–1634. [Google Scholar] [CrossRef] [PubMed]
- Leleu, X.; Karlin, L.; Macro, M.; Hulin, C.; Garderet, L.; Roussel, M.; Arnulf, B.; Pegourie, B.; Kolb, B.; Stoppa, A.M.; et al. Pomalidomide plus low-dose dexamethasone in multiple myeloma with deletion 17p and/or translocation (4;14): IFM 2010-02 trial results. Blood 2015, 125, 1411–1417. [Google Scholar] [CrossRef]
- Mikhael, J.; Richter, J.; Vij, R.; Cole, C.; Zonder, J.; Kaufman, J.L.; Bensinger, W.; Dimopoulos, M.; Lendvai, N.; Hari, P.; et al. A dose-finding phase 2 study of single agent isatuximab (anti-CD38 MAb) in relapsed/refractory multiple myeloma. Leukemia 2020, 34, 3298–3309. [Google Scholar] [CrossRef] [PubMed]
- Chng, W.J.; Dispenzieri, A.; Chim, C.-S.; Fonseca, R.; Goldschmidt, H.; Lentzsch, S.; Munshi, N.; Palumbo, A.; Miguel, J.S.; Sonneveld, P.; et al. IMWG consensus on risk stratification in multiple myeloma. Leukemia 2014, 28, 269–277. [Google Scholar] [CrossRef] [PubMed]
- Weinhold, N.; Ashby, C.; Rasche, L.; Chavan, S.S.; Stein, C.; Stephens, O.W.; Tytarenko, R.; Bauer, M.A.; Meissner, T.; Deshpande, S.; et al. Clonal selection and double-hit events involving tumor suppressor genes underlie relapse in myeloma. Blood 2016, 128, 1735–1744. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, X.; Han, C.; Wan, G.; Huang, X.; Ivan, C.; Jiang, D.; Rodriguez-Aguayo, C.; Lopez-Berestein, G.; Rao, P.H.; et al. TP53 loss creates therapeutic vulnerability in colorectal cancer. Nature 2015, 520, 697–701. [Google Scholar] [CrossRef] [PubMed]
- Pahl, A.; Lutz, C.; Hechler, T. Amanitins and their development as a payload for antibody-drug conjugates. Drug Discov. Today Technol. 2018, 30, 85–89. [Google Scholar] [CrossRef] [PubMed]
- Figueroa-Vazquez, V.; Ko, J.; Breunig, C.; Baumann, A.; Giesen, N.; Pálfi, A.; Müller, C.; Lutz, C.; Hechler, T.; Kulke, M.; et al. HDP-101, anti-BCMA antibody-drug conjugate, safely delivers amanitin to induce cell death in proliferating and resting multiple myeloma cells. Mol. Cancer Ther. 2020. [Google Scholar] [CrossRef]
- Li, Y.; Liu, Y.; Xu, H.; Jiang, G.; der Jeught, K.; Fang, Y.; Zhou, Z.; Zhang, L.; Frieden, M.; Wang, L.; et al. Heterozygous deletion of chromosome 17p renders prostate cancer vulnerable to inhibition of RNA polymerase II. Nat. Commun. 2018, 9, 4394. [Google Scholar] [CrossRef]
- Moreau, P.; Cavo, M.; Sonneveld, P.; Rosinol, L.; Attal, M.; Pezzi, A.; Goldschmidt, H.; Lahuerta, J.J.; Marit, G.; Palumbo, A.; et al. Combination of international scoring system 3, high lactate dehydrogenase, and t(4;14) and/or del(17p) identifies patients with multiple myeloma (MM) treated with front-line autologous stem-cell transplantation at high risk of early MM progression-related death. J. Clin. Oncol. 2014, 32, 2173–2180. [Google Scholar] [CrossRef]
- Lancman, G.; Tremblay, D.; Barley, K.; Barlogie, B.; Cho, H.J.; Jagannath, S.; Madduri, D.; Moshier, E.; Parekh, S.; Chari, A. The effect of novel therapies in high-molecular-risk multiple myeloma. Clin. Adv. Hematol. Oncol. 2017, 15, 870–879. [Google Scholar]
- Avet-Loiseau, H.; Fonseca, R.; Siegel, D.; Dimopoulos, M.A.; Spicka, I.; Masszi, T.; Hájek, R.; Rosiñol, L.; Goranova-Marinova, V.; Mihaylov, G.; et al. Efficacy and safety of carfilzomib, lenalidomide, and dexamethasone vs. lenalidomide and dexamethasone in patients with relapsed multiple myeloma based on cytogenetic risk status: Subgroup analysis from the phase 3 study aspire (NCT01080391). Blood 2015, 126, 731. [Google Scholar] [CrossRef]
- Larocca, A.; Mina, R.; Offidani, M.; Liberati, A.M.; Ledda, A.; Patriarca, F.; Evangelista, A.; Spada, S.; Benevolo, G.; Oddolo, D.; et al. First-line therapy with either bortezomib-melphalan-prednisone or lenalidomide-dexamethasone followed by lenalidomide for transplant-ineligible multiple myeloma patients: A pooled analysis of two randomized trials. Haematologica 2019. [Google Scholar] [CrossRef]
- Shah, V.; Sherborne, A.L.; Walker, B.A.; Johnson, D.C.; Boyle, E.M.; Ellis, S.; Begum, D.B.; Proszek, P.Z.; Jones, J.R.; Pawlyn, C.; et al. Prediction of outcome in newly diagnosed myeloma: A meta-analysis of the molecular profiles of 1905 trial patients. Leukemia 2018, 32, 102–110. [Google Scholar] [CrossRef]
- Cook, G.; Royle, K.; O’Connor, S.; Cairns, D.A.; Ashcroft, A.J.; Williams, C.D.; Hockaday, A.; Cavenagh, J.D.; Snowden, J.A.; Ademokun, D.; et al. The impact of cytogenetics on duration of response and overall survival in patients with relapsed multiple myeloma (long-term follow-up results from BSBMT/UKMF myeloma X relapse [intensive]): A randomised, open-label, phase 3 trial. Br. J. Haematol. 2019, 185, 450–467. [Google Scholar] [CrossRef]
- Corre, J.; Cleynen, A.; Robiou du Pont, S.; Buisson, L.; Bolli, N.; Attal, M.; Munshi, N.; Avet-Loiseau, H. Multiple myeloma clonal evolution in homogeneously treated patients. Leukemia 2018, 32, 2636–2647. [Google Scholar] [CrossRef]
- Maura, F.; Bolli, N.; Angelopoulos, N.; Dawson, K.J.; Leongamornlert, D.; Martincorena, I.; Mitchell, T.J.; Fullam, A.; Gonzalez, S.; Szalat, R.; et al. Genomic landscape and chronological reconstruction of driver events in multiple myeloma. Nat. Commun. 2019, 10, 3835. [Google Scholar] [CrossRef] [PubMed]
- Lohr, J.G.; Stojanov, P.; Carter, S.L.; Cruz-Gordillo, P.; Lawrence, M.S.; Auclair, D.; Sougnez, C.; Knoechel, B.; Gould, J.; Saksena, G.; et al. Widespread Genetic heterogeneity in multiple myeloma: Implications for targeted therapy. Cancer Cell 2014, 25, 91–101. [Google Scholar] [CrossRef] [PubMed]
- Bolli, N.; Avet-Loiseau, H.; Wedge, D.C.; Van Loo, P.; Alexandrov, L.B.; Martincorena, I.; Dawson, K.J.; Iorio, F.; Nik-Zainal, S.; Bignell, G.R.; et al. Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nat. Commun. 2014, 5, 2997. [Google Scholar] [CrossRef]
- Jones, J.R.; Weinhold, N.; Ashby, C.; Walker, B.A.; Wardell, C.; Pawlyn, C.; Rasche, L.; Melchor, L.; Cairns, D.A.; Gregory, W.M.; et al. Clonal evolution in myeloma: The impact of maintenance lenalidomide and depth of response on the genetics and sub-clonal structure of relapsed disease in uniformly treated newly diagnosed patients. Haematologica 2019, 104, 1440–1450. [Google Scholar] [CrossRef]
- Lionetti, M.; Neri, A. utilizing next-generation sequencing in the management of multiple myeloma. Expert. Rev. Mol. Diagn. 2017, 17, 653–663. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.K.; Rajkumar, S.V. The multiple myelomas - current concepts in cytogenetic classification and therapy. Nat. Rev. Clin. Oncol. 2018, 15, 409–421. [Google Scholar] [CrossRef]
- Mulligan, G.; Lichter, D.I.; Di Bacco, A.; Blakemore, S.J.; Berger, A.; Koenig, E.; Bernard, H.; Trepicchio, W.; Li, B.; Neuwirth, R.; et al. Mutation of NRAS but not KRAS significantly reduces myeloma sensitivity to single-agent bortezomib therapy. Blood 2014, 123, 632–639. [Google Scholar] [CrossRef]
- Boyle, E.M.; Ashby, C.; Tytarenko, R.; Deshpande, S.; Wang, Y.; Sawyer, J.; Tian, E.; Johnson, S.; Rutherford, M.W.; Wardell, C.P.; et al. BRAF and DIS3 mutations associate with adverse outcome in a long-term follow-up of patients with multiple myeloma. Clin. Cancer Res. 2020. [Google Scholar] [CrossRef]
- Keats, J.J.; Fonseca, R.; Chesi, M.; Schop, R.; Baker, A.; Chng, W.-J.; Van Wier, S.; Tiedemann, R.; Shi, C.-X.; Sebag, M.; et al. Promiscuous mutations activate the noncanonical NF-kappaB pathway in multiple myeloma. Cancer Cell 2007, 12, 131–144. [Google Scholar] [CrossRef]
- Mroczek, S.; Chlebowska, J.; Kuliński, T.M.; Gewartowska, O.; Gruchota, J.; Cysewski, D.; Liudkovska, V.; Borsuk, E.; Nowis, D.; Dziembowski, A. The non-canonical poly(A) polymerase FAM46C acts as an onco-suppressor in multiple myeloma. Nat. Commun. 2017, 8, 619. [Google Scholar] [CrossRef]
- Ruiz-Heredia, Y.; Sánchez-Vega, B.; Onecha, E.; Barrio, S.; Alonso, R.; Martínez-Ávila, J.C.; Cuenca, I.; Agirre, X.; Braggio, E.; Hernández, M.-T.; et al. Mutational screening of newly diagnosed multiple myeloma patients by deep targeted sequencing. Haematologica 2018, 103, e544–e548. [Google Scholar] [CrossRef] [PubMed]
- Kortuem, K.M.; Braggio, E.; Bruins, L.; Barrio, S.; Shi, C.S.; Zhu, Y.X.; Tibes, R.; Viswanatha, D.; Votruba, P.; Ahmann, G.; et al. Panel sequencing for clinically oriented variant screening and copy number detection in 142 untreated multiple myeloma patients. Blood Cancer J. 2016, 6, e397. [Google Scholar] [CrossRef]
- Walker, B.A.; Mavrommatis, K.; Wardell, C.P.; Ashby, T.C.; Bauer, M.; Davies, F.E.; Rosenthal, A.; Wang, H.; Qu, P.; Hoering, A.; et al. Identification of novel mutational drivers reveals oncogene dependencies in multiple myeloma. Blood 2018, 132, 587–597. [Google Scholar] [CrossRef] [PubMed]
- Niesvizky, R.; Badros, A.Z.; Costa, L.J.; Ely, S.A.; Singhal, S.B.; Stadtmauer, E.A.; Haideri, N.A.; Yacoub, A.; Hess, G.; Lentzsch, S.; et al. Phase 1/2 study of cyclin-dependent kinase (CDK)4/6 inhibitor palbociclib (PD-0332991) with bortezomib and dexamethasone in relapsed/refractory multiple myeloma. Leuk. Lymphoma 2015, 56, 3320–3328. [Google Scholar] [CrossRef]
- Kumar, S.K.; LaPlant, B.; Chng, W.J.; Zonder, J.; Callander, N.; Fonseca, R.; Fruth, B.; Roy, V.; Erlichman, C.; Stewart, A.K.; et al. Dinaciclib, a novel CDK inhibitor, demonstrates encouraging single-agent activity in patients with relapsed multiple myeloma. Blood 2015, 125, 443–448. [Google Scholar] [CrossRef] [PubMed]
- Mey, U.J.M.; Renner, C.; von Moos, R. Vemurafenib in combination with cobimetinib in relapsed and refractory extramedullary multiple myeloma harboring the BRAF V600E mutation. Hematol. Oncol. 2017, 35, 890–893. [Google Scholar] [CrossRef] [PubMed]
- Čepulytė, R.; Žučenka, A.; Pečeliūnas, V. Combination of dabrafenib and trametinib for the treatment of relapsed and refractory multiple myeloma harboring BRAF V600E mutation. Case Rep. Hematol. 2020, 2020, 8894031. [Google Scholar] [CrossRef]
- Blanden, A.R.; Yu, X.; Loh, S.N.; Levine, A.J.; Carpizo, D.R. Reactivating mutant P53 using small molecules as zinc metallochaperones: A wakening a sleeping giant in cancer. Drug Discov. Today 2015, 20, 1391–1397. [Google Scholar] [CrossRef] [PubMed]
- Lambert, J.M.R.; Gorzov, P.; Veprintsev, D.B.; Söderqvist, M.; Segerbäck, D.; Bergman, J.; Fersht, A.R.; Hainaut, P.; Wiman, K.G.; Bykov, V.J.N. PRIMA-1 reactivates mutant P53 by covalent binding to the core domain. Cancer Cell 2009, 15, 376–388. [Google Scholar] [CrossRef]
- Deneberg, S.; Cherif, H.; Lazarevic, V.; Andersson, P.-O.; von Euler, M.; Juliusson, G.; Lehmann, S. An open-label phase I dose-finding study of APR-246 in hematological malignancies. Blood Cancer J. 2016, 6, e447. [Google Scholar] [CrossRef]
- Saha, M.N.; Jiang, H.; Chang, H. Molecular mechanisms of nutlin-induced apoptosis in multiple myeloma: Evidence for P53-transcription-dependent and -independent pathways. Cancer Biol. Ther. 2010, 10, 567–578. [Google Scholar] [CrossRef]
- Issaeva, N.; Bozko, P.; Enge, M.; Protopopova, M.; Verhoef, L.G.G.C.; Masucci, M.; Pramanik, A.; Selivanova, G. Small molecule RITA binds to P53, blocks P53-HDM-2 interaction and activates P53 function in tumors. Nat. Med. 2004, 10, 1321–1328. [Google Scholar] [CrossRef]
- Grasberger, B.L.; Lu, T.; Schubert, C.; Parks, D.J.; Carver, T.E.; Koblish, H.K.; Cummings, M.D.; LaFrance, L.V.; Milkiewicz, K.L.; Calvo, R.R.; et al. Discovery and cocrystal structure of benzodiazepinedione HDM2 antagonists that activate P53 in cells. J. Med. Chem. 2005, 48, 909–912. [Google Scholar] [CrossRef]
- Ding, K.; Lu, Y.; Nikolovska-Coleska, Z.; Wang, G.; Qiu, S.; Shangary, S.; Gao, W.; Qin, D.; Stuckey, J.; Krajewski, K.; et al. Structure-based design of spiro-oxindoles as potent, specific small-molecule inhibitors of the MDM2−p53 interaction. J. Med. Chem. 2006, 49, 3432–3435. [Google Scholar] [CrossRef]
- Rao, B.; Lain, S.; Thompson, A.M. P53-based cyclotherapy: Exploiting the “guardian of the genome” to protect normal cells from cytotoxic therapy. Br. J. Cancer 2013, 109, 2954–2958. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, Y.; Han, Z.-G.; He, K.-Y. Malignancy of cancers and synthetic lethal interactions associated with mutations of cancer driver genes. Med. Baltim 2016, 95, e2697. [Google Scholar] [CrossRef]
- Pasca, S.; Tomuleasa, C.; Teodorescu, P.; Ghiaur, G.; Dima, D.; Moisoiu, V.; Berce, C.; Stefan, C.; Ciechanover, A.; Einsele, H. KRAS/NRAS/BRAF mutations as potential targets in multiple myeloma. Front. Oncol. 2019, 9, 1137. [Google Scholar] [CrossRef]
- Hyman, D.M.; Puzanov, I.; Subbiah, V.; Faris, J.E.; Chau, I.; Blay, J.-Y.; Wolf, J.; Raje, N.S.; Diamond, E.L.; Hollebecque, A.; et al. Vemurafenib in multiple nonmelanoma cancers with BRAF V600 mutations. N. Engl. J. Med. 2015, 373, 726–736. [Google Scholar] [CrossRef] [PubMed]
- Tolcher, A.W.; Patnaik, A.; Papadopoulos, K.P.; Rasco, D.W.; Becerra, C.R.; Allred, A.J.; Orford, K.; Aktan, G.; Ferron-Brady, G.; Ibrahim, N.; et al. Phase I study of the MEK inhibitor trametinib in combination with the AKT inhibitor afuresertib in patients with solid tumors and multiple myeloma. Cancer Chemother. Pharmacol. 2015, 75, 183–189. [Google Scholar] [CrossRef] [PubMed]
- Salama, A.K.S.; Li, S.; Macrae, E.R.; Park, J.-I.; Mitchell, E.P.; Zwiebel, J.A.; Chen, H.X.; Gray, R.J.; McShane, L.M.; Rubinstein, L.V.; et al. Dabrafenib and trametinib in patients with tumors with BRAFV600E mutations: Results of the NCI-MATCH trial subprotocol H. J. Clin. Oncol. 2020, 38, 3895–3904. [Google Scholar] [CrossRef]
- Holkova, B.; Zingone, A.; Kmieciak, M.; Bose, P.; Badros, A.Z.; Voorhees, P.M.; Baz, R.; Korde, N.; Lin, H.-Y.; Chen, J.-Q.; et al. A phase II trial of AZD6244 (selumetinib, ARRY-142886), an oral MEK1/2 inhibitor, in relapsed/refractory multiple myeloma. Clin. Cancer Res. 2016, 22, 1067–1075. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishnan, V.G.; Miller, K.C.; Macon, E.P.; Kimlinger, T.K.; Haug, J.; Kumar, S.; Gonsalves, W.I.; Rajkumar, S.V.; Kumar, S.K. Histone deacetylase inhibition in combination with MEK or BCL-2 inhibition in multiple myeloma. Haematologica 2019, 104, 2061–2074. [Google Scholar] [CrossRef] [PubMed]
- Díaz, T.; Rodríguez, V.; Lozano, E.; Mena, M.-P.; Calderón, M.; Rosiñol, L.; Martínez, A.; Tovar, N.; Pérez-Galán, P.; Bladé, J.; et al. The BET bromodomain inhibitor CPI203 improves lenalidomide and dexamethasone activity in vitro and in vivo models of multiple myeloma by blockade of ikaros and MYC signaling. Haematologica 2017, 102, 1776–1784. [Google Scholar] [CrossRef]
- Amorim, S.; Stathis, A.; Gleeson, M.; Iyengar, S.; Magarotto, V.; Leleu, X.; Morschhauser, F.; Karlin, L.; Broussais, F.; Rezai, K.; et al. Bromodomain inhibitor OTX015 in patients with lymphoma or multiple myeloma: A dose-escalation, open-label, pharmacokinetic, phase 1 study. Lancet Haematol. 2016, 3, e196–e204. [Google Scholar] [CrossRef]
- Corre, J.; Munshi, N.C.; Avet-Loiseau, H. Risk factors in multiple myeloma: Is it time for a revision? Blood 2021, 137, 16–19. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).