Osteocytic Connexin43 Channels Regulate Bone–Muscle Crosstalk
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Muscle Mass Measurement
2.3. Histology and Immunohistochemistry
2.4. RNA Extraction and Real-Time PCR (qPCR)
2.5. Western Blot Analyses
2.6. Ex vivo Muscle Force Measurement
2.7. Primary Osteocytes CM (PO CM) Preparation and Cell Culture Conditions
2.8. Treatment of C2C12 Cells with PO CMs
2.9. Intracellular Calcium (Ca2+) Signal Measurement
2.10. Enzyme-Linked Immunosorbent Assay (ELISA)
2.11. Statistical Analysis
3. Results
3.1. Impairment of Osteocytic HCs Reduces Fast-Twitch Muscle Mass
3.2. Impairment of GJs Alters the Fiber-Type Composition and Muscle Contractile Force
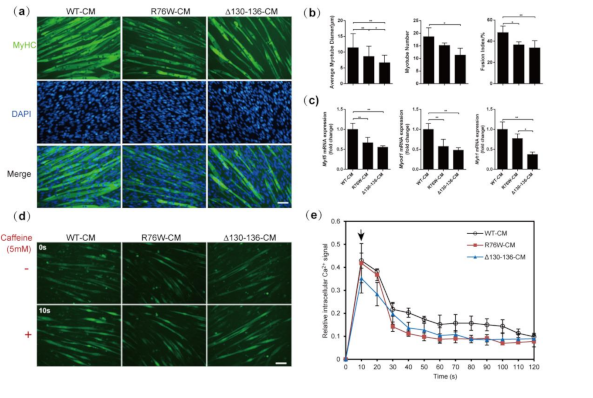
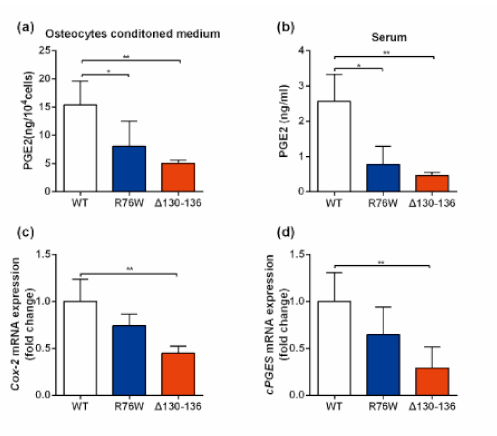
3.3. Primary Osteocytes Conditioned Media (PO CM) Derived From R76W and Δ130–136 Inhibit Myogenic Differentiation of C2C12 Cells
3.4. Impaired Cx43 Channels Reduces Caffeine-Induced Sarcoplasmic Reticulum Calcium Release in Myotubes
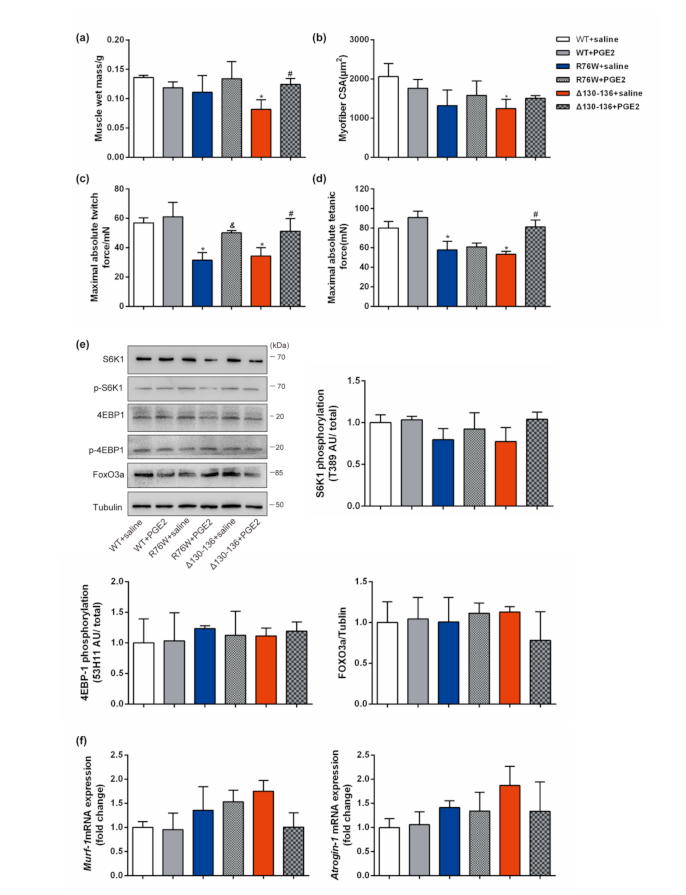
3.5. PGE2 Treatment Augments Skeletal Muscle Mass and Contractile Force
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brotto, M.; Bonewald, L. Bone and muscle: Interactions beyond mechanical. Bone 2015, 80, 109–114. [Google Scholar] [CrossRef]
- Bonewald, L. Use it or lose it to age: A review of bone and muscle communication. Bone 2019, 120, 212–218. [Google Scholar] [CrossRef]
- Brotto, M.; Johnson, M.L. Endocrine Crosstalk between Muscle and Bone. Curr. Osteoporos. Rep. 2014, 12, 135–141. [Google Scholar] [CrossRef]
- Hamrick, P.L.; Patterson, S.L. Role of muscle-derived growth factors in bone formation. J. Musculoskelet. Neuronal Interact. 2010, 10, 64–70. [Google Scholar]
- Qin, Y.; Peng, Y.; Zhao, W.; Pan, J.; Ksiezak-Reding, H.; Cardozo, C.; Wu, Y.; Pajevic, P.D.; Bonewald, L.F.; Bauman, W.A.; et al. Myostatin inhibits osteoblastic differentiation by suppressing osteocyte-derived exosomal microRNA-218: A novel mechanism in muscle-bone communication. J. Biol. Chem. 2017, 292, 11021–11033. [Google Scholar] [CrossRef]
- Kitase, Y.; Vallejo, J.A.; Gutheil, W.; Vemula, H.; Jähn, K.; Yi, J.; Zhou, J.; Brotto, M.; Bonewald, L. β-aminoisobutyric Acid, l-BAIBA, Is a Muscle-Derived Osteocyte Survival Factor. Cell Rep. 2018, 22, 1531–1544. [Google Scholar] [CrossRef]
- Kim, H.; Wrann, C.D.; Jedrychowski, M.; Vidoni, S.; Kitase, Y.; Nagano, K.; Zhou, C.; Chou, J.; Parkman, V.-J.A.; Novick, S.J.; et al. Irisin Mediates Effects on Bone and Fat via αV Integrin Receptors. Cell 2018, 175, 1756–1768.e17. [Google Scholar] [CrossRef]
- Mera, P.; Laue, K.; Ferron, M.; Confavreux, C.; Wei, J.; Galan-Diez, M.; Lacampagne, A.; Mitchell, S.J.; Mattison, J.A.; Chen, Y.; et al. Os-teocalcin Signaling in Myofibers Is Necessary and Sufficient for Optimum Adaptation to Exercise. Cell Metab. 2016, 23, 1078–1092. [Google Scholar]
- Chenglin, M.S.-S.; Lynda, B.; Mark, J.; Marco, B. Prostaglandin E2 from clinical applications to its potential role in bone- muscle crosstalk and myogenic differentiation. Recent. Pat. Biotechnol. 2012, 6, 223–229. [Google Scholar]
- Huang, J.; Romero-Suarez, S.; Lara, N.; Mo, C.; Kaja, S.; Brotto, L.; Dallas, S.L.; Johnson, M.L.; Jahn, K.; Bonewald, L.F.; et al. Cross-talk between MLO-Y4 osteocytes and C2C12 muscle cells is mediated by the Wnt/beta-catenin pathway. JBMR Plus 2017, 1, 86–100. [Google Scholar]
- Kawao, N.; Kaji, H. Interactions between Muscle Tissues and Bone Metabolism. J. Cell. Biochem. 2015, 116, 687–695. [Google Scholar] [CrossRef]
- Dallas, S.L.; Prideaux, M.; Bonewald, L.F. The osteocyte: An endocrine cell and more. Endocr. Rev. 2013, 34, 658–690. [Google Scholar]
- Beno, T.; Yoon, Y.-J.; Cowin, S.C.; Fritton, S.P. Estimation of bone permeability using accurate microstructural measurements. J. Biomech. 2006, 39, 2378–2387. [Google Scholar] [CrossRef]
- Mo, C.; Zhao, R.; Vallejo, J.; Igwe, O.; Bonewald, L.; Wetmore, L.; Brotto, M. Prostaglandin E2 promotes proliferation of skeletal mus-cle myoblasts via EP4 receptor activation. Cell Cycle 2015, 14, 1507–1516. [Google Scholar]
- Batra, N.; Kar, R.; Jiang, J.X. Gap junctions and hemichannels in signal transmission, function and development of bone. Biochim. et Biophys. Acta (BBA) Biomembr. 2012, 1818, 1909–1918. [Google Scholar] [CrossRef]
- Genetos, D.C.; Kephart, C.J.; Zhang, Y.; Yellowley, C.E.; Donahue, H.J. Oscillating fluid flow activation of gap junction hemichan-nels induces ATP release from MLO-Y4 osteocytes. J. Cell. Physiol. 2007, 212, 207–214. [Google Scholar]
- Shen, H.; Grimston, S.; Civitelli, R.; Thomopoulos, S. Deletion of Connexin43 in Osteoblasts/Osteocytes Leads to Impaired Muscle Formation in Mice. J. Bone Miner. Res. 2015, 30, 596–605. [Google Scholar] [CrossRef]
- Xu, H.; Gu, S.; Riquelme, M.A.; Burra, S.; Callaway, D.; Cheng, H.; Guda, T.; Schmitz, J.; Fajardo, R.J.; Werner, S.L.; et al. Connexin 43 Channels Are Essential for Normal Bone Structure and Osteocyte Viability. J. Bone Miner. Res. 2015, 30, 436–448. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, M.; Xue, T.; Li, G.; Wang, D.; Shang, P.; Jiang, J.X.; Xu, H. Osteocytic connexin 43 channels affect fracture healing. J. Cell. Physiol. 2019, 234, 19824–19832. [Google Scholar] [CrossRef]
- Zhao, D.; Liu, R.; Li, G.; Chen, M.; Shang, P.; Yang, H.; Jiang, J.X.; Xu, H. Connexin 43 Channels in Osteocytes Regulate Bone Re-sponses to Mechanical Unloading. Front. Physiol. 2020, 11, 299. [Google Scholar]
- Ho, A.T.V.; Palla, A.R.; Blake, M.R.; Yucel, N.D.; Wang, Y.X.; Magnusson, K.E.G.; Holbrook, C.A.; Kraft, P.E.; Delp, S.L.; Blau, H.M. Prosta-glandin E2 is essential for efficacious skeletal muscle stem-cell function, augmenting regeneration and strength. Proc. Natl. Acad. Sci. USA 2017, 114, 6675–6684. [Google Scholar]
- Brooks, S.V.; Faulkner, J.A. Contractile properties of skeletal muscles from young, adult and aged mice. J. Physiol. 1988, 404, 71–82. [Google Scholar]
- Beaulieu, D.; Thebault, P.; Pelletier, R.; Chapdelaine, P.; Tarnopolsky, M.; Furling, D.; Puymirat, J. Abnormal prostaglandin E2 pro-duction blocks myogenic differentiation in myotonic dystrophy. Neurobiol. Dis. 2012, 45, 122–129. [Google Scholar]
- Colditz, J.; Thiele, S.; Baschant, U.; Niehrs, C.; Bonewald, L.F.; Hofbauer, L.C.; Rauner, M. Postnatal Skeletal Deletion of Dickkopf-1 Increases Bone Formation and Bone Volume in Male and Female Mice, Despite Increased Sclerostin Expression. J. Bone Miner. Res. 2018, 33, 1698–1707. [Google Scholar] [CrossRef]
- Liu, J.; Peng, Y.; Cui, Z.; Wu, Z.; Qian, A.; Shang, P.; Qu, L.; Li, Y.; Liu, J.; Long, J. Depressed mitochondrial biogenesis and dynamic re-modeling in mouse tibialis anterior and gastrocnemius induced by 4-week hindlimb unloading. IUBMB Life 2012, 64, 901–910. [Google Scholar]
- Schiaffino, S.; Reggiani, C. Fiber types in mammalian skeletal muscles. Physiol. Rev. 2011, 91, 1447–1531. [Google Scholar]
- Oláh, T.; Bodnár, D.; Tóth, A.; Vincze, J.; Fodor, J.; Reischl, B.; Kovács, A.; Ruzsnavszky, O.; Dienes, B.; Szentesi, P.; et al. Cannabinoid signalling inhibits sarcoplasmic Ca2+release and regulates excitation-contraction coupling in mammalian skeletal muscle. J. Physiol. 2016, 594, 7381–7398. [Google Scholar] [CrossRef]
- Zhao, X.; Weisleder, N.; Thornton, A.; Oppong, Y.; Campbell, R.; Ma, J.; Brotto, M. Compromised store-operated Ca2+ entry in aged skeletal muscle. Aging Cell 2008, 7, 561–568. [Google Scholar]
- Zhou, J.Z.; Riquelme, M.A.; Gu, S.; Kar, R.; Gao, X.; Sun, L.; Jiang, J.X. Osteocytic connexin hemichannels suppress breast cancer growth and bone metastasis. Oncogene 2016, 35, 5597–5607. [Google Scholar] [CrossRef]
- Goll, D.E.; Neti, G.; Mares, S.W.; Thompson, V.F. Myofibrillar protein turnover: The proteasome and the calpains1. J. Anim. Sci. 2008, 86, E19–E35. [Google Scholar] [CrossRef]
- Bodine, S.C.; Latres, E.; Baumhueter, S.; Lai, V.K.; Nunez, L.; Clarke, B.A.; Poueymirou, W.T.; Panaro, F.J.; Na, E.; Dharmara-jan, K.; et al. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science 2001, 294, 1704–1708. [Google Scholar]
- Sanchez, A.M.; Csibi, A.; Raibon, A.; Cornille, K.; Gay, S.; Bernardi, H.; Candau, R. AMPK promotes skeletal muscle autophagy through activation of forkhead FoxO3a and interaction with Ulk. J. Cell. Biochem. 2012, 113, 695–710. [Google Scholar] [CrossRef]
- Bonewald, L.F.; Kiel, D.P.; Clemens, T.L.; Esser, K.; Orwoll, E.S.; O’Keefe, R.J.; Fielding, R.A. Forum on bone and skeletal muscle interac-tions: Summary of the proceedings of an ASBMR workshop. J. Bone Miner. Res. 2013, 28, 1857–1865. [Google Scholar]
- Greising, S.M.; Call, J.A.; Lund, T.C.; Blazar, B.R.; Tolar, J.; Lowe, D.A. Skeletal muscle contractile function and neuromuscular per-formance in Zmpste24 −/− mice, a murine model of human progeria. Age 2012, 34, 805–819. [Google Scholar]
- Jähn, K.L.-C.; Brotto, L.M.; Johnson, M.L.; Brotto, M.; Bonewald, L.F. Skeletal muscle secreted factors prevent glucocor-ticoid induced osteocyte apoptosis through activation of β-catenin. Eur. Cells Mater. 2012, 24, 197–210. [Google Scholar]
- Wood, C.L.; Pajevic, P.D.; Gooi, J. Osteocyte secreted factors inhibit skeletal muscle differentiation. Bone Rep. 2017, 6, 74–80. [Google Scholar] [CrossRef]
- Cherian, P.P.; Siller-Jackson, A.J.; Gu, S.; Wang, X.; Bonewald, L.F.; Sprague, E.; Jiang, J.X. Mechanical strain opens connexin 43 hemi-channels in osteocytes: A novel mechanism for the release of prostaglandin. Mol. Biol. Cell 2005, 16, 3100–3106. [Google Scholar]
- Cheng, B.X.K.; Zhao, S.J.; Luo, J.; Sprague, E.; Bonewald, L.F.J. PGE2 is Essential for Gap Junction-Mediated Intercellular Communication between Osteocyte-Like MLO-Y4 Cells in Response to Mechanical Strain. Endocrinology 2001, 142, 3464–3473. [Google Scholar]
- Stains, J.P.; Lecanda, F.; Screen, J.; Towler, D.A.; Civitelli, R. Gap junctional communication modulates gene transcription by alter-ing the recruitment of Sp1 and Sp3 to connexin-response elements in osteoblast promoters. J. Biol. Chem. 2003, 278, 24377–24387. [Google Scholar]
- Grimston, S.K.; Brodt, M.D.; Silva, M.J.; Civitelli, R. Attenuated Response to In Vivo Mechanical Loading in Mice With Conditional Osteoblast Ablation of the Connexin43 Gene (Gja1). J. Bone Miner. Res. 2008, 23, 879–886. [Google Scholar] [CrossRef]
- Zhou, J.Z.; Jiang, J.X. Gap junction and hemichannel-independent actions of connexins on cell and tissue functions--an update. FEBS Lett. 2014, 588, 1186–1192. [Google Scholar] [CrossRef]
- Rodemann, H.; Goldberg, A. Arachidonic acid, prostaglandin E2 and F2 alpha influence rates of protein turnover in skeletal and cardiac muscle. J. Biol. Chem. 1982, 257, 1632–1638. [Google Scholar] [CrossRef]
- Barnett, J.G.; Ellis, S. Prostaglandin E2 and the regulation of protein degradation in skeletal muscle. Muscle Nerve 1987, 10, 556–559. [Google Scholar] [CrossRef]
- Garber, A.J.E.; Karl, I.; Kipnis, D.M. Alanine and glutamine synthesis and release from skeletal muscle. IV. beta-Adrenergic inhibition of amino acid release. J. Biol. Chem. 1976, 251, 851–857. [Google Scholar] [CrossRef]



| Gene Name | Primer Sequences (5′ to 3′) |
|---|---|
| mCx43 | Forward: CGGAAGCACCATCTCCAACT |
| Reverse: CCACGATAGCTAAGGGCTGG | |
| mCx50 | Forward: CAAGGGCTGTCTGCTGAGAA |
| Reverse: AGATCATCTGACCTGGCCCT | |
| Myh1 | Forward: GCTTCAAGTTTGGACCCACG |
| Reverse: TTCTGAGCCTCGATTCGCTC | |
| Myh4 | Forward: GGAGGCTGAGGAACAATCCA |
| Reverse: TCTCCTGTCACCTCTCAACAG | |
| Myh7 | Forward: CAACCTGTCCAAGTTCCGCA |
| Reverse: TACTCCTCATTCAGGCCCTTG | |
| Myod1 | Forward: TACGACACCGCCTACTACAGTG |
| Reverse: GTGGTGCATCTGCCAAAAG | |
| Myf5 | Forward: CTGTCTGGTCCCGAAAGAAC |
| Reverse: TGGAGAGAGGGAAGCTGTGT | |
| Cox-2 | Forward: TGAGCAACTATTCCAAACCAGC |
| Reverse: GCACGTAGTCTTCGATCACTATC | |
| cPGEs | Forward: AAGGAGAATCTGGCCAGTCA |
| Reverse: ATCCTCATCACCACCCATGT | |
| MuRF1 | Forward: GCCATCCTGGACGAGAAGAA |
| Reverse: CAGCTGGCAGCCCTTGGA | |
| Atrogin-1 | Forward: AGACCGGCTACTGTGGAAGAG |
| Reverse: CCGTGCATGGATGGTCAGTG | |
| GAPDH | Forward: TCAACAGCAACTCCCACTCTTCCA |
| Reverse: ACCCTGTTGCTGTAGCCGTATTCA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, G.; Zhang, L.; Ning, K.; Yang, B.; Acosta, F.M.; Shang, P.; Jiang, J.X.; Xu, H. Osteocytic Connexin43 Channels Regulate Bone–Muscle Crosstalk. Cells 2021, 10, 237. https://doi.org/10.3390/cells10020237
Li G, Zhang L, Ning K, Yang B, Acosta FM, Shang P, Jiang JX, Xu H. Osteocytic Connexin43 Channels Regulate Bone–Muscle Crosstalk. Cells. 2021; 10(2):237. https://doi.org/10.3390/cells10020237
Chicago/Turabian StyleLi, Guobin, Lan Zhang, Kaiting Ning, Baoqiang Yang, Francisca M. Acosta, Peng Shang, Jean X. Jiang, and Huiyun Xu. 2021. "Osteocytic Connexin43 Channels Regulate Bone–Muscle Crosstalk" Cells 10, no. 2: 237. https://doi.org/10.3390/cells10020237
APA StyleLi, G., Zhang, L., Ning, K., Yang, B., Acosta, F. M., Shang, P., Jiang, J. X., & Xu, H. (2021). Osteocytic Connexin43 Channels Regulate Bone–Muscle Crosstalk. Cells, 10(2), 237. https://doi.org/10.3390/cells10020237







