Abstract
Understanding of pancreatic islet biology has greatly increased over the past few decades based in part on an increased understanding of the transcription factors that guide this process. One such transcription factor that has been increasingly tied to both β-cell development and the development of diabetes in humans is GLIS3. Genetic deletion of GLIS3 in mice and humans induces neonatal diabetes, while single nucleotide polymorphisms (SNPs) in GLIS3 have been associated with both Type 1 and Type 2 diabetes. As a significant progress has been made in understanding some of GLIS3’s roles in pancreas development and diabetes, we sought to compare current knowledge on GLIS3 within the pancreas to that of other islet enriched transcription factors. While GLIS3 appears to regulate similar genes and pathways to other transcription factors, its unique roles in β-cell development and maturation make it a key target for future studies and therapy.
1. Introduction
The pancreas serves a dual-function role within the body. Acinar cells produce and secrete enzymes involved in digestion through the pancreatic ductal network into the duodenum to aid in digestion, while pancreatic endocrine cells play a critical role in the regulation of glycemia via hormone secretion into the blood stream. Endocrine cells cluster into islets of Langerhans, and despite their critical importance, make up only 1–4% of the pancreas [1,2]. Islets are comprised primarily of β-cells, which secrete insulin in response to elevated levels of blood glucose. Insulin insufficiency can have several causes, including insulin resistance coupled with β-cell dysfunction (Type 2 diabetes), autoimmune destruction of the β-cells (Type 1 Diabetes), a variety of monogenic causes of diabetes, as well as pregnancy induced gestational diabetes. With the exception of monogenic diabetes, where β-cell or pancreatic dysfunction is linked to mutations in one particular gene [3], diabetes mostly results from a combination of genetic and environmental factors. A variety of environmental factors (e.g., diet) have been identified that play a critical role in both Type 1 diabetes [4,5,6,7,8], and Type 2 diabetes [9,10], while genetic associations have been incredibly varied, often with small effects [11]. To better understand the interaction between environmental and genetic factors, a more detailed understanding of pancreas development and function is necessary.
The identification and characterization of a variety of transcription factors, including those associated with monogenic diabetes (GLIS3, PDX1, PTF1A, HNF1A, HNF1B, HNF4A, FOXP3, PAX4, RFX6, GATA4, GATA6, NGN3, NEUROD1, PAX6, MNX1, NKX2.2) [12], have greatly contributed to our understanding of pancreatic development. Several reviews have attempted to summarize this research [13,14,15], however new research is constantly expanding our understanding of pancreatic development and the transcription factors driving it. Previous reviews have sought to summarize GLIS3’s role in diabetes, congenital hypothyroidism, as well as a variety of other diseases [16,17,18]. Here, we sought to specifically review our current understanding of the GLIS3 gene as it relates to pancreas development, comparing as well as contrasting its role with that of other transcription factors prominent in the field. We hope to highlight that while mice and humans lacking a functional copy of the GLIS3 gene display many phenotypes similar to other transcription factor knockouts, the timing and features of the phenotypes differ in subtle but distinguishable ways from other transcription factors, highlighting GLIS3’s unique role in pancreatic development.
2. The GLIS3 Gene and Its Encoded Protein
The mouse Glis3 gene was first identified in 2003 as a gene with 5 C2H2-type zinc finger motifs that contain high homology to the Gli and Zic family of genes [19]. In humans, GLIS3 includes 11 exons, and encodes for a protein of 930 amino acids (Figure 1A). The GLIS3 protein contains 3 known domains: an N-terminal Repressive Domain (NRD), a DNA-binding Domain (DBD) made up of the above-mentioned zinc finger motifs, and a C-terminal Transactivation Domain (TAD) (Figure 1B). The N-terminal repressive domain is largely conserved with the GLI family of proteins and contains amino acids that interact with the Suppressor of Fused (SUFU) protein [20]. The HECT E3 ubiquitin ligase ITCH can bind near the NRD domain in the N-terminus and promotes the polyubiquitination and degradation of GLIS3 [21]. Additionally, GLIS3 is SUMOylated on either side of the NRD by PIASy and UBC9, which inhibits its ability to stimulate transcription [22].
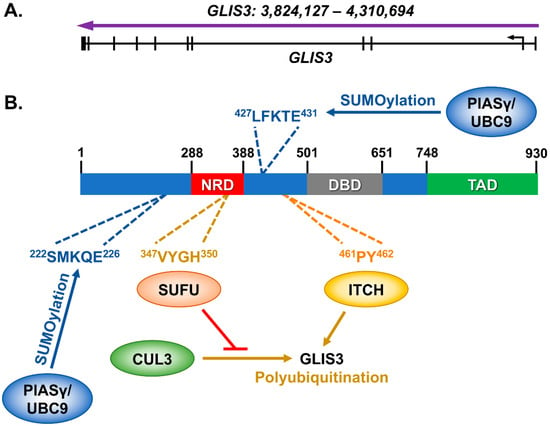
Figure 1.
GLIS3 and its known interacting proteins. (A) The GLIS3 gene (NM_001042413) is composed of 11 exons, which produces (B) a 930 amino acid protein. Interactions have previously been identified with SUFU [20], ITCH [21], and PIASy/UBC9 [22] with either mouse or human GLIS3 protein. SUFU interaction inhibits GLIS3 polyubiquitination by the E3 ubiquitin ligase CUL3. The GLIS3 amino acids that are interacted with or modified are indicated.
Despite these known interactions, there is still a significant gap in our knowledge about GLIS3 protein functions. For instance, while the C-terminal region of GLIS3 has been shown to stimulate its transcriptional activity, the proteins which interact with this domain remain undiscovered. Likewise, the GLIS3 protein is highly phosphorylated, in both N- and C-terminal domains of the protein, yet the role of these phosphorylation sites remains a mystery. Finally, GLIS3 shares very high homology within its DNA binding domains with the GLIS1 and GLIS2, allowing them to bind to similar if not identical DNA sequences. Glis3 is also frequently co-expressed in several cell types with Glis1 and Glis2. Although their DNA binding domains are highly conserved and their expression patterns regularly overlap, the phenotype of Glis3 knockout mice is quite distinct of that of Glis1 and Glis2 knockout mice. This leads to unanswered questions of how GLIS3 protein is specifically recruited to the promoter and regulatory regions of its target genes and how the distinct binding of these proteins to target genes is coordinated.
3. Early Characterization of the GLIS3 Knockout Mice and Humans
Examination of Glis3 expression in mouse development revealed that it was first expressed in the early notochord, followed by expression in various neural progenitor cells [19]. Glis3 mRNA was also detected in a variety of adult tissues, including brain, thymus, lung, kidney, uterus, skeletal muscle, pancreas, liver, and ovary. Global knockout mice were generated similarly by three different labs [23,24,25], which all reported similar pancreatic phenotypes: Hyperglycemia within the first week of life, reduced insulin expression, and early lethality (presumably due to hyperglycemia). This early characterization established that, while Glis3 is expressed in a variety of tissues, it is likely playing a central role in the development of pancreatic β-cells.
Simultaneously, while characterization of Glis3’s role in the mouse provided some phenotypic information, other studies have been published showing that humans with deletions in the GLIS3 gene also had very similar phenotypes. Affected individuals were identified as suffering from permanent neonatal diabetes and congenital hypothyroidism (NDH), as well as facial dysmorphology [26]. Some of the affected individuals also suffered from congenital glaucoma, hepatic fibrosis, and polycystic kidneys [27]. Genetic sequencing of these individuals identified various frame shift and point mutations, and deletions in GLIS3 that were likely responsible for the observed phenotypes [27]. Additional studies have since reinforced this linkage [28,29,30,31,32,33], firmly connecting GLIS3 to diabetes, hypothyroidism, polycystic kidney disease, as well as a host of additional phenotypes. GLIS3’s connection to diabetes has further been reinforced by genome wide association studies (GWAS), which we have previously reviewed [16,18]. Single Nucleotide Polymorphisms (SNPs) within GLIS3 have been linked to both Type 1 and Type 2 diabetes, as well as gestational diabetes and decreased β-cell function. Interestingly, GLIS3 is one of only a small number of genes that have been linked to both Type 1 and Type 2 diabetes. Taken together, these studies highlight the importance of understanding GLIS3’s role in pancreas development, and how it may differ from other genes linked to diabetes.
4. Early Pancreas Development and Glis3 Expression in Mice
Pancreas development begins with the outgrowth of the foregut endoderm into a dorsal and ventral pancreatic bud around embryonic day 9.5 (e9.5) in mice [34,35]. Pancreas development is generally divided into two stages: a primary transition, during which time the pancreatic epithelium proliferates and undergoes extensive branching resulting in the generation of tip and trunk cells, and the secondary transition, in which via distinct differentiation pathways the three main lineages that make up the mature pancreas are generated [36]. The trunk domain is made up of bipotent progenitor cells, which differentiate into ductal and endocrine cells, and tip domain cells, which primarily form acinar cells, although this domain also contains multipotent progenitor cells capable of producing all pancreatic cell types [36]. While a few glucagon+ and insulin+ cells are observed during the primary transition [37], endocrine cell development primarily occurs during the secondary transition, starting at around e13.5 [38]. Endocrine progenitor cells de-laminate from the bipotent trunk domain, then differentiate into the five different cell types that comprise the islet: α-, β-, δ-, ε-, and pp-cells.
Differentiation of the pancreas has largely been characterized by the stepwise expression of a variety of transcription factors (for full reviews, see [14,35,39,40]). Early dorsal and ventral pancreatic buds are marked by expression of the transcription factor Pdx1 and Ptf1a [41,42,43]. Deletion of Pdx1 results in pancreatic agenesis in mice [41,44], and a similar phenotype was observed in human patients with mutations in PDX1 [45,46,47,48,49]. Similarly, Ptf1a has early expression in the pancreas beginning at e9.5 and is a marker of the multipotent progenitor population within the pancreas, while also playing a role in the acinar cells [50,51]. Deletion of Ptf1a also results in pancreatic agenesis similar to Pdx1 [43]. These two factors mark some of the earliest markers of pancreatic development, and as one might expect, their deletion is detrimental to the early formation of the pancreas.
Additional transcription factors are also expressed early in the primary transition, and like Pdx1 and Ptf1a, their expression changes as the pancreas forms its branched structure of tip and trunk domains [15]. For example, Sox9 is first expressed at low levels at e10.5 in multi-potent progenitor cells (MPCs) [52], and its expression remains high in bipotent progenitors prior to being restricted to ductal cells. Hnf6 and Hnf1b both follow similar patterns of expression in the bipotent progenitor cells, followed by restriction to the ductal lineage later in development, while Foxa2 is expressed earlier than Hnf6 and Hnf1b and is maintained in all 3 lineages. As may be expected based on their expression pattern, deletion of Sox9, Hnf6, or Hnf1b results in a form of pancreatic hypoplasia, or a general loss of pancreatic cells [53,54,55,56,57]. Correspondingly, deletions of HNF1B result in a similar phenotype in humans [58]. Deletion of Foxa2 produces even earlier disruption in notochord development [59,60,61].
Glis3 mRNA expression was first detected at e11.5 in both the dorsal and ventral pancreas [24], but expression of GLIS3 protein was only detected starting at e13.5 [62], indicating it likely does not play a role in early pancreas development. Indeed, analysis of Glis3 global knockout mice failed to detect any effect on overall pancreas morphology or acinar cell development [24], consistent with its lack of expression in early pancreatic progenitor cells.
5. Glis3’s Role in the Secondary Transition and Ductal/Endocrine Lineage Determinations
The secondary transition marks a period of differentiation for acinar, ductal, and endocrine cells. While acinar cells differentiate from the “tip” region of the developing pancreatic branches, the trunk domain is composed of bipotent progenitor cells that can differentiate into either ductal or endocrine lineages. Endocrine progenitor cells are distinguished primarily by their high, transient expression of Ngn3, whereas ductal progenitors express Hes1. A form of lateral inhibition has been suggested to drive these fate decisions, as Ngn3 has been linked to upregulation of Notch signaling, which in turn upregulates Hes1, which itself inhibits Ngn3 [63,64]. This is likely only one mechanism involved in making the ductal/endocrine decision, as many HES1+ cells have been observed lacking neighboring NGN3+ cells.
PDX1+, SOX9+, NKX6.1+ bipotent progenitor cells represent the first stage where GLIS3 protein could be detected in a GFP knockin mouse expressing a fusion GLIS3-GFP protein [62]. Glis3 expression is maintained in subsequent differentiation into both the productal (HNF6+, SOX9+) and proendocrine (NGN3+) cells (Figure 2). This distinguishes Glis3 from many of the other transcription factors expressed during this period. While many are expressed during the bipotent progenitor stage, expression is often limited to either the ductal or endocrine lineage. This expression restriction presumably helps drive the differentiation process, as knockouts for many of these factors leads to impairment of the subsequent cell type differentiation. Interestingly, in Glis3 knockout mice, the endocrine lineage is dramatically affected, whereas the ductal lineage does not appear to be affected prior to duct formation [24]. This indicates that Glis3 likely does not play a role in lineage decision-making in the bipotent progenitor cells, but instead plays important roles during or after allocation to the ductal or endocrine lineages.
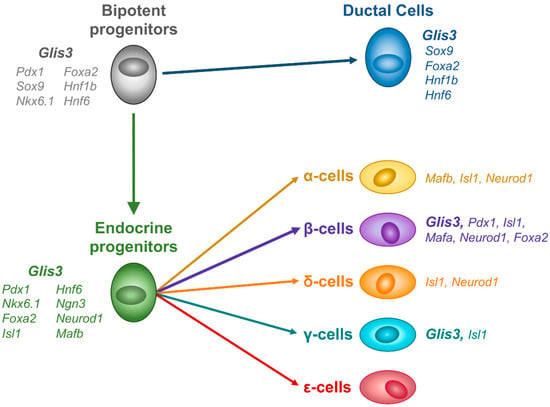
Figure 2.
GLIS3 expression during mouse endocrine development. GLIS3 protein is first detected in bipotent progenitor cells, and remains expressed in both ductal and endocrine lineages, before becoming restricted to β- and γ-cells.
As mentioned, bipotent progenitor cells commit to the endocrine lineage through their expression of Ngn3, Isl1, and Neurod1, as well as several other factors. Ngn3 is a transient marker of endocrine progenitor cells whose expression is mostly lost in mature endocrine cells [38,65], although there is evidence that some Ngn3 expression is required for postnatal β-cells [66]. Isl1 and Neurod1 expression is maintained during the differentiation of endocrine progenitors into α-, β-, δ-, and pp-cells [67,68]. Interestingly, Neurod1 is also expressed earlier in the small number of glucagon+ cells present in the primary transition [68], although its function in these cells is unclear. Consistent with transcription factors expressed during the primary transition, transcription factors expressed predominantly in the endocrine lineage during the secondary transition play a critical role in the regulation of endocrine cell differentiation. Deletion of Ngn3 or Neurod1 results in similar phenotypes, with postnatal pancreas lacking endocrine cells and mice dying of hyperglycemia due to a lack of insulin [65,68]. Pancreas-specific deletion of Isl1 produces a similar phenotype [69], although global Isl1 knockouts die earlier due to heart defects.
The phenotype of Glis3 mice is therefore most similar to that of other transcription factors controlling the endocrine lineage. Glis3 global knockouts display decreased gene expression and staining for all endocrine hormones, and pups die within the first 10 days, likely due to hyperglycemia [23,24,25]. One of the genes that is decreased in Glis3 knockout embryonic pancreas is Ngn3, providing a potential mechanism for the decrease in endocrine cell number. Conditional knockout of Glis3 using a pancreas specific (Pdx1-cre) produced a different phenotype, with islets present well after birth, and α/δ- cells appearing relatively unaffected [70]. This could potentially be due to a slightly later deletion of Glis3 allowing for more Ngn3 expression during development. Alternatively, as the Pdx1-cre line is known to be mosaic (deletion efficiency ranged from about 50% to about 80%), it is possible that a small but significant number of GLIS3+ cells persisted during embryonic development, allowing for the establishment of a sufficient endocrine progenitor population. A more in-depth analysis of GLIS3’s role during endocrine development could help distinguish what role it plays in guiding this process.
6. Links between Glis3 and β-Cell Maturation
Following embryonic and early postnatal development, β-cells within pancreatic islets undergo a still-poorly defined process known as maturation [71]. This occurs roughly around weaning in mice, when pups transition from a primarily milk-fat based diet to a carbohydrate diet. Mature islets not only secrete more insulin in response to glucose, but more tightly regulate their insulin secretion [72]. This is due to a variety of changes in the metabolism of β-cells (reviewed in [71]). In mice, the expression of two transcription factors, Mafa and Mafb [73], is often used for marking β-cell maturation, where Mafb expression is repressed, while Mafa is exclusively expressed in mature pancreatic β-cells.
Glis3’s expression is maintained in pancreatic β-cells, from the immature to mature state [62]. Similarly, Mafa is expressed starting at around e13.5 in insulin+ cells and its expression is maintained in mature β-cells [73]. Mafa expression appears to be directly regulated by Glis3 within pancreatic islets [70], in line with the observed decreased expression of Mafa in β-cells from Glis3 knockout mice, and rising hyperglycemia. Glis3 also directly regulates Ins2 expression, which is significantly down across all Glis3 knockout models [23,24,25,70]. This correlates well with what is seen in humans (discussed in the next section), where GLIS3 appears to be a critical regulator of INS expression [74].
The phenotypes of Mafa and Mafb knockout mice are different from that of the transcription factors mentioned in the previous section (Isl1, Neurod1, and Ngn3). Mafa knockout mice produce insulin+ β-cells but exhibit impaired insulin secretion in response to glucose challenge as early as three to four weeks postnatally [75,76,77]. While islet β-cell mass is modestly reduced in these mice, defects in glucose tolerance are thought to be driven primarily by defects in insulin secretion [76,77]. Pancreas-specific Mafb knockout mice, unlike Mafa knockout mice, appear to have normal glucose clearance in 3-week glucose tolerance tests [78]. In addition, unlike Mafa knockout mice, Mafb knockout mice exhibit an embryonic phenotype, with reduced numbers of α- and β-cells during prenatal development. Mafa/Mafb double knockouts die shortly after birth, presumably due to hyperglycemia from a lack of islet β-cells [78]. Glis3 potentially plays a role in directly regulating both Mafa and Mafb expression, as it binds to a presumptive enhancer and promoter region, respectively, and their expression is downregulated in Glis3 pancreas-specific knockouts [70]. Thus, Glis3 may act upstream of both Mafa and Mafb during β-cell development and affect β-cell maturation via their regulation.
Research over the past decade has suggested that not only is the activation of many genes required for β-cell differentiation and maturation, but also the repression of certain genes. These latter genes have been termed “disallowed” genes, in that their downregulation is correlated with β-cell identity and function [79,80]. These genes include Acot7, Cox5a, Fam59a, Gas6, Itih5, Ldha, Lmo4, Mgst1, Nfib, Pdgfra, Plec1, Rpl36, Tgm2, Tst, and Zdhhc9, which are upregulated in Type 2 diabetic islets [80]. Other studies have identified a de-differentiation pathway in response to extreme cellular stress, which involves the upregulation of genes expressed primarily during β-cell development and subsequently silenced, such as Ngn3, Oct4, Nanog, and L-myc [81]. Of note, none of these genes were upregulated in Glis3 knockout mice, highlighting the unique role that Glis3 is likely playing in guiding β-cell maturation and function [70].
7. GLIS3 in Human β-Cell Development
Pancreas development in the human seems to largely follow a similar pattern to that of mice, with some key differences [82]. One key difference between mice and humans is the apparent lack of a first wave of INS+ or GCG+ cells observed early in mouse pancreas development [83]. This is possibly due to subtle delays in human pancreas development compared to mice, preventing early differentiation of endocrine cells. Additionally, human endocrine progenitor cells lack the expression of NKX2.2, a transcription factor that is critical for beta cell development in mice [84]. Unfortunately, due to a lack of available antibodies, we do not know the expression of GLIS3 during human pancreatic development. It is therefore possible that GLIS3 expression and function in human pancreatic development differs somewhat from that of its role in mouse described above, although humans with deletions in GLIS3 develop neonatal diabetes similar to mice due to a lack of insulin (reviewed in [16]).
A useful tool in studying human pancreatic development has been the differentiation of human embryonic stem cells (hESCs). A significant amount of research has been devoted to the differentiation of human pancreatic β-cells from hESCs, with the hope of developing a potential therapy for people with Type 1 Diabetes [85]. While this differentiation does not exactly mimic human pancreatic development, many of the differentiation stages obtained do express the appropriate marker genes similar to what has been observed in humans. The advent of CRISPR technology and its use in hESCs allows for the study of many of the pancreatic genes identified in mice to be studied in humans [86,87]. Disruption of the GLIS3 gene function by deletions within the DNA binding domain revealed that, in the absence of a functional copy of GLIS3, hESCs were able to differentiate into a similar number of PDX1+ and C-PEPTIDE+ cells as normal hESCs [87]. Defects were observed in PDX1, RFX6, and NGN3 disrupted hESCs, suggesting that in humans GLIS3 may play a later role in human β-cell development than in mice. Interestingly, a subsequent study using a CRISPR knockout of GLIS3 function saw a reduction in INS+ cells, as well as reductions in the expression of several critical transcription factors, such as PDX1, MAFA, NKX6.1, and NEUROD1 [88]. The authors attributed this phenotype to an increase in cell death due to activation of the TGFβ pathway, a finding not previously observed in mice [70], but supported by a study using cell lines [89].
Of note, the differentiation protocols initially used by Zhu et al. showed minimal upregulation of GLIS3, although their protocol produced largely INS+GCG+ cells [87]. The subsequent differentiation protocol used by Amin et al. produced significantly more INS+GCG− cells and saw a greater increase in GLIS3 expression [88]. This could suggest that, in humans, GLIS3 functions as a crucial regulator at a relatively later stage of β-cell differentiation or maturation. Additionally, GLIS3 protein seems to coordinate this function with other transcription factors via co-binding of genomic loci, as has been reported for ISL1 and LDB1 [70,90]. Similar results have been observed in human islets, with GLIS3 binding to the human INS promoter region together with other transcription factors [74]. Not only does GLIS3 appear to bind and activate the INS promoter, but it appears to be required for binding of PDX1 and NEUROD1 to the INS promoter as well [74]. This evidence further supports a model of β-cell transcription factor co-binding and coordination where multiple transcription factors bind to overlapping regions and are required for proper gene regulation.
Studies in a rat insulinoma cell line (INS-1E cells) and dissociated human islets have indicated that GLIS3 may play a role in preventing β-cell apoptosis, including in response to cytokine treatment [89]. The proposed mechanism of action involves GLIS3 regulation of SRSF6 (also known as SRP55), a splicing factor that is down-regulated in human islets upon cytokine treatment [91], and is involved in the splicing of a variety of critical genes in a human β-cell line [92]. By regulating the alternative splicing of Bim, down-regulation of SRSF6 promotes apoptosis in β-cells by increasing the generation of the proapoptotic isoform BIM S [93,94,95]. Alternatively, GLIS3 has also been proposed to regulate apoptosis through the TGF-β pathway during differentiation of hESCs to β-like cells [88]. Unfortunately, these studies offer confusing and sometimes conflicting results for GLIS3 function in humans, as we have previously highlighted [16]. GLIS3 regulation of SRSF6 does not appear to be transcriptional, as hESCs and Glis3 knockout mice show normal expression of SRSF6 mRNA [70,88]. This highlights one of the difficulties of the human models that currently exist: Apoptosis is dramatically higher in cell lines and in vitro culture systems than has been observed in vivo.
A separate study also linked Glis3 to regulation of the Manf gene, an anti-apoptotic gene upregulated in β-cells during unfolded protein stress response (UPR) [96]. However, this study did not examine apoptosis in their model (Glis3 heterozygous mice undergoing β-cell specific UPR), but instead relied on previous reports from cell lines [89]. Manf was not downregulated in Glis3 knockout mice, but GLIS3 protein does appear to bind to a regulatory region within the mouse Manf gene, alongside other islet-enriched transcription factors [70]. This raises the possibility that GLIS3 regulation of certain genes may be context specific and be dependent on signaling pathways activated by certain conditions, such as cellular stress or cytokine exposure. Clearly, more work is needed to address whether GLIS3 is linked to apoptosis in humans or mice in vivo and under what circumstances.
8. Newly Identified Human Mutations within GLIS3
GLIS3 has been linked to both Type 1 and Type 2 diabetes, gestational diabetes, and β-cell function in many GWAS (previously reviewed in [16]). Many of the SNPs identified in these studies reside within the first few introns of the GLIS3 gene, likely regulating GLIS3 expression through enhancer elements. Additional reports identified a link between several deletions and mutations within GLIS3 and neonatal diabetes with congenital hypothyroidism (NDH) syndrome (reviewed in [18]). Recently, novel mutations within the coding region of GLIS3 gene have been identified. One such novel homozygous mutation created a premature stop codon within the C-terminus of GLIS3 (c.2392C>T; p.Gln798Ter) and caused a syndrome characterized by neonatal diabetes, congenital hypothyroidism, congenital glaucoma, and cystic kidney disease [97]. The premature stop codon lies within the known transactivation of the protein (see Figure 1B), thus confirming the functional conservation of the domain from mice [98]. Separately, a novel heterozygous mutation was identified in the N-terminal region (c.589G>T; Asp197Tyr) of GLIS3, in a Turkish patient diagnosed with maturity onset diabetes of the young (MODY) [99]. However, this N-terminal mutation (Asp197Tyr) lies outside any of the previously identified domains of GLIS3. This mutation could therefore provide interesting insights into the previously unknown functional domains within GLIS3. Hopefully, as genetic sequencing of patients increases in both frequency and thoroughness, novel mutations could allow us not only to extend our knowledge of GLIS3 function in humans but may also point us toward which regions of the gene to explore in mice and in vitro human models.
9. Conclusions
A significant body of research now exists tying GLIS3 to the regulation of pancreatic β-cells and diabetes. GLIS3 deficiency significantly reduces the generation of endocrine cells, particularly β-cells, causing severe hyperglycemia in both mice and humans. GLIS3’s expression pattern and phenotype within the pancreas of Glis3 knockout mice most closely resembles that of transcription factors that play a role in endocrine differentiation, such as NGN3, ISL1, and NEUROD1. However, GLIS3 appears to play a distinct role from these factors in that, while total endocrine cell numbers are reduced, β- and γ-cell numbers are more severely affected in Glis3 knockout mice. Moreover, GLIS3 appears to be necessary for the binding of additional transcription factors to the human INS promoter [74]. And GLIS3 is one of only a few genes that have been linked through GWAS to both Type 1 and Type 2 diabetes.
Significant questions remain as to the exact role of GLIS3 in the human β-cells and how closely it mimics the function in mice. For instance, Ngn3 expression has been reported to be downregulated in global Glis3 knockout mice at e13.5 and e15.5 [24,25]. GLIS3 binding does overlap with that of PDX1 in a region downstream from the Ngn3 gene consistent with the concept that it is directly regulated by GLIS3 [25]. However, the differentiation of human ESCs into β-cells in which GLIS3 was deleted appeared to have no effect, whereas NGN3 deletion has a dramatic effect on β-cell generation. This lack of a GLIS3 phenotype might be attributed to the in vitro and artificial nature of the protocol failing to recapitulate human development or to inherent differences in GLIS3 function between human and mouse. Additional studies using primary human islets might establish a more physiologically relevant assessment of GLIS3 function and provide greater insights into the potential of GLIS3 as a therapeutic target in the management of diabetes.
Funding
This work is supported by the Intramural Research Program of the NIEHS, the NIH Z01-ES-100485.
Acknowledgments
The authors would like to thank Hong Soon Kang and Laura Miller for their contributions to the study of GLIS3 in pancreatic β-cells.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ionescu-Tirgoviste, C.; Gagniuc, P.A.; Gubceac, E.; Mardare, L.; Popescu, I.; Dima, S.; Militaru, M. A 3D map of the islet routes throughout the healthy human pancreas. Sci. Rep. 2015, 5, 14634. [Google Scholar] [CrossRef]
- Brissova, M.; Fowler, M.J.; Nicholson, W.E.; Chu, A.; Hirshberg, B.; Harlan, D.M.; Powers, A.C. Assessment of human pancreatic islet architecture and composition by laser scanning confocal microscopy. J. Histochem. Cytochem. 2005, 53, 1087–1097. [Google Scholar] [CrossRef] [PubMed]
- Murphy, R.; Ellard, S.; Hattersley, A.T. Clinical implications of a molecular genetic classification of monogenic beta-cell diabetes. Nat. Clin. Pract. Endocrinol. Metab. 2008, 4, 200–213. [Google Scholar] [CrossRef]
- Schneider, D.A.; von Herrath, M.G. Potential viral pathogenic mechanism in human type 1 diabetes. Diabetologia 2014, 57, 2009–2018. [Google Scholar] [CrossRef]
- Roivainen, M.; Klingel, K. Virus infections and type 1 diabetes risk. Curr. Diab. Rep. 2010, 10, 350–356. [Google Scholar] [CrossRef] [PubMed]
- Giongo, A.; Gano, K.A.; Crabb, D.B.; Mukherjee, N.; Novelo, L.L.; Casella, G.; Drew, J.C.; Ilonen, J.; Knip, M.; Hyoty, H.; et al. Toward defining the autoimmune microbiome for type 1 diabetes. ISME J. 2011, 5, 82–91. [Google Scholar] [CrossRef]
- Norris, J.M.; Barriga, K.; Klingensmith, G.; Hoffman, M.; Eisenbarth, G.S.; Erlich, H.A.; Rewers, M. Timing of initial cereal exposure in infancy and risk of islet autoimmunity. JAMA 2003, 290, 1713–1720. [Google Scholar] [CrossRef] [PubMed]
- Ziegler, A.G.; Schmid, S.; Huber, D.; Hummel, M.; Bonifacio, E. Early infant feeding and risk of developing type 1 diabetes-associated autoantibodies. JAMA 2003, 290, 1721–1728. [Google Scholar] [CrossRef]
- Chan, J.M.; Rimm, E.B.; Colditz, G.A.; Stampfer, M.J.; Willett, W.C. Obesity, fat distribution, and weight gain as risk factors for clinical diabetes in men. Diabetes Care 1994, 17, 961–969. [Google Scholar] [CrossRef]
- Colditz, G.A.; Willett, W.C.; Rotnitzky, A.; Manson, J.E. Weight gain as a risk factor for clinical diabetes mellitus in women. Ann. Intern. Med. 1995, 122, 481–486. [Google Scholar] [CrossRef]
- Prasad, R.B.; Groop, L. Genetics of type 2 diabetes-pitfalls and possibilities. Genes 2015, 6, 87–123. [Google Scholar] [CrossRef] [PubMed]
- Hattersley, A.T.; Greeley, S.A.W.; Polak, M.; Rubio-Cabezas, O.; Njolstad, P.R.; Mlynarski, W.; Castano, L.; Carlsson, A.; Raile, K.; Chi, D.V.; et al. ISPAD Clinical Practice Consensus Guidelines 2018: The diagnosis and management of monogenic diabetes in children and adolescents. Pediatr. Diabetes 2018, 19, 47–63. [Google Scholar] [CrossRef] [PubMed]
- Jensen, J. Gene regulatory factors in pancreatic development. Dev. Dyn. 2004, 229, 176–200. [Google Scholar] [CrossRef] [PubMed]
- Murtaugh, L.C.; Melton, D.A. Genes, signals, and lineages in pancreas development. Annu. Rev. Cell Dev. Biol. 2003, 19, 71–89. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Law, A.C.; Rajagopal, J.; Anderson, W.J.; Gray, P.A.; Melton, D.A. A multipotent progenitor domain guides pancreatic organogenesis. Dev. Cell 2007, 13, 103–114. [Google Scholar] [CrossRef] [PubMed]
- Scoville, D.W.; Kang, H.S.; Jetten, A.M. Transcription factor GLIS3: Critical roles in thyroid hormone biosynthesis, hypothyroidism, pancreatic beta cells and diabetes. Pharmacol. Ther. 2020, 215, 107632. [Google Scholar] [CrossRef] [PubMed]
- Scoville, D.W.; Kang, H.S.; Jetten, A.M. GLIS1-3: Emerging roles in reprogramming, stem and progenitor cell differentiation and maintenance. Stem Cell Investig. 2017, 4, 80. [Google Scholar] [CrossRef]
- Jetten, A.M. GLIS1-3 transcription factors: Critical roles in the regulation of multiple physiological processes and diseases. Cell Mol. Life Sci. 2018, 75, 3473–3494. [Google Scholar] [CrossRef]
- Kim, Y.S.; Nakanishi, G.; Lewandoski, M.; Jetten, A.M. GLIS3, a novel member of the GLIS subfamily of Kruppel-like zinc finger proteins with repressor and activation functions. Nucleic Acids Res. 2003, 31, 5513–5525. [Google Scholar] [CrossRef]
- ZeRuth, G.T.; Yang, X.P.; Jetten, A.M. Modulation of the transactivation function and stability of Kruppel-like zinc finger protein Gli-similar 3 (Glis3) by Suppressor of Fused. J. Biol. Chem. 2011, 286, 22077–22089. [Google Scholar] [CrossRef]
- ZeRuth, G.T.; Williams, J.G.; Cole, Y.C.; Jetten, A.M. HECT E3 Ubiquitin Ligase Itch Functions as a Novel Negative Regulator of Gli-Similar 3 (Glis3) Transcriptional Activity. PLoS ONE 2015, 10, e0131303. [Google Scholar] [CrossRef]
- Hoard, T.M.; Yang, X.P.; Jetten, A.M.; ZeRuth, G.T. PIAS-family proteins negatively regulate Glis3 transactivation function through SUMO modification in pancreatic beta cells. Heliyon 2018, 4, e00709. [Google Scholar] [CrossRef]
- Watanabe, N.; Hiramatsu, K.; Miyamoto, R.; Yasuda, K.; Suzuki, N.; Oshima, N.; Kiyonari, H.; Shiba, D.; Nishio, S.; Mochizuki, T.; et al. A murine model of neonatal diabetes mellitus in Glis3-deficient mice. FEBS Lett. 2009, 583, 2108–2113. [Google Scholar] [CrossRef]
- Kang, H.S.; Kim, Y.S.; ZeRuth, G.; Beak, J.Y.; Gerrish, K.; Kilic, G.; Sosa-Pineda, B.; Jensen, J.; Pierreux, C.E.; Lemaigre, F.P.; et al. Transcription factor Glis3, a novel critical player in the regulation of pancreatic beta-cell development and insulin gene expression. Mol. Cell. Biol. 2009, 29, 6366–6379. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Chang, B.H.; Yechoor, V.; Chen, W.; Li, L.; Tsai, M.J.; Chan, L. The Kruppel-like zinc finger protein GLIS3 transactivates neurogenin 3 for proper fetal pancreatic islet differentiation in mice. Diabetologia 2011, 54, 2595–2605. [Google Scholar] [CrossRef]
- Taha, D.; Barbar, M.; Kanaan, H.; Williamson Balfe, J. Neonatal diabetes mellitus, congenital hypothyroidism, hepatic fibrosis, polycystic kidneys, and congenital glaucoma: A new autosomal recessive syndrome? Am. J. Med. Genet. A 2003, 122, 269–273. [Google Scholar] [CrossRef] [PubMed]
- Senee, V.; Chelala, C.; Duchatelet, S.; Feng, D.; Blanc, H.; Cossec, J.C.; Charon, C.; Nicolino, M.; Boileau, P.; Cavener, D.R.; et al. Mutations in GLIS3 are responsible for a rare syndrome with neonatal diabetes mellitus and congenital hypothyroidism. Nat. Genet. 2006, 38, 682–687. [Google Scholar] [CrossRef]
- Dimitri, P.; Warner, J.T.; Minton, J.A.; Patch, A.M.; Ellard, S.; Hattersley, A.T.; Barr, S.; Hawkes, D.; Wales, J.K.; Gregory, J.W. Novel GLIS3 mutations demonstrate an extended multisystem phenotype. Eur. J. Endocrinol. 2011, 164, 437–443. [Google Scholar] [CrossRef]
- Dimitri, P.; Habeb, A.M.; Gurbuz, F.; Millward, A.; Wallis, S.; Moussa, K.; Akcay, T.; Taha, D.; Hogue, J.; Slavotinek, A.; et al. Expanding the Clinical Spectrum Associated With GLIS3 Mutations. J. Clin. Endocrinol. Metab. 2015, 100, 1362–1369. [Google Scholar] [CrossRef]
- Dimitri, P.; De Franco, E.; Habeb, A.M.; Gurbuz, F.; Moussa, K.; Taha, D.; Wales, J.K.; Hogue, J.; Slavotinek, A.; Shetty, A.; et al. An emerging, recognizable facial phenotype in association with mutations in GLI-similar 3 (GLIS3). Am. J. Med. Genet. A 2016, 170, 1918–1923. [Google Scholar] [CrossRef]
- Fu, C.; Luo, S.; Long, X.; Li, Y.; She, S.; Hu, X.; Mo, M.; Wang, Z.; Chen, Y.; He, C.; et al. Mutation screening of the GLIS3 gene in a cohort of 592 Chinese patients with congenital hypothyroidism. Clin. Chim. Acta 2018, 476, 38–43. [Google Scholar] [CrossRef]
- Alghamdi, K.A.; Alsaedi, A.B.; Aljasser, A.; Altawil, A.; Kamal, N.M. Extended clinical features associated with novel Glis3 mutation: A case report. BMC Endocr. Disord. 2017, 17, 14. [Google Scholar] [CrossRef]
- Fu, C.; Luo, S.; Zhang, Y.; Fan, X.; D’Gama, A.M.; Zhang, X.; Zheng, H.; Su, J.; Li, C.; Luo, J.; et al. Chromosomal microarray and whole exome sequencing identify genetic causes of congenital hypothyroidism with extra-thyroidal congenital malformations. Clin. Chim. Acta 2019, 489, 103–108. [Google Scholar] [CrossRef]
- Gittes, G.K. Developmental biology of the pancreas: A comprehensive review. Dev. Biol. 2009, 326, 4–35. [Google Scholar] [CrossRef] [PubMed]
- Pan, F.C.; Wright, C. Pancreas organogenesis: From bud to plexus to gland. Dev. Dyn. 2011, 240, 530–565. [Google Scholar] [CrossRef]
- Villasenor, A.; Chong, D.C.; Henkemeyer, M.; Cleaver, O. Epithelial dynamics of pancreatic branching morphogenesis. Development 2010, 137, 4295–4305. [Google Scholar] [CrossRef]
- Herrera, P.L. Adult insulin- and glucagon-producing cells differentiate from two independent cell lineages. Development 2000, 127, 2317–2322. [Google Scholar] [CrossRef] [PubMed]
- Gu, G.; Dubauskaite, J.; Melton, D.A. Direct evidence for the pancreatic lineage: NGN3+ cells are islet progenitors and are distinct from duct progenitors. Development 2002, 129, 2447–2457. [Google Scholar] [CrossRef]
- Doyle, M.J.; Sussel, L. Engineering islets: Lessons from stem cells and embryonic development. Endocrinol. Metab. Clin. N. Am. 2004, 33, 149–162. [Google Scholar] [CrossRef]
- Guney, M.A.; Gannon, M. Pancreas cell fate. Birth Defects Res. C Embryo Today 2009, 87, 232–248. [Google Scholar] [CrossRef]
- Offield, M.F.; Jetton, T.L.; Labosky, P.A.; Ray, M.; Stein, R.W.; Magnuson, M.A.; Hogan, B.L.; Wright, C.V. PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development 1996, 122, 983–995. [Google Scholar] [CrossRef] [PubMed]
- Ohlsson, H.; Karlsson, K.; Edlund, T. IPF1, a homeodomain-containing transactivator of the insulin gene. EMBO J. 1993, 12, 4251–4259. [Google Scholar] [CrossRef] [PubMed]
- Kawaguchi, Y.; Cooper, B.; Gannon, M.; Ray, M.; MacDonald, R.J.; Wright, C.V. The role of the transcriptional regulator Ptf1a in converting intestinal to pancreatic progenitors. Nat. Genet. 2002, 32, 128–134. [Google Scholar] [CrossRef]
- Jonsson, J.; Carlsson, L.; Edlund, T.; Edlund, H. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature 1994, 371, 606–609. [Google Scholar] [CrossRef] [PubMed]
- Stoffers, D.A.; Zinkin, N.T.; Stanojevic, V.; Clarke, W.L.; Habener, J.F. Pancreatic agenesis attributable to a single nucleotide deletion in the human IPF1 gene coding sequence. Nat. Genet. 1997, 15, 106–110. [Google Scholar] [CrossRef]
- Schwitzgebel, V.M.; Mamin, A.; Brun, T.; Ritz-Laser, B.; Zaiko, M.; Maret, A.; Jornayvaz, F.R.; Theintz, G.E.; Michielin, O.; Melloul, D.; et al. Agenesis of human pancreas due to decreased half-life of insulin promoter factor 1. J. Clin. Endocrinol. Metab. 2003, 88, 4398–4406. [Google Scholar] [CrossRef]
- Thomas, I.H.; Saini, N.K.; Adhikari, A.; Lee, J.M.; Kasa-Vubu, J.Z.; Vazquez, D.M.; Menon, R.K.; Chen, M.; Fajans, S.S. Neonatal diabetes mellitus with pancreatic agenesis in an infant with homozygous IPF-1 Pro63fsX60 mutation. Pediatr. Diabetes 2009, 10, 492–496. [Google Scholar] [CrossRef]
- Nicolino, M.; Claiborn, K.C.; Senee, V.; Boland, A.; Stoffers, D.A.; Julier, C. A novel hypomorphic PDX1 mutation responsible for permanent neonatal diabetes with subclinical exocrine deficiency. Diabetes 2010, 59, 733–740. [Google Scholar] [CrossRef]
- Caetano, L.A.; Santana, L.S.; Costa-Riquetto, A.D.; Lerario, A.M.; Nery, M.; Nogueira, G.F.; Ortega, C.D.; Rocha, M.S.; Jorge, A.A.L.; Teles, M.G. PDX1 -MODY and dorsal pancreatic agenesis: New phenotype of a rare disease. Clin. Genet. 2018, 93, 382–386. [Google Scholar] [CrossRef] [PubMed]
- Rose, S.D.; Kruse, F.; Swift, G.H.; MacDonald, R.J.; Hammer, R.E. A single element of the elastase I enhancer is sufficient to direct transcription selectively to the pancreas and gut. Mol. Cell. Biol. 1994, 14, 2048–2057. [Google Scholar] [CrossRef]
- Krapp, A.; Knofler, M.; Frutiger, S.; Hughes, G.J.; Hagenbuchle, O.; Wellauer, P.K. The p48 DNA-binding subunit of transcription factor PTF1 is a new exocrine pancreas-specific basic helix-loop-helix protein. EMBO J. 1996, 15, 4317–4329. [Google Scholar] [CrossRef] [PubMed]
- Akiyama, H.; Kim, J.E.; Nakashima, K.; Balmes, G.; Iwai, N.; Deng, J.M.; Zhang, Z.; Martin, J.F.; Behringer, R.R.; Nakamura, T.; et al. Osteo-chondroprogenitor cells are derived from Sox9 expressing precursors. Proc. Natl. Acad. Sci. USA 2005, 102, 14665–14670. [Google Scholar] [CrossRef]
- Jacquemin, P.; Lemaigre, F.P.; Rousseau, G.G. The Onecut transcription factor HNF-6 (OC-1) is required for timely specification of the pancreas and acts upstream of Pdx-1 in the specification cascade. Dev. Biol. 2003, 258, 105–116. [Google Scholar] [CrossRef]
- Pierreux, C.E.; Poll, A.V.; Kemp, C.R.; Clotman, F.; Maestro, M.A.; Cordi, S.; Ferrer, J.; Leyns, L.; Rousseau, G.G.; Lemaigre, F.P. The transcription factor hepatocyte nuclear factor-6 controls the development of pancreatic ducts in the mouse. Gastroenterology 2006, 130, 532–541. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Ables, E.T.; Pope, C.F.; Washington, M.K.; Hipkens, S.; Means, A.L.; Path, G.; Seufert, J.; Costa, R.H.; Leiter, A.B.; et al. Multiple, temporal-specific roles for HNF6 in pancreatic endocrine and ductal differentiation. Mech. Dev. 2009, 126, 958–973. [Google Scholar] [CrossRef]
- Haumaitre, C.; Barbacci, E.; Jenny, M.; Ott, M.O.; Gradwohl, G.; Cereghini, S. Lack of TCF2/vHNF1 in mice leads to pancreas agenesis. Proc. Natl. Acad. Sci. USA 2005, 102, 1490–1495. [Google Scholar] [CrossRef]
- Seymour, P.A.; Freude, K.K.; Tran, M.N.; Mayes, E.E.; Jensen, J.; Kist, R.; Scherer, G.; Sander, M. SOX9 is required for maintenance of the pancreatic progenitor cell pool. Proc. Natl. Acad. Sci. USA 2007, 104, 1865–1870. [Google Scholar] [CrossRef]
- Haumaitre, C.; Fabre, M.; Cormier, S.; Baumann, C.; Delezoide, A.L.; Cereghini, S. Severe pancreas hypoplasia and multicystic renal dysplasia in two human fetuses carrying novel HNF1beta/MODY5 mutations. Hum. Mol. Genet. 2006, 15, 2363–2375. [Google Scholar] [CrossRef]
- Ang, S.L.; Rossant, J. HNF-3 beta is essential for node and notochord formation in mouse development. Cell 1994, 78, 561–574. [Google Scholar] [CrossRef]
- Dufort, D.; Schwartz, L.; Harpal, K.; Rossant, J. The transcription factor HNF3beta is required in visceral endoderm for normal primitive streak morphogenesis. Development 1998, 125, 3015–3025. [Google Scholar] [CrossRef]
- Weinstein, D.C.; Ruiz i Altaba, A.; Chen, W.S.; Hoodless, P.; Prezioso, V.R.; Jessell, T.M.; Darnell, J.E., Jr. The winged-helix transcription factor HNF-3 beta is required for notochord development in the mouse embryo. Cell 1994, 78, 575–588. [Google Scholar] [CrossRef]
- Kang, H.S.; Takeda, Y.; Jeon, K.; Jetten, A.M. The Spatiotemporal Pattern of Glis3 Expression Indicates a Regulatory Function in Bipotent and Endocrine Progenitors during Early Pancreatic Development and in Beta, PP and Ductal Cells. PLoS ONE 2016, 11, e0157138. [Google Scholar] [CrossRef] [PubMed]
- Norgaard, G.A.; Jensen, J.N.; Jensen, J. FGF10 signaling maintains the pancreatic progenitor cell state revealing a novel role of Notch in organ development. Dev. Biol. 2003, 264, 323–338. [Google Scholar] [CrossRef] [PubMed]
- Hart, A.; Papadopoulou, S.; Edlund, H. Fgf10 maintains notch activation, stimulates proliferation, and blocks differentiation of pancreatic epithelial cells. Dev. Dyn. 2003, 228, 185–193. [Google Scholar] [CrossRef]
- Gradwohl, G.; Dierich, A.; LeMeur, M.; Guillemot, F. neurogenin3 is required for the development of the four endocrine cell lineages of the pancreas. Proc. Natl. Acad. Sci. USA 2000, 97, 1607–1611. [Google Scholar] [CrossRef]
- Wang, S.; Jensen, J.N.; Seymour, P.A.; Hsu, W.; Dor, Y.; Sander, M.; Magnuson, M.A.; Serup, P.; Gu, G. Sustained Neurog3 expression in hormone-expressing islet cells is required for endocrine maturation and function. Proc. Natl. Acad. Sci. USA 2009, 106, 9715–9720. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Asa, S.L.; Drucker, D.J. Islet cell and extrapancreatic expression of the LIM domain homeobox gene isl-1. Mol. Endocrinol. 1991, 5, 1633–1641. [Google Scholar] [CrossRef][Green Version]
- Naya, F.J.; Huang, H.P.; Qiu, Y.; Mutoh, H.; DeMayo, F.J.; Leiter, A.B.; Tsai, M.J. Diabetes, defective pancreatic morphogenesis, and abnormal enteroendocrine differentiation in BETA2/neuroD-deficient mice. Genes Dev. 1997, 11, 2323–2334. [Google Scholar] [CrossRef]
- Du, A.; Hunter, C.S.; Murray, J.; Noble, D.; Cai, C.L.; Evans, S.M.; Stein, R.; May, C.L. Islet-1 is required for the maturation, proliferation, and survival of the endocrine pancreas. Diabetes 2009, 58, 2059–2069. [Google Scholar] [CrossRef] [PubMed]
- Scoville, D.; Lichti-Kaiser, K.; Grimm, S.; Jetten, A. GLIS3 binds pancreatic beta cell regulatory regions alongside other islet transcription factors. J. Endocrinol. 2019, 243, 1–14. [Google Scholar] [CrossRef]
- Wortham, M.; Sander, M. Transcriptional mechanisms of pancreatic beta-cell maturation and functional adaptation. Trends Endocrinol. Metab. 2021, 32, 474–487. [Google Scholar] [CrossRef] [PubMed]
- Blum, B.; Hrvatin, S.; Schuetz, C.; Bonal, C.; Rezania, A.; Melton, D.A. Functional beta-cell maturation is marked by an increased glucose threshold and by expression of urocortin 3. Nat. Biotechnol. 2012, 30, 261–264. [Google Scholar] [CrossRef]
- Hang, Y.; Stein, R. MafA and MafB activity in pancreatic beta cells. Trends Endocrinol. Metab. 2011, 22, 364–373. [Google Scholar] [CrossRef]
- Akerman, I.; Maestro, M.A.; De Franco, E.; Grau, V.; Flanagan, S.; Garcia-Hurtado, J.; Mittler, G.; Ravassard, P.; Piemonti, L.; Ellard, S.; et al. Neonatal diabetes mutations disrupt a chromatin pioneering function that activates the human insulin gene. Cell Rep. 2021, 35, 108981. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Moriguchi, T.; Kajihara, M.; Esaki, R.; Harada, A.; Shimohata, H.; Oishi, H.; Hamada, M.; Morito, N.; Hasegawa, K.; et al. MafA is a key regulator of glucose-stimulated insulin secretion. Mol. Cell. Biol. 2005, 25, 4969–4976. [Google Scholar] [CrossRef]
- Hang, Y.; Yamamoto, T.; Benninger, R.K.; Brissova, M.; Guo, M.; Bush, W.; Piston, D.W.; Powers, A.C.; Magnuson, M.; Thurmond, D.C.; et al. The MafA transcription factor becomes essential to islet beta-cells soon after birth. Diabetes 2014, 63, 1994–2005. [Google Scholar] [CrossRef] [PubMed]
- Scoville, D.W.; Cyphert, H.A.; Liao, L.; Xu, J.; Reynolds, A.; Guo, S.; Stein, R. MLL3 and MLL4 Methyltransferases Bind to the MAFA and MAFB Transcription Factors to Regulate Islet beta-Cell Function. Diabetes 2015, 64, 3772–3783. [Google Scholar] [CrossRef]
- Conrad, E.; Dai, C.; Spaeth, J.; Guo, M.; Cyphert, H.A.; Scoville, D.; Carroll, J.; Yu, W.M.; Goodrich, L.V.; Harlan, D.M.; et al. The MAFB transcription factor impacts islet alpha-cell function in rodents and represents a unique signature of primate islet beta-cells. Am. J. Physiol. Endocrinol. Metab. 2016, 310, 91–102. [Google Scholar] [CrossRef]
- Rutter, G.A.; Pullen, T.J.; Hodson, D.J.; Martinez-Sanchez, A. Pancreatic beta-cell identity, glucose sensing and the control of insulin secretion. Biochem. J. 2015, 466, 203–218. [Google Scholar] [CrossRef] [PubMed]
- Pullen, T.J.; Rutter, G.A. When less is more: The forbidden fruits of gene repression in the adult beta-cell. Diabetes Obes. Metab. 2013, 15, 503–512. [Google Scholar] [CrossRef]
- Talchai, C.; Xuan, S.; Lin, H.V.; Sussel, L.; Accili, D. Pancreatic beta cell dedifferentiation as a mechanism of diabetic beta cell failure. Cell 2012, 150, 1223–1234. [Google Scholar] [CrossRef]
- Jennings, R.E.; Berry, A.A.; Strutt, J.P.; Gerrard, D.T.; Hanley, N.A. Human pancreas development. Development 2015, 142, 3126–3137. [Google Scholar] [CrossRef]
- Jennings, R.E.; Berry, A.A.; Kirkwood-Wilson, R.; Roberts, N.A.; Hearn, T.; Salisbury, R.J.; Blaylock, J.; Piper Hanley, K.; Hanley, N.A. Development of the human pancreas from foregut to endocrine commitment. Diabetes 2013, 62, 3514–3522. [Google Scholar] [CrossRef]
- Sussel, L.; Kalamaras, J.; Hartigan-O’Connor, D.J.; Meneses, J.J.; Pedersen, R.A.; Rubenstein, J.L.; German, M.S. Mice lacking the homeodomain transcription factor Nkx2.2 have diabetes due to arrested differentiation of pancreatic beta cells. Development 1998, 125, 2213–2221. [Google Scholar] [CrossRef]
- Pagliuca, F.W.; Melton, D.A. How to make a functional beta-cell. Development 2013, 140, 2472–2483. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, F.; Zhu, Z.; Shi, Z.D.; Lelli, K.; Verma, N.; Li, Q.V.; Huangfu, D. An iCRISPR platform for rapid, multiplexable, and inducible genome editing in human pluripotent stem cells. Cell Stem Cell 2014, 15, 215–226. [Google Scholar] [CrossRef]
- Zhu, Z.; Li, Q.V.; Lee, K.; Rosen, B.P.; Gonzalez, F.; Soh, C.L.; Huangfu, D. Genome Editing of Lineage Determinants in Human Pluripotent Stem Cells Reveals Mechanisms of Pancreatic Development and Diabetes. Cell Stem Cell 2016, 18, 755–768. [Google Scholar] [CrossRef] [PubMed]
- Amin, S.; Cook, B.; Zhou, T.; Ghazizadeh, Z.; Lis, R.; Zhang, T.; Khalaj, M.; Crespo, M.; Perera, M.; Xiang, J.Z.; et al. Discovery of a drug candidate for GLIS3-associated diabetes. Nat. Commun. 2018, 9, 2681. [Google Scholar] [CrossRef]
- Nogueira, T.C.; Paula, F.M.; Villate, O.; Colli, M.L.; Moura, R.F.; Cunha, D.A.; Marselli, L.; Marchetti, P.; Cnop, M.; Julier, C.; et al. GLIS3, a susceptibility gene for type 1 and type 2 diabetes, modulates pancreatic beta cell apoptosis via regulation of a splice variant of the BH3-only protein Bim. PLoS Genet. 2013, 9, e1003532. [Google Scholar] [CrossRef]
- Ediger, B.N.; Lim, H.W.; Juliana, C.; Groff, D.N.; Williams, L.T.; Dominguez, G.; Liu, J.H.; Taylor, B.L.; Walp, E.R.; Kameswaran, V.; et al. LIM domain-binding 1 maintains the terminally differentiated state of pancreatic beta cells. J. Clin. Investig. 2017, 127, 215–229. [Google Scholar] [CrossRef] [PubMed]
- Eizirik, D.L.; Sammeth, M.; Bouckenooghe, T.; Bottu, G.; Sisino, G.; Igoillo-Esteve, M.; Ortis, F.; Santin, I.; Colli, M.L.; Barthson, J.; et al. The human pancreatic islet transcriptome: Expression of candidate genes for type 1 diabetes and the impact of pro-inflammatory cytokines. PLoS Genet. 2012, 8, e1002552. [Google Scholar] [CrossRef] [PubMed]
- Alvelos, M.I.; Bruggemann, M.; Sutandy, F.R.; Juan-Mateu, J.; Colli, M.L.; Busch, A.; Lopes, M.; Castela, A.; Aartsma-Rus, A.; Konig, J.; et al. The RNA-binding profile of the splicing factor SRSF6 in immortalized human pancreatic beta-cells. Life Sci. Alliance 2021, 4, e202000825. [Google Scholar] [CrossRef]
- Santin, I.; Moore, F.; Colli, M.L.; Gurzov, E.N.; Marselli, L.; Marchetti, P.; Eizirik, D.L. PTPN2, a candidate gene for type 1 diabetes, modulates pancreatic beta-cell apoptosis via regulation of the BH3-only protein Bim. Diabetes 2011, 60, 3279–3288. [Google Scholar] [CrossRef]
- Barthson, J.; Germano, C.M.; Moore, F.; Maida, A.; Drucker, D.J.; Marchetti, P.; Gysemans, C.; Mathieu, C.; Nunez, G.; Jurisicova, A.; et al. Cytokines tumor necrosis factor-alpha and interferon-gamma induce pancreatic beta-cell apoptosis through STAT1-mediated Bim protein activation. J. Biol. Chem. 2011, 286, 39632–39643. [Google Scholar] [CrossRef]
- Moore, F.; Santin, I.; Nogueira, T.C.; Gurzov, E.N.; Marselli, L.; Marchetti, P.; Eizirik, D.L. The transcription factor C/EBP delta has anti-apoptotic and anti-inflammatory roles in pancreatic beta cells. PLoS ONE 2012, 7, e31062. [Google Scholar] [CrossRef]
- Dooley, J.; Tian, L.; Schonefeldt, S.; Delghingaro-Augusto, V.; Garcia-Perez, J.E.; Pasciuto, E.; Di Marino, D.; Carr, E.J.; Oskolkov, N.; Lyssenko, V.; et al. Genetic predisposition for beta cell fragility underlies type 1 and type 2 diabetes. Nat. Genet. 2016, 48, 519–527. [Google Scholar] [CrossRef] [PubMed]
- London, S.; De Franco, E.; Elias-Assad, G.; Barhoum, M.N.; Felszer, C.; Paniakov, M.; Weiner, S.A.; Tenenbaum-Rakover, Y. Case Report: Neonatal Diabetes Mellitus Caused by a Novel GLIS3 Mutation in Twins. Front. Endocrinol. 2021, 12, 673755. [Google Scholar] [CrossRef]
- ZeRuth, G.T.; Takeda, Y.; Jetten, A.M. The Kruppel-like protein Gli-similar 3 (Glis3) functions as a key regulator of insulin transcription. Mol. Endocrinol. 2013, 27, 1692–1705. [Google Scholar] [CrossRef] [PubMed]
- Yalcintepe, S.; Ozguc Comlek, F.; Gurkan, H.; Demir, S.; Atli, E.I.; Atli, E.; Eker, D.; Tutunculer Kokenli, F. The Application of Next Generation Sequencing Maturity Onset Diabetes of the Young Gene Panel in Turkish Patients from Trakya Region. J. Clin. Res. Pediatr. Endocrinol. 2021, 13, 320–331. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).