When Down Is Up: Heterochromatin, Nuclear Organization and X Upregulation
Abstract
1. Maintaining Appropriate Ratios of Gene Dosage Is Vital for Cells and Organisms
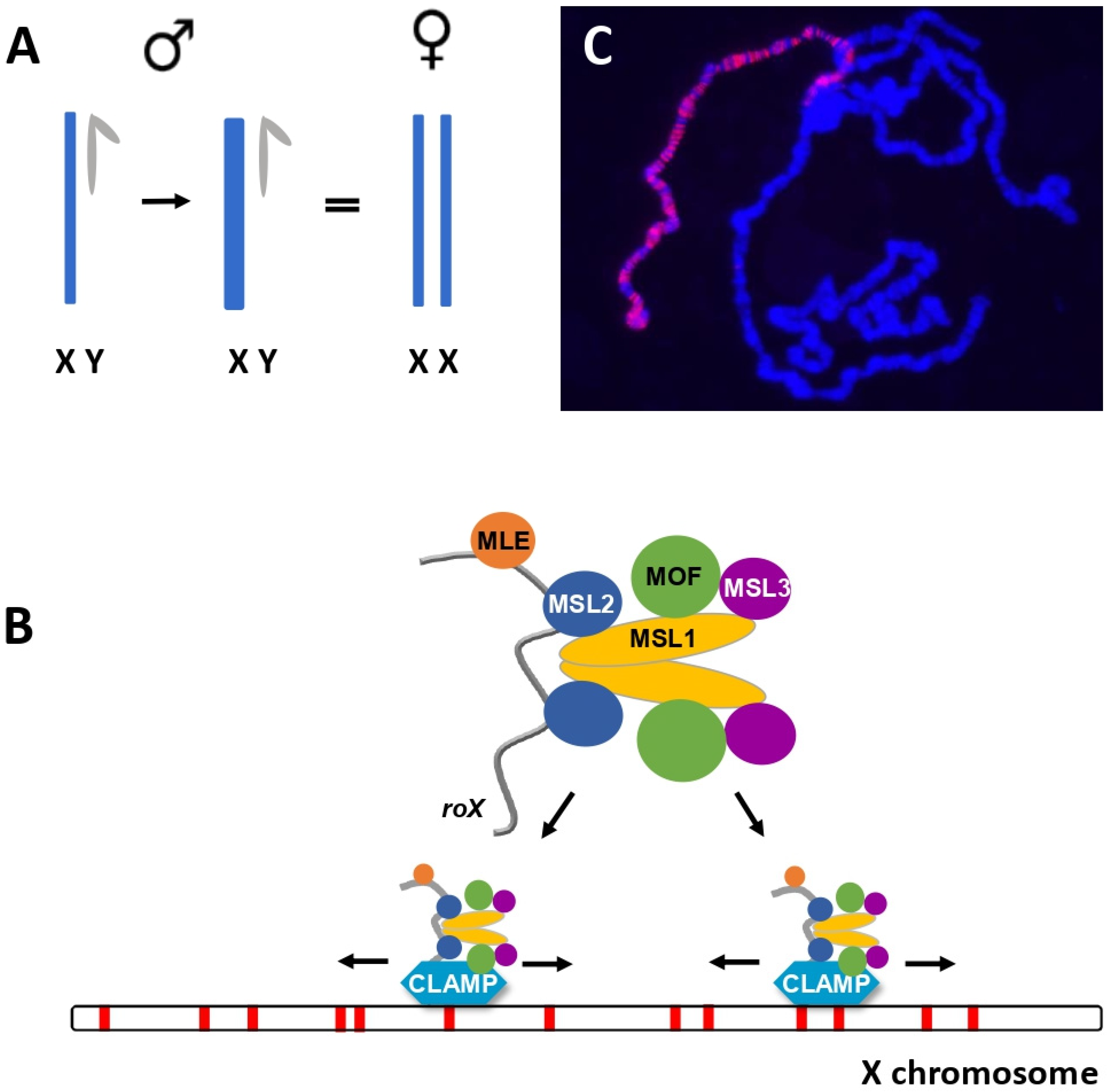
2. Subunits of the MSL Complex Determine Localization and Chromatin Modification
3. Compensated X Chromosomes Establish Distinct Nuclear Compartments
4. How Does the MSL Complex Find the X?
5. The roX Genes Have Multiple, Intertwined Roles in X Recognition
6. Satellite Repeats, Epigenetic Modifications and X Recognition
7. Proteins That Bind RNA Interact Genetically with roX1 roX2
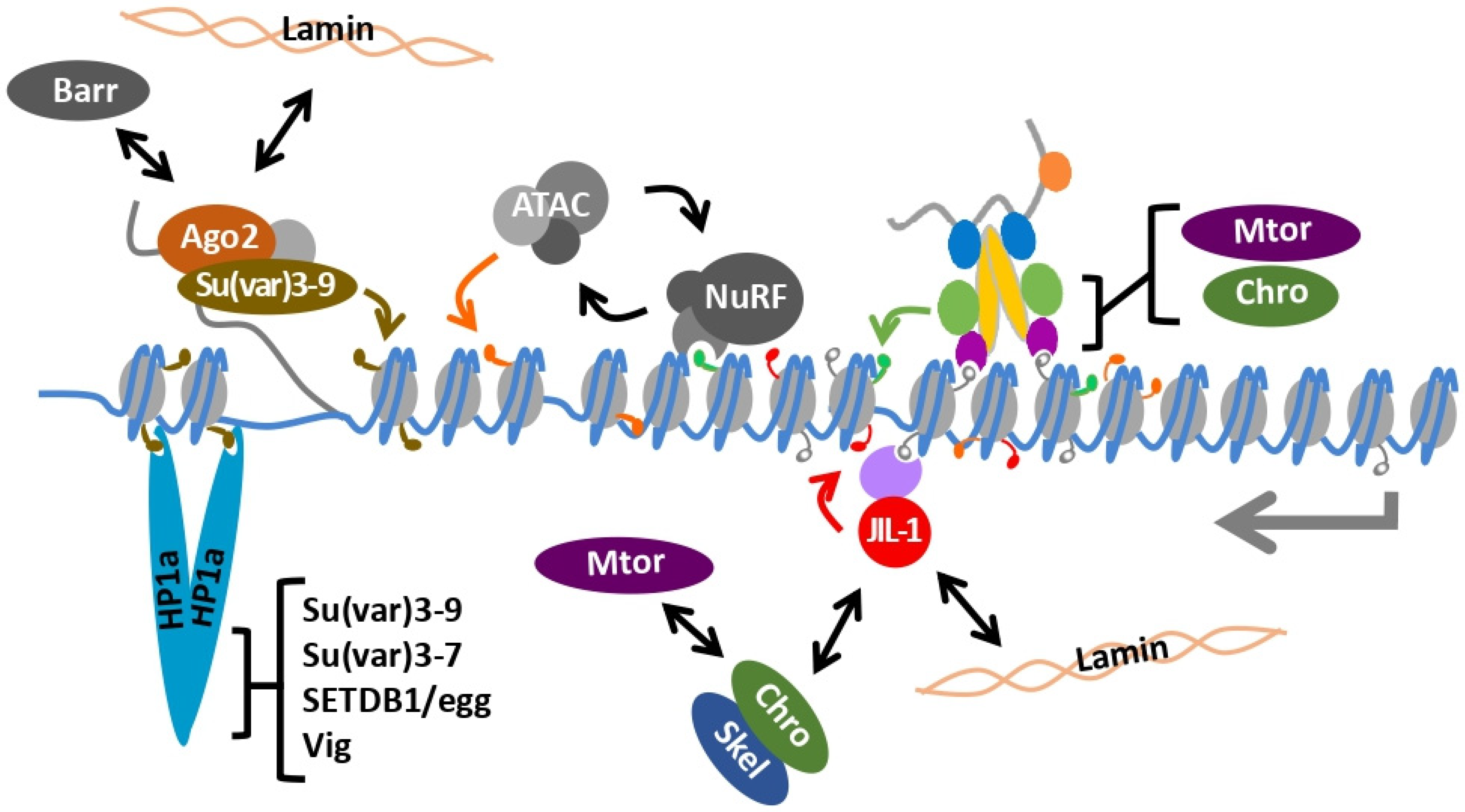
8. Heterochromatin and the Male X Chromosome
9. Chromatin Remodeling and the Male X Chromosome
10. The Dual kinase JIL-1 Maintains Interphase Chromatin Structure
11. A Governor to Limit over Activation?
12. Heterochromatin and Dosage Compensation Are Integrated on the 4th Chromosome
13. Genetic Interactions between roX Genes and Male Heterochromatin
14. Full Compensation Involves Multiple Mechanisms
15. Systems of Dosage Compensation Converge on Nuclear Organization
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Brennan, C.M.; Vaites, L.P.; Wells, J.N.; Santaguida, S.; Paulo, J.A.; Storchova, Z.; Harper, J.W.; Marsh, J.A.; Amon, A. Protein Aggregation Mediates Stoichiometry of Protein Complexes in Aneuploid Cells. Genes Dev. 2019, 33, 1031–1047. [Google Scholar] [CrossRef]
- Gupta, V.; Parisi, M.; Sturgill, D.; Nuttall, R.; Doctolero, M.; Dudko, O.; Malley, J.; Eastman, P.S.; Oliver, B. Global analysis of X-chromosome dosage compensation. J. Biol. 2006, 5, 3. [Google Scholar] [CrossRef]
- Deng, X.; Hiatt, J.B.; Nguyen, D.K.; Ercan, S.; Sturgill, D.; Hillier, L.W.; Schlesinger, F.; Davis, C.A.; Reinke, V.J.; Gingeras, T.R.; et al. Evidence for Compensatory Upregulation of Expressed X-Linked Genes in Mammals, Caenorhabditis elegans and Drosophila melanogaster. Nat. Genet. 2011, 43, 1179–1185. [Google Scholar] [CrossRef]
- Lee, J.T.; Jaenisch, R. The (epi)genetic control of mammalian X-chromosome inactivation. Curr. Opin. Genet. Dev. 1997, 7, 274–280. [Google Scholar] [CrossRef]
- Heard, E.; Disteche, C.M. Dosage Compensation in Mammals: Fine-Tuning the Expression of the X Chromosome. Genes Dev. 2006, 20, 1848–1867. [Google Scholar] [CrossRef]
- Żylicz, J.J.; Bousard, A.; Žumer, K.; Dossin, F.; Mohammad, E.; da Rocha, S.T.; Schwalb, B.; Syx, L.; Dingli, F.; Loew, D.; et al. The Implication of Early Chromatin Changes in X Chromosome Inactivation. Cell 2019, 176, 182–197. [Google Scholar] [CrossRef] [PubMed]
- Strehle, M.; Guttman, M. Xist Drives Spatial Compartmentalization of DNA and Protein to Orchestrate Initiation and Maintenance of X Inactivation. Curr. Opin. Cell Biol. 2020, 64, 139–147. [Google Scholar] [CrossRef] [PubMed]
- Chaumeil, J.; Le Baccon, P.; Wutz, A.; Heard, E. A Novel Role for Xist RNA in the Formation of a Repressive Nuclear Compartment into Which Genes Are Recruited When Silenced. Genes Dev. 2006, 20, 2223–2237. [Google Scholar] [CrossRef]
- Chen, C.-K.; Blanco, M.; Jackson, C.; Aznauryan, E.; Ollikainen, N.; Surka, C.; Chow, A.; Cerase, A.; McDonel, P.; Guttman, M. Xist Recruits the X Chromosome to the Nuclear Lamina to Enable Chromosome-Wide Silencing. Science 2016, 354, 468–472. [Google Scholar] [CrossRef] [PubMed]
- Meyer, B.J.; Casson, L.P. Caenorhabditis elegans Compensates for the Difference in X Chromosome Dosage between the Sexes by Regulating Transcript Levels. Cell 1986, 47, 871–881. [Google Scholar] [CrossRef]
- Csankovszki, G.; McDonel, P.; Meyer, B.J. Recruitment and Spreading of the C. Elegans Dosage Compensation Complex Along X Chromosomes. Science 2004, 303, 1182–1185. [Google Scholar] [CrossRef]
- Albritton, S.E.; Ercan, S. Caenorhabditis elegans Dosage Compensation: Insights into Condensin-Mediated Gene Regulation. Trends Genet. 2018, 34, 41–53. [Google Scholar] [CrossRef]
- Sass, G.L.; Pannuti, A.; Lucchesi, J.C. Male-Specific Lethal Complex of Drosophila Targets Activated Regions of the X Chromosome for Chromatin Remodeling. Proc. Natl. Acad. Sci. USA 2003, 100, 8287–8291. [Google Scholar] [CrossRef] [PubMed]
- Alekseyenko, A.A. High-Resolution ChIP-Chip Analysis Reveals That the Drosophila MSL Complex Selectively Identifies Active Genes on the Male X Chromosome. Genes Dev. 2006, 20, 848–857. [Google Scholar] [CrossRef]
- Larschan, E.; Bishop, E.P.; Kharchenko, P.V.; Core, L.J.; Lis, J.T.; Park, P.J.; Kuroda, M.I. X Chromosome Dosage Compensation via Enhanced Transcriptional Elongation in Drosophila. Nature 2011, 471, 115–118. [Google Scholar] [CrossRef] [PubMed]
- Shogren-Knaak, M.; Ishii, H.; Sun, J.-M.; Pazin, M.J.; Davie, J.R.; Peterson, C.L. Histone H4-K16 Acetylation Controls Chromatin Structure and Protein Interactions. Science 2006, 311, 844–847. [Google Scholar] [CrossRef]
- Buscaino, A.; Legube, G.; Akhtar, A. X-chromosome Targeting and Dosage Compensation Are Mediated by Distinct Domains in MSL-3. EMBO Rep. 2006, 7, 531–538. [Google Scholar] [CrossRef]
- Ferrari, F.; Plachetka, A.; Alekseyenko, A.A.; Jung, Y.L.; Ozsolak, F.; Kharchenko, P.V.; Park, P.J.; Kuroda, M.I. “Jump Start and Gain” Model for Dosage Compensation in Drosophila Based on Direct Sequencing of Nascent Transcripts. Cell Rep. 2013, 5, 629–636. [Google Scholar] [CrossRef]
- Conrad, T.; Cavalli, F.M.G.; Holz, H.; Hallacli, E.; Kind, J.; Ilik, I.; Vaquerizas, J.M.; Luscombe, N.M.; Akhtar, A. The MOF Chromobarrel Domain Controls Genome-Wide H4K16 Acetylation and Spreading of the MSL Complex. Dev. Cell 2012, 22, 610–624. [Google Scholar] [CrossRef]
- Copur, Ö.; Gorchakov, A.; Finkl, K.; Kuroda, M.I.; Müller, J. Sex-Specific Phenotypes of Histone H4 Point Mutants Establish Dosage Compensation as the Critical Function of H4K16 Acetylation in Drosophila. Proc. Natl. Acad. Sci. USA 2018, 115, 13336–13341. [Google Scholar] [CrossRef]
- Lyman, L.M.; Copps, K.; Rastelli, L.; Kelly, R.L.; Kuroda, M.I. Drosophila male-specific lethal-4 protein: Structure/function analysis and dependence on MSL-1 for chromosome association. Genetics 1997, 147, 1743–1753. [Google Scholar] [CrossRef]
- Kelley, R.L.; Solovyeva, I.; Lyman, L.M.; Richman, R.; Solovyev, V.; Kuroda, M.I. Expression of Msl-2 Causes Assembly of Dosage Compensation Regulators on the X Chromosomes and Female Lethality in Drosophila. Cell 1995, 81, 867–877. [Google Scholar] [CrossRef]
- Zhou, S.; Yang, Y.; Scott, M.J.; Pannuti, A.; Fehr, K.C.; Eisen, A.; Koonin, E.V.; Fouts, D.L.; Wrightsman, R.; Manning, J.E. Male-Specific Lethal 2, a Dosage Compensation Gene of Drosophila, Undergoes Sex-Specific Regulation and Encodes a Protein with a RING Finger and a Metallothionein-like Cysteine Cluster. EMBO J. 1995, 14, 2884–2895. [Google Scholar] [CrossRef]
- Bashaw, G.J.; Baker, B.S. The MSL-2 dosage compensation gene of drosophila encodes a putative DNA-binding protein whose expression is sex specifically regulated by sex-lethal. Development 1995, 121, 3245–3258. [Google Scholar] [CrossRef]
- Rastelli, L.; Richman, R.; Kuroda, M.I. The Dosage Compensation Regulators MLE, MSL-1 and MSL-2 Are Interdependent since Early Embryogenesis in Drosophila. Mech. Dev. 1995, 53, 223–233. [Google Scholar] [CrossRef]
- Scott, M.J. MSL1 Plays a Central Role in Assembly of the MSL Complex, Essential for Dosage Compensation in Drosophila. EMBO J. 2000, 19, 144–155. [Google Scholar] [CrossRef]
- Li, F.; Parry, D.A.D.; Scott, M.J. The Amino-Terminal Region of Drosophila MSL1 Contains Basic, Glycine-Rich, and Leucine Zipper-Like Motifs That Promote X Chromosome Binding, Self-Association, and MSL2 Binding, Respectively. Mol. Cell Biol. 2005, 25, 8913–8924. [Google Scholar] [CrossRef]
- Copps, K.; Richman, R.; Lyman, L.M.; Chang, K.A.; Rampersad-Ammons, J.; Kuroda, M.I. Complex Formation by the Drosophila MSL Proteins: Role of the MSL2 RING Finger in Protein Complex Assembly. EMBO J. 1998, 17, 5409–5417. [Google Scholar] [CrossRef]
- Hallacli, E.; Lipp, M.; Georgiev, P.; Spielman, C.; Cusack, S.; Akhtar, A.; Kadlec, J. Msl1-Mediated Dimerization of the Dosage Compensation Complex Is Essential for Male X-Chromosome Regulation in Drosophila. Mol. Cell 2012, 48, 587–600. [Google Scholar] [CrossRef]
- Sural, T.H.; Peng, S.; Li, B.; Workman, J.L.; Park, P.J.; Kuroda, M.I. The MSL3 Chromodomain Directs a Key Targeting Step for Dosage Compensation of the Drosophila melanogaster X Chromosome. Nat. Struct. Mol. Biol. 2008, 15, 1318–1325. [Google Scholar] [CrossRef]
- Hilfiker, A. MOF, a putative acetyl transferase gene related to the tip60 and moz human genes and to the SAS genes of yeast, is required for dosage compensation in drosophila. EMBO J. 1997, 16, 2054–2060. [Google Scholar] [CrossRef]
- Akhtar, A.; Becker, P.B. Activation of Transcription through Histone H4 Acetylation by MOF, an Acetyltransferase Essential for Dosage Compensation in Drosophila. Mol. Cell 2000, 5, 367–375. [Google Scholar] [CrossRef]
- Smith, E.R.; Pannuti, A.; Gu, W.; Steurnagel, A.; Cook, R.G.; Allis, C.D.; Lucchesi, J.C. The Drosophila MSL Complex Acetylates Histone H4 at Lysine 16, a Chromatin Modification Linked to Dosage Compensation. Mol. Cell Biol. 2000, 20, 312–318. [Google Scholar] [CrossRef]
- Lee, C.-G. The NTPase/Helicase Activities of Drosophila Maleless, an Essential Factor in Dosage Compensation. EMBO J. 1997, 16, 2671–2681. [Google Scholar] [CrossRef]
- Lv, M.; Yao, Y.; Li, F.; Xu, L.; Yang, L.; Gong, Q.; Xu, Y.-Z.; Shi, Y.; Fan, Y.-J.; Tang, Y. Structural Insights Reveal the Specific Recognition of RoX RNA by the DsRNA-Binding Domains of the RNA Helicase MLE and Its Indispensable Role in Dosage Compensation in Drosophila. Nucleic Acids Res. 2019, 47, 3142–3157. [Google Scholar] [CrossRef]
- Meller, V.H.; Rattner, B.P. The RoX Genes Encode Redundant Male-Specifc Lethal Transcripts Required for Targeting of the MSL Complex. EMBO J. 2002, 21, 1084–1091. [Google Scholar] [CrossRef]
- Deng, X.; Meller, V.H. RoX RNAs Are Required for Increased Expression of X-Linked Genes in Drosophila melanogaster Males. Genetics 2006, 174, 1859–1866. [Google Scholar] [CrossRef] [PubMed]
- Figueiredo, M.L.A.; Kim, M.; Philip, P.; Allgardsson, A.; Stenberg, P.; Larsson, J. Non-Coding RoX RNAs Prevent the Binding of the MSL-Complex to Heterochromatic Regions. PLoS Genet. 2014, 10, e1004865. [Google Scholar] [CrossRef] [PubMed]
- Kelley, R.L.; Lee, O.-K.; Shim, Y.-K. Transcription Rate of Noncoding RoX1 RNA Controls Local Spreading of the Drosophila MSL Chromatin Remodeling Complex. Mech. Dev. 2008, 125, 1009–1019. [Google Scholar] [CrossRef]
- Stuckenholz, C.; Meller, V.H.; Kuroda, M.I. Functional Redundancy Within RoX1, a Noncoding RNA Involved in Dosage Compensation in Drosophila melanogaster. Genetics 2003, 164, 1003–1014. [Google Scholar] [CrossRef] [PubMed]
- Müller, M.; Schauer, T.; Krause, S.; Villa, R.; Thomae, A.W.; Becker, P.B. Two-Step Mechanism for Selective Incorporation of LncRNA into a Chromatin Modifier. Nucleic Acids Res. 2020, 48, 7483–7501. [Google Scholar] [CrossRef] [PubMed]
- Ilik, I.A.; Maticzka, D.; Georgiev, P.; Gutierrez, N.M.; Backofen, R.; Akhtar, A. A Mutually Exclusive Stem-Loop Arrangement in RoX2 RNA Is Essential for X-Chromosome Regulation in Drosophila. Genes Dev. 2017, 31, 1973–1987. [Google Scholar] [CrossRef] [PubMed]
- Maenner, S.; Müller, M.; Fröhlich, J.; Langer, D.; Becker, P.B. ATP-Dependent RoX RNA Remodeling by the Helicase Maleless Enables Specific Association of MSL Proteins. Mol. Cell 2013, 51, 174–184. [Google Scholar] [CrossRef] [PubMed]
- Morales, V.; Straub, T.; Neumann, M.F.; Mengus, G.; Akhtar, A.; Becker, P.B. Functional Integration of the Histone Acetyltransferase MOF into the Dosage Compensation Complex. EMBO J. 2004, 23, 2258–2268. [Google Scholar] [CrossRef]
- Nielsen, P.R.; Nietlispach, D.; Buscaino, A.; Warner, R.J.; Akhtar, A.; Murzin, A.G.; Murzina, N.V.; Laue, E.D. Structure of the Chromo Barrel Domain from the MOF Acetyltransferase. J. Biol. Chem. 2005, 280, 32326–32331. [Google Scholar] [CrossRef] [PubMed]
- Buscaino, A.; Köcher, T.; Kind, J.H.; Holz, H.; Taipale, M.; Wagner, K.; Wilm, M.; Akhtar, A. MOF-Regulated Acetylation of MSL-3 in the Drosophila Dosage Compensation Complex. Mol. Cell 2003, 11, 1265–1277. [Google Scholar] [CrossRef]
- Valsecchi, C.I.K.; Basilicata, M.F.; Georgiev, P.; Gaub, A.; Seyfferth, J.; Kulkarni, T.; Panhale, A.; Semplicio, G.; Manjunath, V.; Holz, H.; et al. RNA Nucleation by MSL2 Induces Selective X Chromosome Compartmentalization. Nature 2021, 589, 137–142. [Google Scholar] [CrossRef]
- Grimaud, C.; Becker, P.B. The Dosage Compensation Complex Shapes the Conformation of the X Chromosome in Drosophila. Genes Dev. 2009, 23, 2490–2495. [Google Scholar] [CrossRef]
- Gorchakov, A.A.; Alekseyenko, A.A.; Kharchenko, P.; Park, P.J.; Kuroda, M.I. Long-Range Spreading of Dosage Compensation in Drosophila Captures Transcribed Autosomal Genes Inserted on X. Genes Dev. 2009, 23, 2266–2271. [Google Scholar] [CrossRef]
- Mendjan, S.; Taipale, M.; Kind, J.; Holz, H.; Gebhardt, P.; Schelder, M.; Vermeulen, M.; Buscaino, A.; Duncan, K.; Mueller, J.; et al. Nuclear Pore Components Are Involved in the Transcriptional Regulation of Dosage Compensation in Drosophila. Mol. Cell 2006, 21, 811–823. [Google Scholar] [CrossRef]
- Galupa, R.; Heard, E. X-Chromosome Inactivation: A Crossroads Between Chromosome Architecture and Gene Regulation. Annu. Rev. Genet. 2018, 52, 535–566. [Google Scholar] [CrossRef]
- Belmont, A.S.; Bignone, F.; Ts’O, P. The Relative Lntranuclear Positions of Barr Bodies in XXX Non-Transformed Human Fibroblasts. Exp. Cell Res. 1986, 15, 165–179. [Google Scholar] [CrossRef]
- Zhang, L.-F.; Huynh, K.D.; Lee, J.T. Perinucleolar Targeting of the Inactive X during S Phase: Evidence for a Role in the Maintenance of Silencing. Cell 2007, 129, 693–706. [Google Scholar] [CrossRef]
- Splinter, E.; de Wit, E.; Nora, E.P.; Klous, P.; van de Werken, H.J.G.; Zhu, Y.; Kaaij, L.J.T.; van IJcken, W.; Gribnau, J.; Heard, E.; et al. The Inactive X Chromosome Adopts a Unique Three-Dimensional Conformation That Is Dependent on Xist RNA. Genes Dev. 2011, 25, 1371–1383. [Google Scholar] [CrossRef] [PubMed]
- Eils, R.; Dietzel, S.; Bertin, E.; Schröck, E.; Speicher, M.R.; Ried, T.; Robert-Nicoud, M.; Cremer, C.; Cremer, T. Three-Dimensional Reconstruction of Painted Human Interphase Chromosomes: Active and Inactive X Chromosome Territories Have Similar Volumes but Differ in Shape and Surface Structure. J. Cell Biol. 1996, 135, 1427–1440. [Google Scholar] [CrossRef] [PubMed]
- Teller, K.; Illner, D.; Thamm, S.; Casas-Delucchi, C.S.; Versteeg, R.; Indemans, M.; Cremer, T.; Cremer, M. A Top-down Analysis of Xa- and Xi-Territories Reveals Differences of Higher Order Structure at ≥ 20 Mb Genomic Length Scales. Nucleus 2011, 2, 465–477. [Google Scholar] [CrossRef] [PubMed]
- Cerase, A.; Armaos, A.; Neumayer, C.; Avner, P.; Guttman, M.; Tartaglia, G.G. Phase Separation Drives X-Chromosome Inactivation: A Hypothesis. Nat. Struct. Mol. Biol. 2019, 26, 331–334. [Google Scholar] [CrossRef]
- Sharma, R.; Jost, D.; Kind, J.; Gómez-Saldivar, G.; van Steensel, B.; Askjaer, P.; Vaillant, C.; Meister, P. Differential Spatial and Structural Organization of the X Chromosome Underlies Dosage Compensation in C. Elegans. Genes Dev. 2014, 28, 2591–2596. [Google Scholar] [CrossRef]
- Lau, A.C.; Nabeshima, K.; Csankovszki, G. The C. Elegans Dosage Compensation Complex Mediates Interphase X Chromosome Compaction. Epigenet. Chromatin 2014, 7, 31. [Google Scholar] [CrossRef]
- Ikegami, K.; Egelhofer, T.A.; Strome, S.; Lieb, J.D. Caenorhabditis elegans Chromosome Arms Are Anchored to the Nuclear Membrane via Discontinuous Association with LEM-2. Genome Biol. 2010, 11, R120. [Google Scholar] [CrossRef]
- Towbin, B.D.; González-Aguilera, C.; Sack, R.; Gaidatzis, D.; Kalck, V.; Meister, P.; Askjaer, P.; Gasser, S.M. Step-Wise Methylation of Histone H3K9 Positions Heterochromatin at the Nuclear Periphery. Cell 2012, 150, 934–947. [Google Scholar] [CrossRef]
- Crane, E.; Bian, Q.; McCord, R.P.; Lajoie, B.R.; Wheeler, B.S.; Ralston, E.J.; Uzawa, S.; Dekker, J.; Meyer, B.J. Condensin-Driven Remodelling of X Chromosome Topology during Dosage Compensation. Nature 2015, 523, 240–244. [Google Scholar] [CrossRef]
- Sharma, R.; Meister, P. Linking Dosage Compensation and X Chromosome Nuclear Organization in C. Elegans. Nucleus 2015, 6, 266–272. [Google Scholar] [CrossRef]
- Snyder, M.J.; Lau, A.C.; Brouhard, E.A.; Davis, M.B.; Jiang, J.; Sifuentes, M.H.; Csankovszki, G. Anchoring of Heterochromatin to the Nuclear Lamina Reinforces Dosage Compensation-Mediated Gene Repression. PLoS Genet. 2016, 12, e1006341. [Google Scholar] [CrossRef] [PubMed]
- Dahlsveen, I.K.; Gilfillan, G.D.; Shelest, V.I.; Lamm, R.; Becker, P.B. Targeting Determinants of Dosage Compensation in Drosophila. PLoS Genet. 2006, 2, e5. [Google Scholar] [CrossRef]
- Park, Y.; Mengus, G.; Bai, X.; Kageyama, Y.; Meller, V.H.; Becker, P.B.; Kuroda, M.I. Sequence-Specific Targeting of Drosophila RoX Genes by the MSL Dosage Compensation Complex. Mol. Cell 2003, 11, 977–986. [Google Scholar] [CrossRef]
- Alekseyenko, A.A.; Peng, S.; Larschan, E.; Gorchakov, A.A.; Lee, O.-K.; Kharchenko, P.; McGrath, S.D.; Wang, C.I.; Mardis, E.R.; Park, P.J.; et al. A Sequence Motif within Chromatin Entry Sites Directs MSL Establishment on the Drosophila X Chromosome. Cell 2008, 134, 599–609. [Google Scholar] [CrossRef] [PubMed]
- Straub, T.; Grimaud, C.; Gilfillan, G.D.; Mitterweger, A.; Becker, P.B. The Chromosomal High-Affinity Binding Sites for the Drosophila Dosage Compensation Complex. PLoS Genet. 2008, 4, e1000302. [Google Scholar] [CrossRef] [PubMed]
- Villa, R.; Schauer, T.; Smialowski, P.; Straub, T.; Becker, P.B. PionX Sites Mark the X Chromosome for Dosage Compensation. Nature 2016, 537, 244–248. [Google Scholar] [CrossRef]
- Larschan, E.; Soruco, M.M.L.; Lee, O.-K.; Peng, S.; Bishop, E.; Chery, J.; Goebel, K.; Feng, J.; Park, P.J.; Kuroda, M.I. Identification of Chromatin-Associated Regulators of MSL Complex Targeting in Drosophila Dosage Compensation. PLoS Genet. 2012, 8, e1002830. [Google Scholar] [CrossRef] [PubMed]
- Soruco, M.M.L.; Chery, J.; Bishop, E.P.; Siggers, T.; Tolstorukov, M.Y.; Leydon, A.R.; Sugden, A.U.; Goebel, K.; Feng, J.; Xia, P.; et al. The CLAMP Protein Links the MSL Complex to the X Chromosome during Drosophila Dosage Compensation. Genes Dev. 2013, 27, 1551–1556. [Google Scholar] [CrossRef]
- Park, Y.; Kelley, R.L.; Oh, H.; Kuroda, M.I.; Meller, V.H. Extent of Chromatin Spreading Determined by RoX RNA Recruitment of MSL Proteins. Science 2002, 298, 1620–1623. [Google Scholar] [CrossRef] [PubMed]
- Kelley, R.L.; Meller, V.H.; Gordadze, P.R.; Roman, G.; Davis, R.L.; Kuroda, M.I. Epigenetic Spreading of the Drosophila Dosage Compensation Complex from RoX RNA Genes into Flanking Chromatin. Cell 1999, 98, 513–522. [Google Scholar] [CrossRef]
- Henry, R.A.; Tews, B.; Li, X.; Scott, M.J. Recruitment of the Male-Specific Lethal (MSL) Dosage Compensation Complex to an Autosomally Integrated RoXChromatin Entry Site Correlates with an Increased Expression of an Adjacent Reporter Gene in Male Drosophila. J. Biol. Chem. 2001, 276, 31953–31958. [Google Scholar] [CrossRef] [PubMed]
- Fauth, T.; Müller-Planitz, F.; König, C.; Straub, T.; Becker, P.B. The DNA Binding CXC Domain of MSL2 Is Required for Faithful Targeting the Dosage Compensation Complex to the X Chromosome. Nucleic Acids Res. 2010, 38, 3209–3221. [Google Scholar] [CrossRef]
- Tikhonova, E.; Fedotova, A.; Bonchuk, A.; Mogila, V.; Larschan, E.N.; Georgiev, P.; Maksimenko, O. The Simultaneous Interaction of MSL2 with CLAMP and DNA Provides Redundancy in the Initiation of Dosage Compensation in Drosophila Males. Development 2019, 146, dev179663. [Google Scholar] [CrossRef] [PubMed]
- Ramírez, F.; Lingg, T.; Toscano, S.; Lam, K.C.; Georgiev, P.; Chung, H.-R.; Lajoie, B.R.; de Wit, E.; Zhan, Y.; de Laat, W.; et al. High-Affinity Sites Form an Interaction Network to Facilitate Spreading of the MSL Complex across the X Chromosome in Drosophila. Mol. Cell 2015, 60, 146–162. [Google Scholar] [CrossRef] [PubMed]
- Oh, H. Local Spreading of MSL Complexes from RoX Genes on the Drosophila X Chromosome. Genes Dev. 2003, 17, 1334–1339. [Google Scholar] [CrossRef]
- Straub, T. The Drosophila MSL Complex Activates the Transcription of Target Genes. Genes Dev. 2005, 19, 2284–2288. [Google Scholar] [CrossRef]
- Larschan, E.; Alekseyenko, A.A.; Gortchakov, A.A.; Peng, S.; Li, B.; Yang, P.; Workman, J.L.; Park, P.J.; Kuroda, M.I. MSL Complex Is Attracted to Genes Marked by H3K36 Trimethylation Using a Sequence-Independent Mechanism. Mol. Cell 2007, 28, 121–133. [Google Scholar] [CrossRef] [PubMed]
- Bell, O.; Conrad, T.; Kind, J.; Wirbelauer, C.; Akhtar, A.; Schübeler, D. Transcription-Coupled Methylation of Histone H3 at Lysine 36 Regulates Dosage Compensation by Enhancing Recruitment of the MSL Complex in Drosophila melanogaster. Mol. Cell Biol. 2008, 28, 3401–3409. [Google Scholar] [CrossRef]
- Hamada, F.N. Global Regulation of X Chromosomal Genes by the MSL Complex in Drosophila melanogaster. Genes Dev. 2005, 19, 2289–2294. [Google Scholar] [CrossRef] [PubMed]
- Joshi, S.S.; Meller, V.H. Satellite Repeats Identify X Chromatin for Dosage Compensation in Drosophila melanogaster Males. Curr. Biol. 2017, 27, 1393–1402. [Google Scholar] [CrossRef]
- Plath, K.; Mlynarczyk-Evans, S.; Nusinow, D.A.; Panning, B. Xist RNA and the Mechanism of X Chromosome Inactivation. Annu. Rev. Genet. 2002, 36, 233–278. [Google Scholar] [CrossRef]
- Waring, G.L.; Pollack, J.C. Cloning and Characterization of a Dispersed, Multicopy, X Chromosome Sequence in Drosophila Melanogaster. Proc. Natl. Acad. Sci. USA 1987, 84, 2843–2847. [Google Scholar] [CrossRef]
- Di Bartolomeis, S.M.; Tartof, K.D.; Rob Jackson, F. A Superfamily of Drosophila Satellite Related (SR) DNA Repeats Restricted to the X Chromosome Euchromatin. Nucl. Acids Res. 1992, 20, 1113–1116. [Google Scholar] [CrossRef] [PubMed]
- Gallach, M. Recurrent Turnover of Chromosome-Specific Satellites in Drosophila. Genome Biol. Evol. 2014, 6, 1279–1286. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, G.C.S.; Küttler, H.; Moreira-Filho, O.; Heslop-Harrison, J.S. The 1.688 Repetitive DNA of Drosophila: Concerted Evolution at Different Genomic Scales and Association with Genes. Mol. Biol. Evol. 2012, 29, 7–11. [Google Scholar] [CrossRef]
- Lohe, A.R.; Hilliker, A.J.; Roberts, P.A. Mapping simple repeated DNA sequences in heterochromatin of drosophila melanogaster. Genetics 1993, 134, 1149–1174. [Google Scholar] [CrossRef]
- Usakin, L.; Abad, J.; Vagin, V.V.; de Pablos, B.; Villasante, A.; Gvozdev, V.A. Transcription of the 1.688 Satellite DNA Family Is Under the Control of RNA Interference Machinery in Drosophila melanogaster Ovaries. Genetics 2007, 176, 1343–1349. [Google Scholar] [CrossRef]
- Menon, D.U.; Coarfa, C.; Xiao, W.; Gunaratne, P.H.; Meller, V.H. SiRNAs from an X-Linked Satellite Repeat Promote X-Chromosome Recognition in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2014, 111, 16460–16465. [Google Scholar] [CrossRef]
- Menon, D.U.; Meller, V.H. A Role for SiRNA in X-Chromosome Dosage Compensation in Drosophila melanogaster. Genetics 2012, 191, 1023–1028. [Google Scholar] [CrossRef]
- Deshpande, N.; Meller, V.H. Chromatin That Guides Dosage Compensation Is Modulated by the SiRNA Pathway in Drosophila melanogaster. Genetics 2018, 209, 1085–1097. [Google Scholar] [CrossRef]
- Rand, T.A.; Petersen, S.; Du, F.; Wang, X. Argonaute2 Cleaves the Anti-Guide Strand of SiRNA during RISC Activation. Cell 2005, 123, 621–629. [Google Scholar] [CrossRef]
- Ishizuka, A. A Drosophila Fragile X Protein Interacts with Components of RNAi and Ribosomal Proteins. Genes Dev. 2002, 16, 2497–2508. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, E.; Caudy, A.A.; Hammond, S.M.; Hannon, G.J. Role for a Bidentate Ribonuclease in the Initiation Step of RNA Interference. Nature 2001, 409, 363–366. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S.; Nakahara, K.; Pham, J.W.; Kim, K.; He, Z.; Sontheimer, E.J.; Carthew, R.W. Distinct Roles for Drosophila Dicer-1 and Dicer-2 in the SiRNA/MiRNA Silencing Pathways. Cell 2004, 117, 69–81. [Google Scholar] [CrossRef]
- Lim, D.H.; Kim, J.; Kim, S.; Carthew, R.W.; Lee, Y.S. Functional Analysis of Dicer-2 Missense Mutations in the SiRNA Pathway of Drosophila. Biochem. Biophys. Res. Commun. 2008, 371, 525–530. [Google Scholar] [CrossRef] [PubMed]
- Lipardi, C.; Paterson, B.M. Identification of an RNA-Dependent RNA Polymerase in Drosophila Involved in RNAi and Transposon Suppression. Proc. Natl. Acad. Sci. USA 2009, 106, 15645–15650. [Google Scholar] [CrossRef] [PubMed]
- Marques, J.T.; Kim, K.; Wu, P.-H.; Alleyne, T.M.; Jafari, N.; Carthew, R.W. Loqs and R2D2 Act Sequentially in the SiRNA Pathway in Drosophila. Nat. Struct. Mol. Biol. 2010, 17, 24–30. [Google Scholar] [CrossRef] [PubMed]
- Gracheva, E.; Dus, M.; Elgin, S.C.R. Drosophila RISC Component VIG and Its Homolog Vig2 Impact Heterochromatin Formation. PLoS ONE 2009, 4, e6182. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhou, R.; Hotta, I.; Denli, A.M.; Hong, P.; Perrimon, N.; Hannon, G.J. Comparative Analysis of Argonaute-Dependent Small RNA Pathways in Drosophila. Mol. Cell 2008, 32, 592–599. [Google Scholar] [CrossRef]
- Caudy, A.A. Fragile X-Related Protein and VIG Associate with the RNA Interference Machinery. Genes Dev. 2002, 16, 2491–2496. [Google Scholar] [CrossRef]
- Pek, J.W.; Kai, T. DEAD-Box RNA Helicase Belle/DDX3 and the RNA Interference Pathway Promote Mitotic Chromosome Segregation. Proc. Natl. Acad. Sci. USA 2011, 108, 12007–12012. [Google Scholar] [CrossRef]
- Luo, H.; Li, X.; Claycomb, J.M.; Lipshitz, H.D. The Smaug RNA-Binding Protein Is Essential for MicroRNA Synthesis During the Drosophila Maternal-to-Zygotic Transition. G3 Genes Genomes Genet. 2016, 6, 3541–3551. [Google Scholar] [CrossRef] [PubMed]
- Dahanukar, A.; Walker, J.A.; Wharton, R.P. Smaug, a Novel RNA-Binding Protein That Operates a Translational Switch in Drosophila. Mol. Cell 1999, 4, 209–218. [Google Scholar] [CrossRef]
- Spierer, A.; Seum, C.; Delattre, M.; Spierer, P. Loss of the Modifiers of Variegation Su(Var)3–7 or HP1 Impacts Male X Polytene Chromosome Morphology and Dosage Compensation. J. Cell Sci. 2005, 118, 5047–5057. [Google Scholar] [CrossRef]
- Delattre, M.; Spierer, A.; Jaquet, Y.; Spierer, P. Increased Expression of Drosophila Su(Var)3–7 Triggers Su(Var)3–9-Dependent Heterochromatin Formation. J. Cell Sci. 2004, 117, 6239–6247. [Google Scholar] [CrossRef][Green Version]
- Spierer, A.; Begeot, F.; Spierer, P.; Delattre, M. SU(VAR)3–7 Links Heterochromatin and Dosage Compensation in Drosophila. PLoS Genet. 2008, 4, e1000066. [Google Scholar] [CrossRef] [PubMed]
- Corona, D.F.V.; Clapier, C.R.; Becker, P.B.; Tamkun, J.W. Modulation of ISWI Function by Site-specific Histone Acetylation. EMBO Rep. 2002, 3, 242–247. [Google Scholar] [CrossRef]
- Bai, X.; Larschan, E.; Kwon, S.Y.; Badenhorst, P.; Kuroda, M.I. Regional Control of Chromatin Organization by Noncoding RoX RNAs and the NURF Remodeling Complex in Drosophila melanogaster. Genetics 2007, 176, 1491–1499. [Google Scholar] [CrossRef]
- Tsukiyama, T.; Daniel, C.; Tamkun, J.; Wu, C. ISWI, a member of the SWL2/SNF2 ATPase family, encodes the 140 KDA subunit of the nucleosome remodeling factor. Cell 1995, 83, 1021–1026. [Google Scholar] [CrossRef]
- Suganuma, T.; Gutiérrez, J.L.; Li, B.; Florens, L.; Swanson, S.K.; Washburn, M.P.; Abmayr, S.M.; Workman, J.L. ATAC Is a Double Histone Acetyltransferase Complex That Stimulates Nucleosome Sliding. Nat. Struct. Mol. Biol. 2008, 15, 364–372. [Google Scholar] [CrossRef]
- Carré, C.; Ciurciu, A.; Komonyi, O.; Jacquier, C.; Fagegaltier, D.; Pidoux, J.; Tricoire, H.; Tora, L.; Boros, I.M.; Antoniewski, C. The Drosophila NURF Remodelling and the ATAC Histone Acetylase Complexes Functionally Interact and Are Required for Global Chromosome Organization. EMBO Rep. 2008, 9, 187–192. [Google Scholar] [CrossRef]
- Lim, C.K.; Kelley, R.L. The Drosophila Over Compensating Males Gene Genetically Inhibits Dosage Compensation in Males. PLoS ONE 2013, 8, e60450. [Google Scholar] [CrossRef]
- Alfieri, C.; Gambetta, M.C.; Matos, R.; Glatt, S.; Sehr, P.; Fraterman, S.; Wilm, M.; Muller, J.; Muller, C.W. Structural Basis for Targeting the Chromatin Repressor Sfmbt to Polycomb Response Elements. Genes Dev. 2013, 27, 2367–2379. [Google Scholar] [CrossRef] [PubMed]
- Aleman, J.R.; Kuhn, T.M.; Pascual-Garcia, P.; Gospocic, J.; Lan, Y.; Bonasio, R.; Little, S.C.; Capelson, M. Correct Dosage of X Chromosome Transcription Is Controlled by a Nuclear Pore Component. Cell Rep. 2021, 35, 109236. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, W.; Jin, Y.; Johansen, J.; Johansen, K.M. The JIL-1 Tandem Kinase Mediates Histone H3 Phosphorylation and Is Required for Maintenance of Chromatin Structure in Drosophila. Cell 2001, 105, 433–443. [Google Scholar] [CrossRef]
- Deng, X.; Rattner, B.P.; Souter, S.; Meller, V.H. The Severity of RoX1 Mutations Is Predicted by MSL Localization on the X Chromosome. Mech. Dev. 2005, 122, 1094–1105. [Google Scholar] [CrossRef]
- Rincon-Arano, H.; Halow, J.; Delrow, J.J.; Parkhurst, S.M.; Groudine, M. UpSET Recruits HDAC Complexes and Restricts Chromatin Accessibility and Acetylation at Promoter Regions. Cell 2012, 151, 1214–1228. [Google Scholar] [CrossRef]
- McElroy, K.A.; Jung, Y.L.; Zee, B.M.; Wang, C.I.; Park, P.J.; Kuroda, M.I. UpSET, the Drosophila Homologue of SET3, Is Required for Viability and the Proper Balance of Active and Repressive Chromatin Marks. G3 Genes Genomes Genet. 2017, 7, 625–635. [Google Scholar] [CrossRef]
- Zee, B.M.; Alekseyenko, A.A.; McElroy, K.A.; Kuroda, M.I. Streamlined Discovery of Cross-Linked Chromatin Complexes and Associated Histone Modifications by Mass Spectrometry. Proc. Natl. Acad. Sci. USA 2016, 113, 1784–1789. [Google Scholar] [CrossRef] [PubMed]
- Pinder, B.D.; Smibert, C.A. MicroRNA-independent Recruitment of Argonaute 1 to Nanos MRNA through the Smaug RNA-binding Protein. EMBO Rep. 2013, 14, 80–86. [Google Scholar] [CrossRef]
- De Wit, E. Genome-Wide HP1 Binding in Drosophila: Developmental Plasticity and Genomic Targeting Signals. Genome Res. 2005, 15, 1265–1273. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.-P.; Ni, J.-Q.; Shi, Y.-D.; Oakeley, E.J.; Sun, F.-L. Sex-Specific Role of Drosophila melanogaster HP1 in Regulating Chromatin Structure and Gene Transcription. Nat. Genet. 2005, 37, 1361–1366. [Google Scholar] [CrossRef]
- Park, A.R.; Liu, N.; Neuenkirchen, N.; Guo, Q.; Lin, H. The Role of Maternal HP1a in Early Drosophila Embryogenesis via Regulation of Maternal Transcript Production. Genetics 2019, 211, 201–217. [Google Scholar] [CrossRef]
- Delattre, M.; Spierer, A.; Tonka, C.H.; Spierer, P. The genomic silencing of position-effect variegation in drosophila melanogaster: Interaction between the heterochromatin-associated proteins SU(VAR)3–7 and HP. J. Cell Sci. 2000, 113, 4253–4261. [Google Scholar] [CrossRef]
- Demakova, O.V.; Kotlikova, I.V.; Gordadze, P.R.; Alekseyenko, A.A.; Kuroda, M.I.; Zhimulev, I.F. The MSL Complex Levels Are Critical for Its Correct Targeting to the Chromosomes in Drosophila melanogaster. Chromosoma 2003, 112, 103–115. [Google Scholar] [CrossRef] [PubMed]
- Badenhorst, P. Biological Functions of the ISWI Chromatin Remodeling Complex NURF. Genes Dev. 2002, 16, 3186–3198. [Google Scholar] [CrossRef] [PubMed]
- Deuring, R.; Fanti, L.; Armstrong, J.A.; Sarte, M.; Papoulas, O.; Prestel, M.; Daubresse, G.; Verardo, M.; Moseley, S.L.; Berloco, M.; et al. The ISWI Chromatin-Remodeling Protein Is Required for Gene Expression and the Maintenance of Higher Order Chromatin Structure In Vivo. Mol. Cell 2000, 5, 355–365. [Google Scholar] [CrossRef]
- Albig, C.; Tikhonova, E.; Krause, S.; Maksimenko, O.; Regnard, C.; Becker, P.B. Factor Cooperation for Chromosome Discrimination in Drosophila. Nucleic Acids Res. 2019, 47, 1706–1724. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.I.; Alekseyenko, A.A.; LeRoy, G.; Elia, A.E.; Gorchakov, A.A.; Britton, L.-M.P.; Elledge, S.J.; Kharchenko, P.V.; Garcia, B.A.; Kuroda, M.I. Chromatin Proteins Captured by ChIP–Mass Spectrometry Are Linked to Dosage Compensation in Drosophila. Nat. Struct. Mol. Biol. 2013, 20, 202–209. [Google Scholar] [CrossRef] [PubMed]
- Rath, U.; Ding, Y.; Deng, H.; Qi, H.; Bao, X.; Zhang, W.; Girton, J.; Johansen, J.; Johansen, K.M. The Chromodomain Protein, Chromator, Interacts with JIL-1 Kinase and Regulates the Structure of Drosophila Polytene Chromosomes. J. Cell Sci. 2006, 119, 2332–2341. [Google Scholar] [CrossRef] [PubMed]
- Rath, U.; Wang, D.; Ding, Y.; Xu, Y.-Z.; Qi, H.; Blacketer, M.J.; Girton, J.; Johansen, J.; Johansen, K.M. Chromator, a Novel and Essential Chromodomain Protein Interacts Directly with the Putative Spindle Matrix Protein Skeletor. J. Cell. Biochem. 2004, 93, 1033–1047. [Google Scholar] [CrossRef]
- Bao, X.; Zhang, W.; Krencik, R.; Deng, H.; Wang, Y.; Girton, J.; Johansen, J.; Johansen, K.M. The JIL-1 Kinase Interacts with Lamin Dm0 and Regulates Nuclear Lamina Morphology of Drosophila Nurse Cells. J. Cell Sci. 2005, 118, 5079–5087. [Google Scholar] [CrossRef]
- Zhang, W.; Deng, H.; Bao, X.; Lerach, S.; Girton, J.; Johansen, J.; Johansen, K.M. The JIL-1 Histone H3S10 Kinase Regulates Dimethyl H3K9 Modifications and Heterochromatic Spreading in Drosophila. Development 2006, 133, 229–235. [Google Scholar] [CrossRef]
- Yao, C.; Rath, U.; Maiato, H.; Sharp, D.; Girton, J.; Johansen, K.M.; Johansen, J. A Nuclear-Derived Proteinaceous Matrix Embeds the Microtubule Spindle Apparatus during Mitosis. Mol. Biol. Cell 2012, 23, 3532–3541. [Google Scholar] [CrossRef]
- Mondal, B.C.; Shim, J.; Evans, C.J.; Banerjee, U. Pvr Expression Regulators in Equilibrium Signal Control and Maintenance of Drosophila Blood Progenitors. eLife 2014, 3, e03626. [Google Scholar] [CrossRef] [PubMed]
- Stofanko, M.; Kwon, S.Y.; Badenhorst, P. A Misexpression Screen to Identify Regulators of Drosophila Larval Hemocyte Development. Genetics 2008, 180, 253–267. [Google Scholar] [CrossRef]
- Vaquerizas, J.M.; Suyama, R.; Kind, J.; Miura, K.; Luscombe, N.M.; Akhtar, A. Nuclear Pore Proteins Nup153 and Megator Define Transcriptionally Active Regions in the Drosophila Genome. PLoS Genet. 2010, 6, e1000846. [Google Scholar] [CrossRef]
- Yao, C.; Wang, C.; Li, Y.; Zavortink, M.; Archambault, V.; Girton, J.; Johansen, K.M.; Johansen, J. Evidence for a Role of Spindle Matrix Formation in Cell Cycle Progression by Antibody Perturbation. PLoS ONE 2018, 13, e0208022. [Google Scholar] [CrossRef]
- Riddle, N.C.; Elgin, S.C.R. The Drosophila Dot Chromosome: Where Genes Flourish Amidst Repeats. Genetics 2018, 210, 757–772. [Google Scholar] [CrossRef]
- Vicoso, B.; Bachtrog, D. Reversal of an Ancient Sex Chromosome to an Autosome in Drosophila. Nature 2013, 499, 332–335. [Google Scholar] [CrossRef] [PubMed]
- Johansson, A.-M.; Stenberg, P.; Bernhardsson, C.; Larsson, J. Painting of Fourth and Chromosome-Wide Regulation of the 4th Chromosome in Drosophila melanogaster. EMBO J. 2007, 26, 2307–2316. [Google Scholar] [CrossRef] [PubMed]
- Larsson, J.; Chen, J.D.; Rasheva, V.; Rasmuson-Lestander, A.; Pirrotta, V. Painting of Fourth, a Chromosome-Specific Protein in Drosophila. Proc. Natl. Acad. Sci. USA 2001, 98, 6273–6278. [Google Scholar] [CrossRef] [PubMed]
- Johansson, A.-M.; Stenberg, P.; Allgardsson, A.; Larsson, J. POF Regulates the Expression of Genes on the Fourth Chromosome in Drosophila melanogaster by Binding to Nascent RNA. Mol. Cell Biol. 2012, 32, 2121–2134. [Google Scholar] [CrossRef] [PubMed]
- Seum, C.; Reo, E.; Peng, H.; Rauscher, F.J.; Spierer, P.; Bontron, S. Drosophila SETDB1 Is Required for Chromosome 4 Silencing. PLoS Genet. 2007, 3, e76. [Google Scholar] [CrossRef] [PubMed]
- Tzeng, T.-Y.; Lee, C.-H.; Chan, L.-W.; Shen, C.-K.J. Epigenetic Regulation of the Drosophila Chromosome 4 by the Histone H3K9 Methyltransferase DSETDB. Proc. Natl. Acad. Sci. USA 2007, 104, 12691–12696. [Google Scholar] [CrossRef]
- Kalashnikova, D.A.; Maksimov, D.A.; Romanov, S.E.; Laktionov, P.P.; Koryakov, D.E. SetDB1 and Su(Var)3–9 Play Non-Overlapping Roles in Somatic Cell Chromosomes of Drosophila melanogaster. J. Cell Sci. 2021, 134, jcs253096. [Google Scholar] [CrossRef]
- Riddle, N.C.; Jung, Y.L.; Gu, T.; Alekseyenko, A.A.; Asker, D.; Gui, H.; Kharchenko, P.V.; Minoda, A.; Plachetka, A.; Schwartz, Y.B.; et al. Enrichment of HP1a on Drosophila Chromosome 4 Genes Creates an Alternate Chromatin Structure Critical for Regulation in This Heterochromatic Domain. PLoS Genet. 2012, 8, e1002954. [Google Scholar] [CrossRef]
- Piacentini, L.; Sergio, P. Positive Regulation of Euchromatic Gene Expression by HP1a. Fly 2010, 4, 299–301. [Google Scholar] [CrossRef]
- Yasuhara, J.; Wakimoto, B. Oxymoron No More: The Expanding World of Heterochromatic Genes. Trends Genet. 2006, 22, 330–338. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Koya, S.K.; Kong, Y.; Meller, V.H. Coordinated Regulation of Heterochromatic Genes in Drosophila Melanogaster Males. Genetics 2009, 182, 481–491. [Google Scholar] [CrossRef] [PubMed]
- Apte, M.S.; Meller, V.H. Sex Differences in Drosophila melanogaster Heterochromatin Are Regulated by Non-Sex Specific Factors. PLoS ONE 2015, 10, e0128114. [Google Scholar] [CrossRef]
- Koya, S.K.; Meller, V.H. Modulation of Heterochromatin by Male Specific Lethal Proteins and RoX RNA in Drosophila Melanogaster Males. PLoS ONE 2015, 10, e0140259. [Google Scholar] [CrossRef] [PubMed]
- Stenberg, P.; Lundberg, L.E.; Johansson, A.-M.; Rydén, P.; Svensson, M.J.; Larsson, J. Buffering of Segmental and Chromosomal Aneuploidies in Drosophila melanogaster. PLoS Genet. 2009, 5, e1000465. [Google Scholar] [CrossRef] [PubMed]
- Lundberg, L.E.; Figueiredo, M.L.A.; Stenberg, P.; Larsson, J. Buffering and Proteolysis Are Induced by Segmental Monosomy in Drosophila melanogaster. Nucleic Acids Res. 2012, 40, 5926–5937. [Google Scholar] [CrossRef]
- Zhang, Y.; Oliver, B. Dosage Compensation Goes Global. Curr. Opin. Genet. Dev. 2007, 17, 113–120. [Google Scholar] [CrossRef]
- Lee, H.; Oliver, B. Non-Canonical Drosophila X Chromosome Dosage Compensation and Repressive Topologically Associated Domains. Epigenet. Chromatin 2018, 11, 62. [Google Scholar] [CrossRef] [PubMed]
- Lott, S.E.; Villalta, J.E.; Schroth, G.P.; Luo, S.; Tonkin, L.A.; Eisen, M.B. Noncanonical Compensation of Zygotic X Transcription in Early Drosophila melanogaster Development Revealed through Single-Embryo RNA-Seq. PLoS Biol. 2011, 9, e1000590. [Google Scholar] [CrossRef]
| Complex or Process | Gene | Functions | Mutant Phenotype | Citations |
|---|---|---|---|---|
| Small RNA production or action | Ago2 | siRNA slicer nuclease | Enhances roX1 roX2 male lethality | [92,93,94] |
| Rm62 | RNA helicase, RNA processing | [93,95] | ||
| Dcr1 | Small RNA processing | [92,93,96,97] | ||
| Dcr2 | Small RNA processing | [92,93,97,98] | ||
| Fmr1 | RNA-binding, translational regulation | [93,95] | ||
| Elp1 | RNAPII elongation, binds Ago2 Dcr-1,-2 | [92,99] | ||
| Loqs | dsRNA-binding, siRNA processing | [92,100] | ||
| vig | Interacts with Ago1, Ago2 and HP1a | [93,101,102,103] | ||
| barr | Interacts with Ago2 and spn-E | [93,104] | ||
| Smg | RNA-binding, translation, mRNA stability, miRNA production | [93,105,106] | ||
| Heterochromatin | Su(var)3-9 | H3K9 methyltransferaseHeterochromatin formation | Polytenized male X disorganized Enhances roX1 roX2 male lethality | [93,107] |
| Su(var)3-7 | Heterochromatin formation | [93,107,108,109] | ||
| HP1a | H3K9me2/3 binding Heterochromatin formation | Polytenized male X disorganized | [107] | |
| NURF complex | ISWI | ATP-dependent nucleosome remodeler | Polytenized male X disorganized Enhances roX1 roX2 male lethality | [110,111,112], Meller lab unpublished |
| Nurf301 | Nucleosome remodeling | Polytenized male X disorganized | [111] | |
| ATAC complex | Gcn5 | Histone acetyltransferase | Polytenized male X disorganized | [113,114] |
| Ada2a | Chromatin binding | [113,114] | ||
| Limit compensation | Ocm | Polycomb group interactions | Suppresses roX1 roX2 male lethality | [115,116] |
| Mtor | Nuclear pore subunit | [117] | ||
| Misc. | JIL-1 | Dual kinase, boundary elementEnriched on male X chromosome | Polytenized male X disorganized | [118,119] |
| upSET | Maintains heterochromatin Binds MSL3, HDAC1 and SIN3-A | Enhances roX1 roX2 male lethality | [93,120,121,122] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Makki, R.; Meller, V.H. When Down Is Up: Heterochromatin, Nuclear Organization and X Upregulation. Cells 2021, 10, 3416. https://doi.org/10.3390/cells10123416
Makki R, Meller VH. When Down Is Up: Heterochromatin, Nuclear Organization and X Upregulation. Cells. 2021; 10(12):3416. https://doi.org/10.3390/cells10123416
Chicago/Turabian StyleMakki, Reem, and Victoria H. Meller. 2021. "When Down Is Up: Heterochromatin, Nuclear Organization and X Upregulation" Cells 10, no. 12: 3416. https://doi.org/10.3390/cells10123416
APA StyleMakki, R., & Meller, V. H. (2021). When Down Is Up: Heterochromatin, Nuclear Organization and X Upregulation. Cells, 10(12), 3416. https://doi.org/10.3390/cells10123416





