Multidimensional Single-Nuclei RNA-Seq Reconstruction of Adipose Tissue Reveals Adipocyte Plasticity Underlying Thermogenic Response
Abstract
1. Introduction
2. Materials and Methods
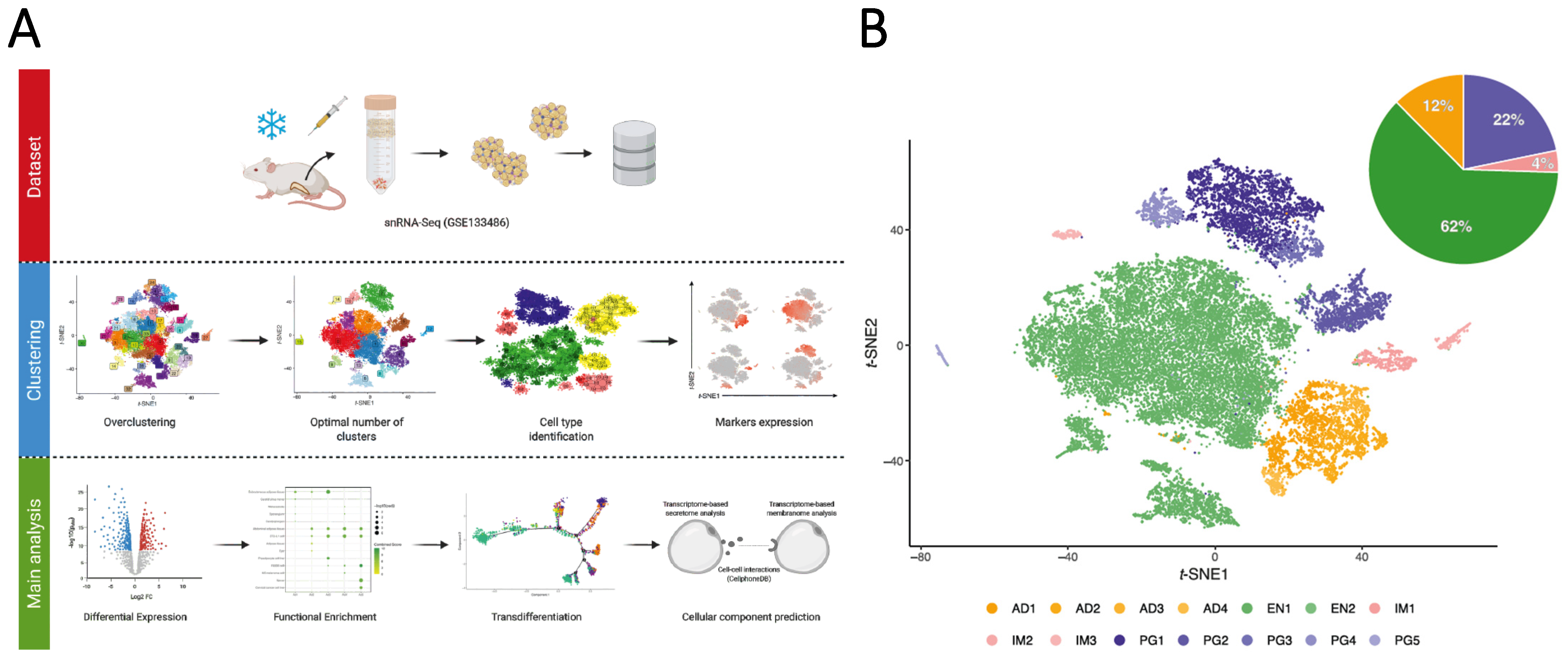
2.1. Single-Cell RNA-Seq Data
2.2. Data Pre-Processing
2.3. Optimal Number of Clusters
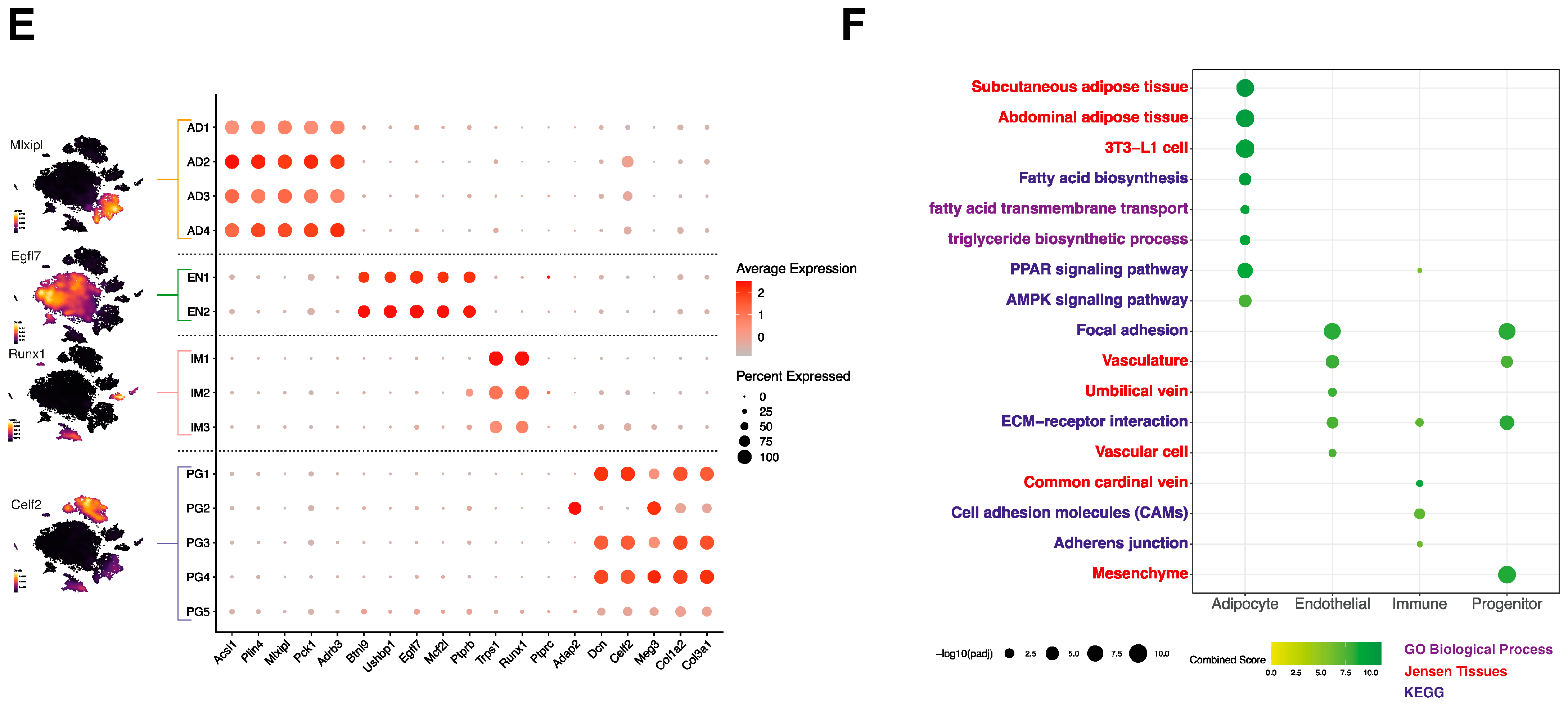
2.4. Cell-Type Classification
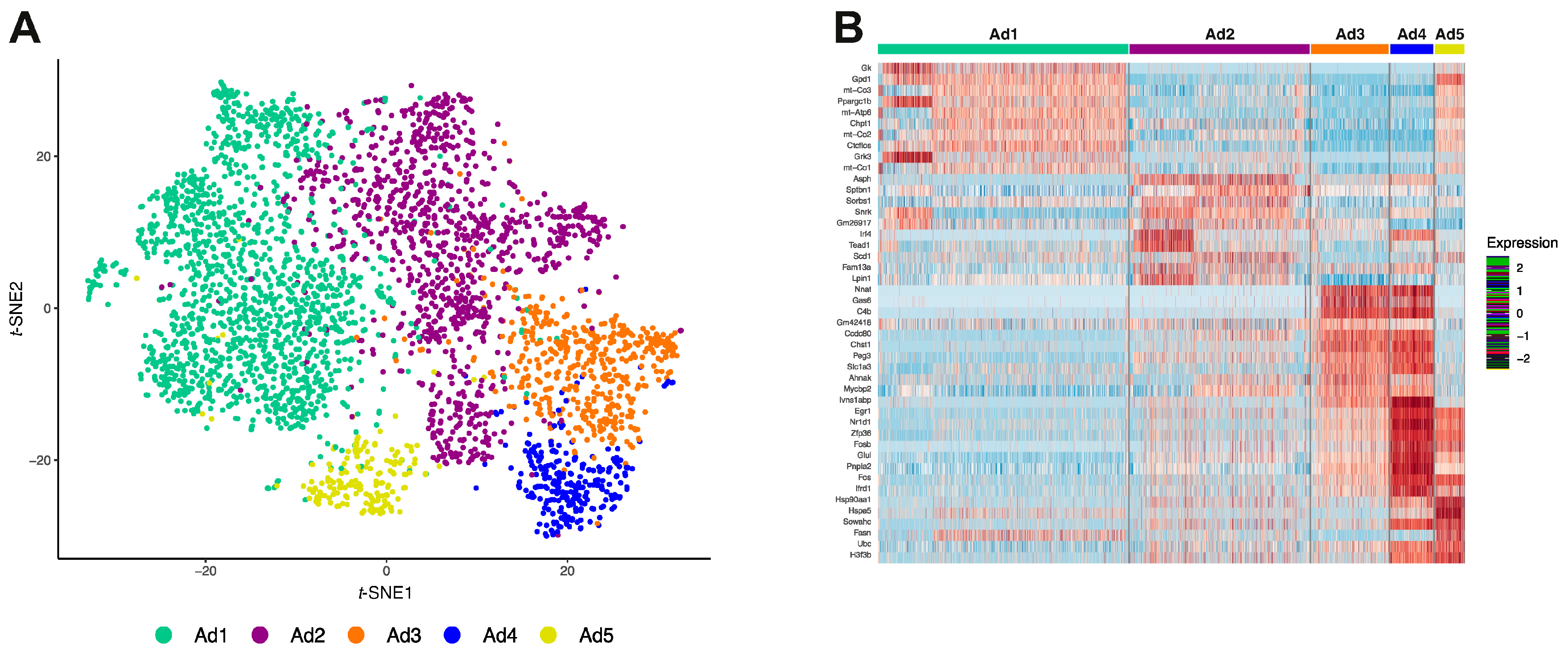
2.5. Optimal Mature Adipocyte Subclusters
2.6. Differential Expression and Enrichment Analysis
2.7. Transcriptome-Based Secretome Analysis
2.8. Membranome Prediction
2.9. Cell-Cell Interaction
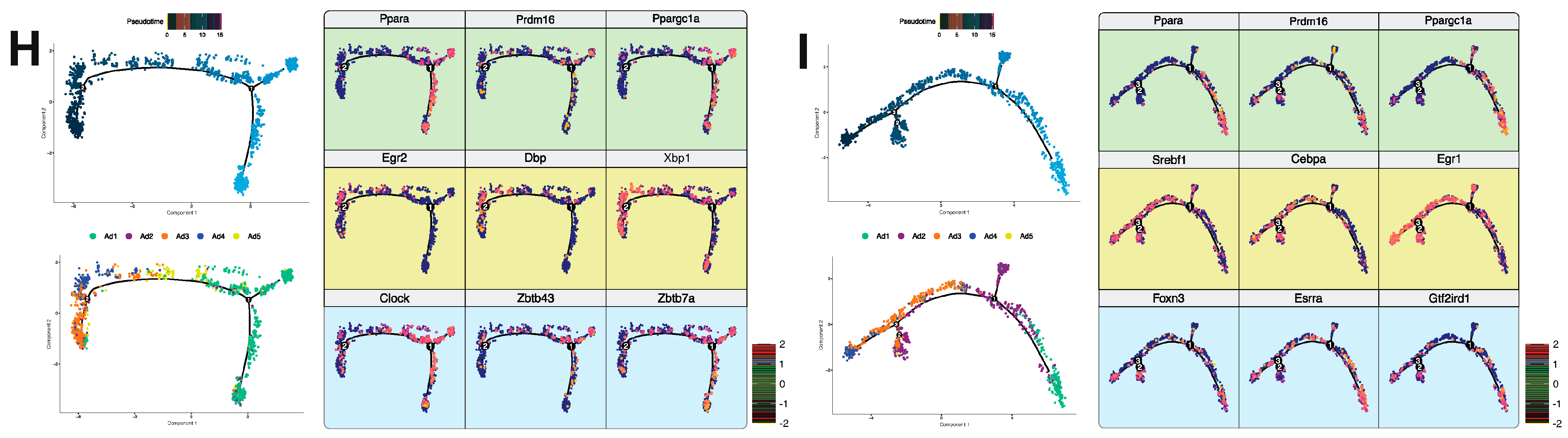
2.10. Pseudotime Analysis
2.11. Transcription Factors (tfs) Enrichment Analysis and Protein-Protein Interaction (PPI)
2.12. Data Representation
2.13. Data and Code Availability
3. Results
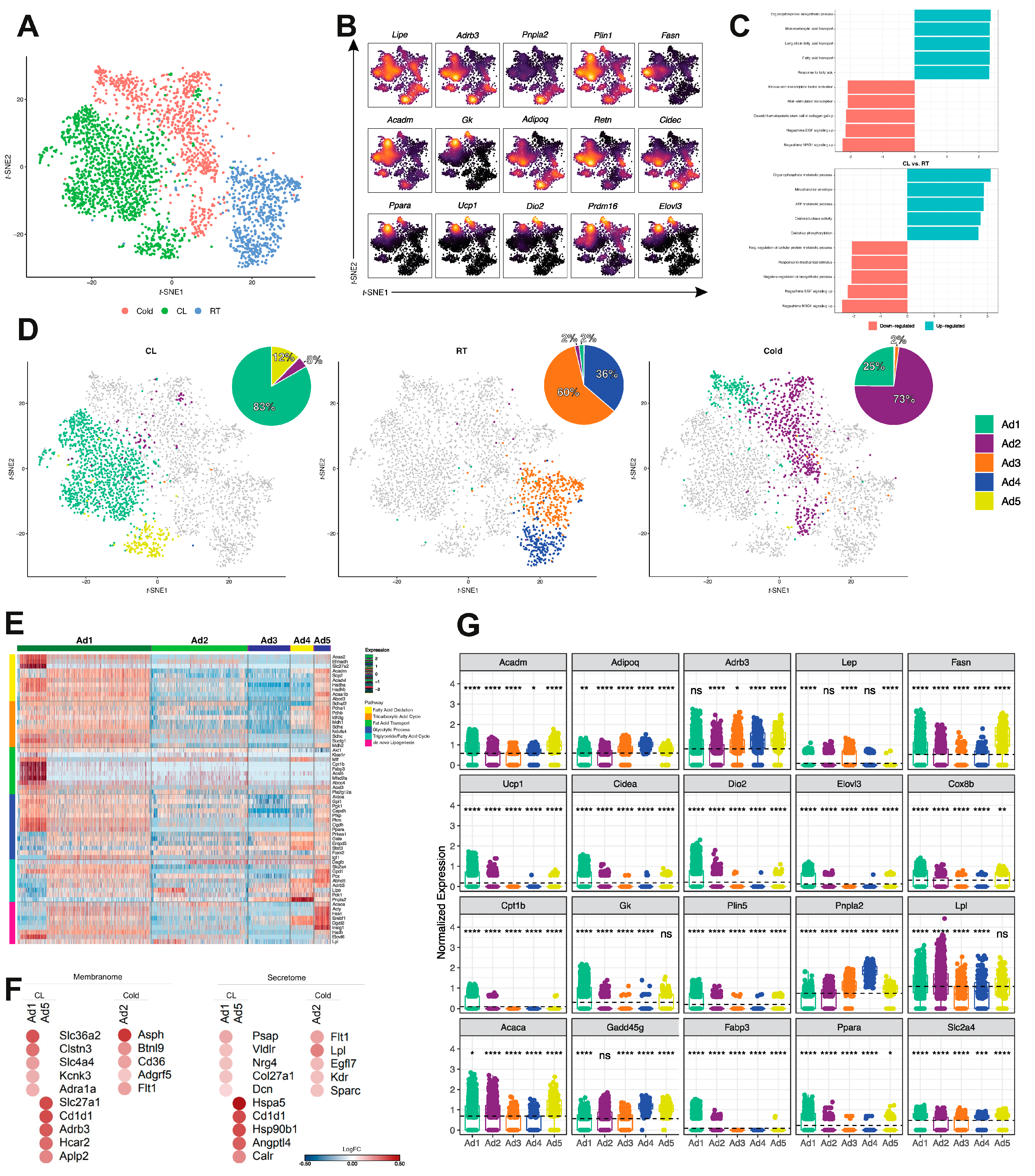
3.1. Multidimensional snRNA-Seq Reconstruction Reveals Distinct Adipocyte Subpopulations Derived from Mouse iWAT
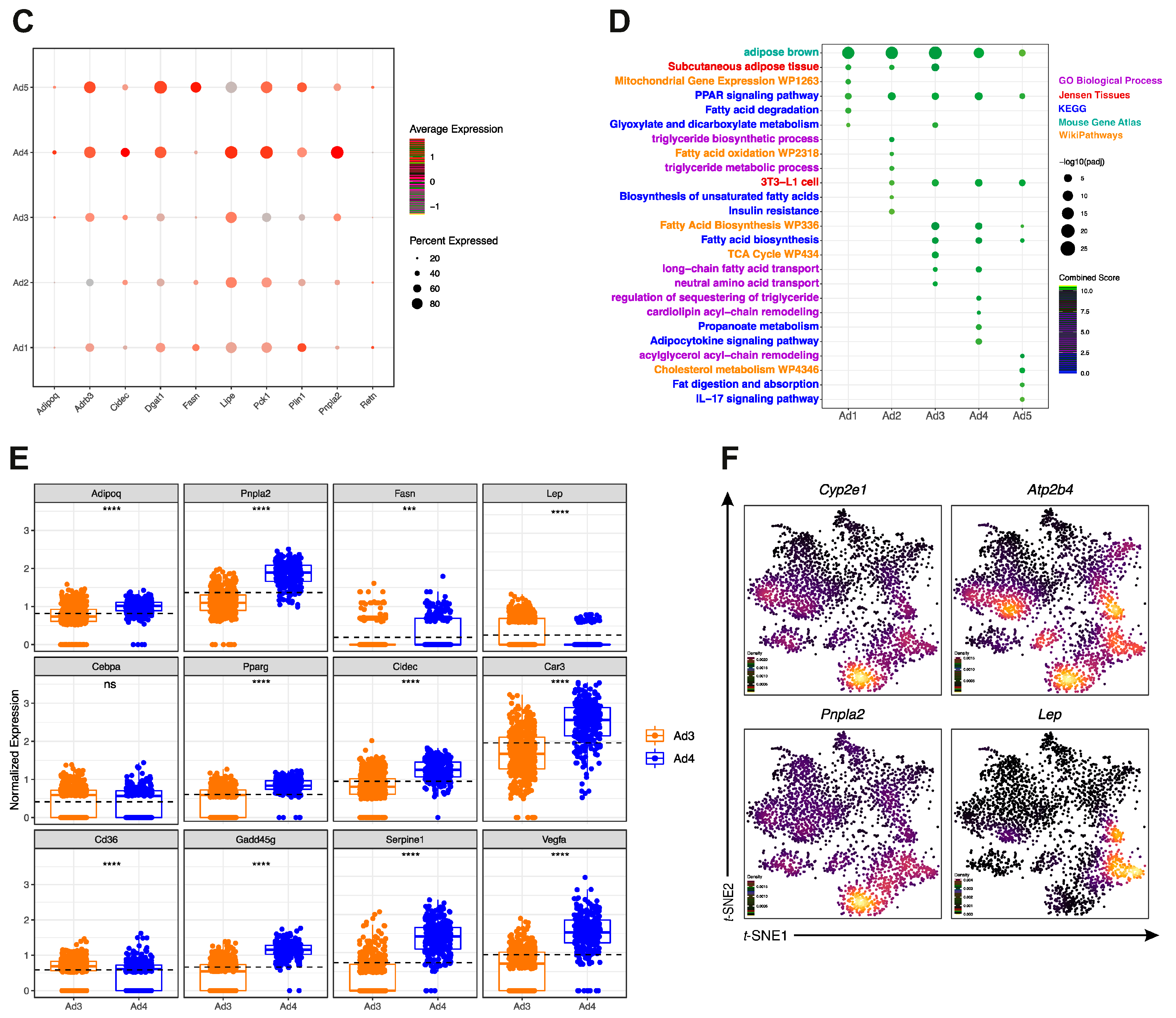
3.2. Reclustering of Adipocyte Clusters Reveals Two Distinct Mature Adipocyte Populations at Room Temperature
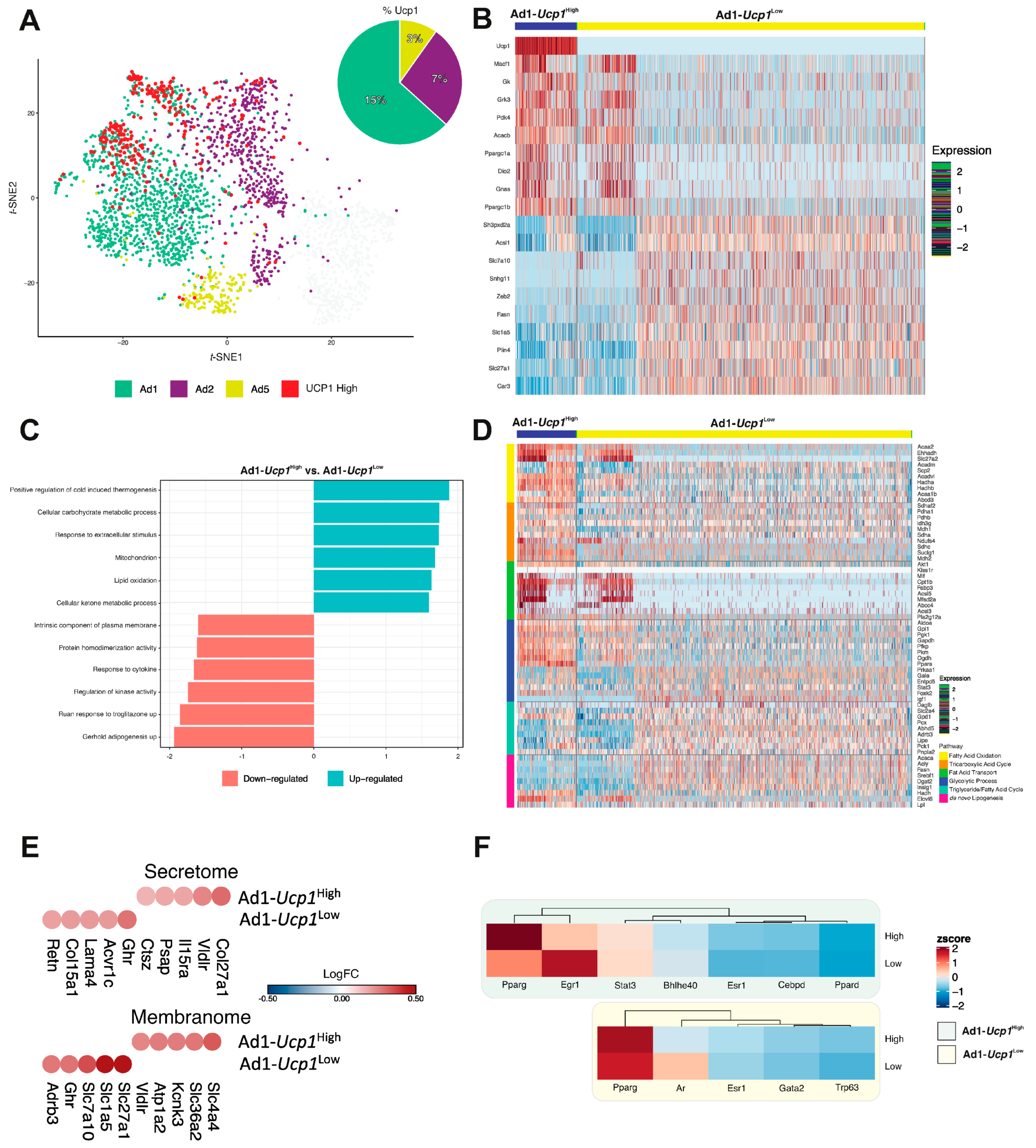
3.3. Identification of a Unique Adipocyte Thermogenic Subpopulation Corresponding to Both Cold and CL-Treatment
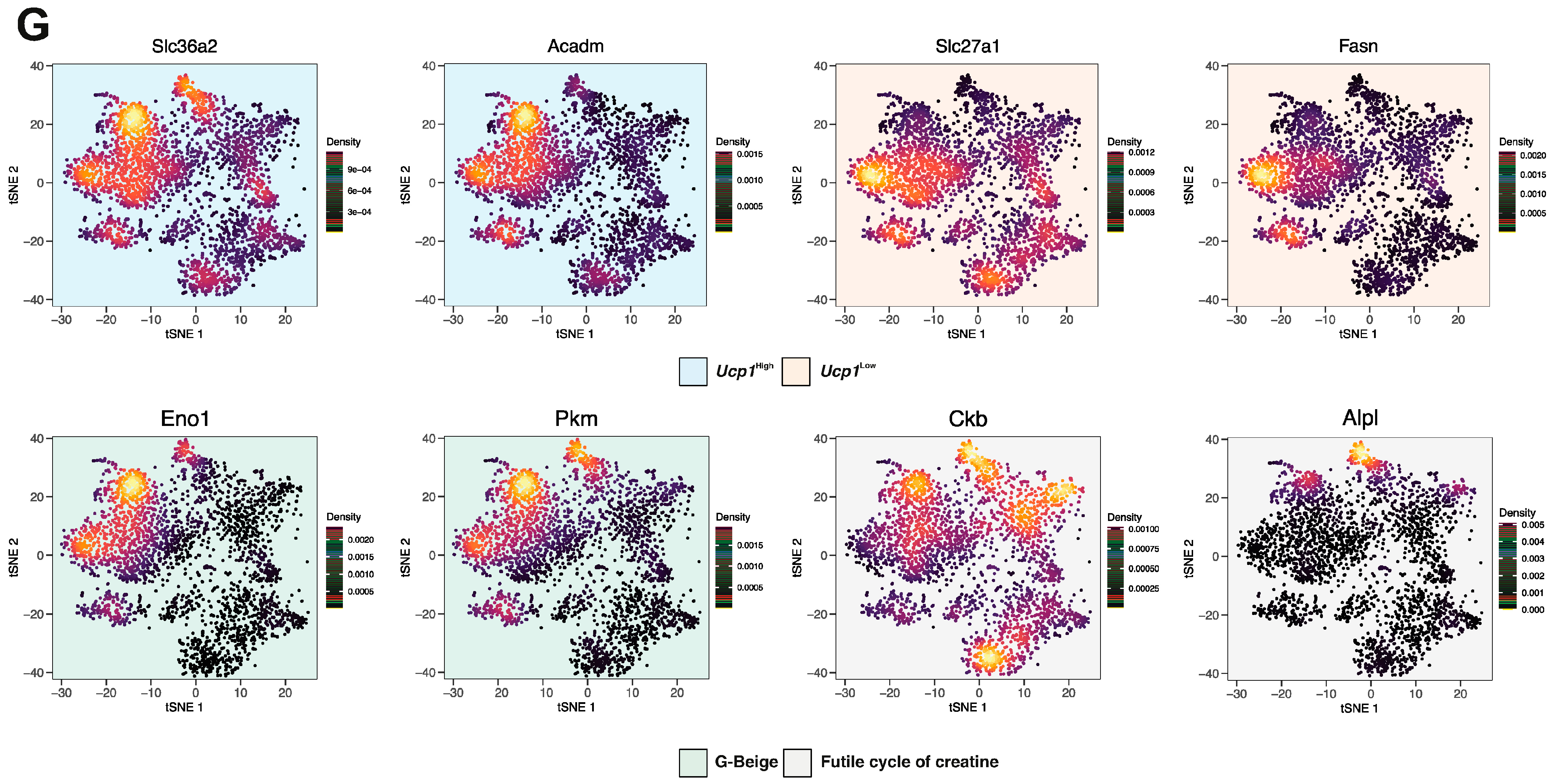
3.4. Ad1-Ucp1High Subpopulation Shows a “Classical” Thermogenic Profile
4. Discussion
5. Limitations Should Be Addressed
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Friedman, J.M.; Leibel, R.L.; Siegel, D.S.; Walsh, J.; Bahary, N. Molecular Mapping of the Mouse Ob Mutation. Genomics 1991, 11, 1054–1062. [Google Scholar] [CrossRef]
- Scherer, P.E. Adipose Tissue: From Lipid Storage Compartment to Endocrine Organ. Diabetes 2006, 55, 1537–1545. [Google Scholar] [CrossRef]
- Lee, M.-J.; Wu, Y.; Fried, S.K. Adipose Tissue Heterogeneity: Implication of Depot Differences in Adipose Tissue for Obesity Complications. Mol. Asp. Med. 2013, 34, 1–11. [Google Scholar] [CrossRef]
- Rosenwald, M.; Wolfrum, C. The Origin and Definition of Brite versus White and Classical Brown Adipocytes. Adipocyte 2014, 3, 4–9. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Liu, L.; Lin, J.Z.; Aprahamian, T.R.; Farmer, S.R. Browning of White Adipose Tissue with Roscovitine Induces a Distinct Population of UCP1 + Adipocytes. Cell Metab. 2016, 24, 835–847. [Google Scholar] [CrossRef]
- Pollard, A.E.; Carling, D. Thermogenic Adipocytes: Lineage, Function and Therapeutic Potential. Biochem. J. 2020, 477, 2071–2093. [Google Scholar] [CrossRef]
- Wu, J.; Boström, P.; Sparks, L.M.; Ye, L.; Choi, J.H.; Giang, A.-H.; Khandekar, M.; Virtanen, K.A.; Nuutila, P.; Schaart, G.; et al. Beige Adipocytes Are a Distinct Type of Thermogenic Fat Cell in Mouse and Human. Cell 2012, 150, 366–376. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Berry, D.C.; Graff, J.M. Distinct Cellular and Molecular Mechanisms for Β3 Adrenergic Receptor-Induced Beige Adipocyte Formation. eLife 2017, 6, e30329. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.-H.; Petkova, A.P.; Granneman, J.G. Identification of an Adipogenic Niche for Adipose Tissue Remodeling and Restoration. Cell Metab. 2013, 18, 355–367. [Google Scholar] [CrossRef] [PubMed]
- Kir, S.; White, J.P.; Kleiner, S.; Kazak, L.; Cohen, P.; Baracos, V.E.; Spiegelman, B.M. Tumour-Derived PTH-Related Protein Triggers Adipose Tissue Browning and Cancer Cachexia. Nature 2014, 513, 100–104. [Google Scholar] [CrossRef]
- Henriques, F.; Lopes, M.A.; Franco, F.O.; Knobl, P.; Santos, K.B.; Bueno, L.L.; Correa, V.A.; Bedard, A.H.; Guilherme, A.; Birbrair, A.; et al. Toll-Like Receptor-4 Disruption Suppresses Adipose Tissue Remodeling and Increases Survival in Cancer Cachexia Syndrome. Sci. Rep. 2018, 8, 18024. [Google Scholar] [CrossRef] [PubMed]
- Chouchani, E.T.; Kazak, L.; Spiegelman, B.M. New Advances in Adaptive Thermogenesis: UCP1 and Beyond. Cell Metab. 2019, 29, 27–37. [Google Scholar] [CrossRef] [PubMed]
- Leal, L.G.; Lopes, M.A.; Batista, M.L. Physical Exercise-Induced Myokines and Muscle-Adipose Tissue Crosstalk: A Review of Current Knowledge and the Implications for Health and Metabolic Diseases. Front. Physiol. 2018, 9, 1307. [Google Scholar] [CrossRef]
- De Jong, J.M.A.; Sun, W.; Pires, N.D.; Frontini, A.; Balaz, M.; Jespersen, N.Z.; Feizi, A.; Petrovic, K.; Fischer, A.W.; Bokhari, M.H.; et al. Human Brown Adipose Tissue Is Phenocopied by Classical Brown Adipose Tissue in Physiologically Humanized Mice. Nat. Metab. 2019, 1, 830–843. [Google Scholar] [CrossRef]
- Rosenwald, M.; Perdikari, A.; Rülicke, T.; Wolfrum, C. Bi-Directional Interconversion of Brite and White Adipocytes. Nat. Cell Biol. 2013, 15, 659–667. [Google Scholar] [CrossRef]
- Zheng, G.X.Y.; Terry, J.M.; Belgrader, P.; Ryvkin, P.; Bent, Z.W.; Wilson, R.; Ziraldo, S.B.; Wheeler, T.D.; McDermott, G.P.; Zhu, J.; et al. Massively Parallel Digital Transcriptional Profiling of Single Cells. Nat. Commun. 2017, 8, 14049. [Google Scholar] [CrossRef] [PubMed]
- Ziegenhain, C.; Vieth, B.; Parekh, S.; Reinius, B.; Guillaumet-Adkins, A.; Smets, M.; Leonhardt, H.; Heyn, H.; Hellmann, I.; Enard, W. Comparative Analysis of Single-Cell RNA Sequencing Methods. Mol. Cell 2017, 65, 631–643.e4. [Google Scholar] [CrossRef]
- Deutsch, A.; Feng, D.; Pessin, J.E.; Shinoda, K. The Impact of Single-Cell Genomics on Adipose Tissue Research. Int. J. Mol. Sci. 2020, 21, 4773. [Google Scholar] [CrossRef]
- Burl, R.B.; Ramseyer, V.D.; Rondini, E.A.; Pique-Regi, R.; Lee, Y.-H.; Granneman, J.G. Deconstructing Adipogenesis Induced by Β3-Adrenergic Receptor Activation with Single-Cell Expression Profiling. Cell Metab. 2018, 28, 300–309.e4. [Google Scholar] [CrossRef] [PubMed]
- Cho, D.S.; Lee, B.; Doles, J.D. Refining the Adipose Progenitor Cell Landscape in Healthy and Obese Visceral Adipose Tissue Using Single-Cell Gene Expression Profiling. Life Sci. Alliance 2019, 2, e201900561. [Google Scholar] [CrossRef] [PubMed]
- Gu, W.; Nowak, W.N.; Xie, Y.; Le Bras, A.; Hu, Y.; Deng, J.; Issa Bhaloo, S.; Lu, Y.; Yuan, H.; Fidanis, E.; et al. Single-Cell RNA-Sequencing and Metabolomics Analyses Reveal the Contribution of Perivascular Adipose Tissue Stem Cells to Vascular Remodeling. Arterioscler. Thromb. Vasc. Biol. 2019, 39, 2049–2066. [Google Scholar] [CrossRef] [PubMed]
- Henriques, F.; Bedard, A.H.; Guilherme, A.; Kelly, M.; Chi, J.; Zhang, P.; Lifshitz, L.M.; Bellvé, K.; Rowland, L.A.; Yenilmez, B.; et al. Single-Cell RNA Profiling Reveals Adipocyte to Macrophage Signaling Sufficient to Enhance Thermogenesis. Cell Rep. 2020, 32, 107998. [Google Scholar] [CrossRef]
- Hepler, C.; Shan, B.; Zhang, Q.; Henry, G.H.; Shao, M.; Vishvanath, L.; Ghaben, A.L.; Mobley, A.B.; Strand, D.; Hon, G.C.; et al. Identification of Functionally Distinct Fibro-Inflammatory and Adipogenic Stromal Subpopulations in Visceral Adipose Tissue of Adult Mice. eLife 2018, 7, e39636. [Google Scholar] [CrossRef] [PubMed]
- Jaitin, D.A.; Adlung, L.; Thaiss, C.A.; Weiner, A.; Li, B.; Descamps, H.; Lundgren, P.; Bleriot, C.; Liu, Z.; Deczkowska, A.; et al. Lipid-Associated Macrophages Control Metabolic Homeostasis in a Trem2-Dependent Manner. Cell 2019, 178, 686–698.e14. [Google Scholar] [CrossRef]
- Merrick, D.; Sakers, A.; Irgebay, Z.; Okada, C.; Calvert, C.; Morley, M.P.; Percec, I.; Seale, P. Identification of a Mesenchymal Progenitor Cell Hierarchy in Adipose Tissue. Science 2019, 364, eaav2501. [Google Scholar] [CrossRef]
- Schwalie, P.C.; Dong, H.; Zachara, M.; Russeil, J.; Alpern, D.; Akchiche, N.; Caprara, C.; Sun, W.; Schlaudraff, K.-U.; Soldati, G.; et al. A Stromal Cell Population That Inhibits Adipogenesis in Mammalian Fat Depots. Nature 2018, 559, 103–108. [Google Scholar] [CrossRef] [PubMed]
- Spallanzani, R.G.; Zemmour, D.; Xiao, T.; Jayewickreme, T.; Li, C.; Bryce, P.J.; Benoist, C.; Mathis, D. Distinct Immunocyte-Promoting and Adipocyte-Generating Stromal Components Coordinate Adipose Tissue Immune and Metabolic Tenors. Sci. Immunol. 2019, 4, eaaw3658. [Google Scholar] [CrossRef]
- The Tabula Muris Consortium; Overall Coordination; Logistical Coordination; Organ Collection and Processing; Library Preparation and Sequencing; Computational Data Analysis; Cell Type Annotation; Writing Group; Supplemental Text Writing Group; Principal Investigators. Single-Cell Transcriptomics of 20 Mouse Organs Creates a Tabula Muris. Nature 2018, 562, 367–372. [Google Scholar] [CrossRef] [PubMed]
- Weinstock, A.; Brown, E.J.; Garabedian, M.L.; Pena, S.; Sharma, M.; Lafaille, J.; Moore, K.J.; Fisher, E.A. Single-Cell RNA Sequencing of Visceral Adipose Tissue Leukocytes Reveals That Caloric Restriction Following Obesity Promotes the Accumulation of a Distinct Macrophage Population with Features of Phagocytic Cells. Immunometabolism 2019, 1, e190008. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Shao, M.; Hepler, C.; Zi, Z.; Zhao, S.; An, Y.A.; Zhu, Y.; Ghaben, A.L.; Wang, M.; Li, N.; et al. Dermal Adipose Tissue Has High Plasticity and Undergoes Reversible Dedifferentiation in Mice. J. Clin. Investig. 2019, 129, 5327–5342. [Google Scholar] [CrossRef] [PubMed]
- Acosta, J.R.; Joost, S.; Karlsson, K.; Ehrlund, A.; Li, X.; Aouadi, M.; Kasper, M.; Arner, P.; Rydén, M.; Laurencikiene, J. Single Cell Transcriptomics Suggest That Human Adipocyte Progenitor Cells Constitute a Homogeneous Cell Population. Stem Cell Res. Ther. 2017, 8, 250. [Google Scholar] [CrossRef]
- Raajendiran, A.; Ooi, G.; Bayliss, J.; O’Brien, P.E.; Schittenhelm, R.B.; Clark, A.K.; Taylor, R.A.; Rodeheffer, M.S.; Burton, P.R.; Watt, M.J. Identification of Metabolically Distinct Adipocyte Progenitor Cells in Human Adipose Tissues. Cell Rep. 2019, 27, 1528–1540.e7. [Google Scholar] [CrossRef] [PubMed]
- Vijay, J.; Gauthier, M.-F.; Biswell, R.L.; Louiselle, D.A.; Johnston, J.J.; Cheung, W.A.; Belden, B.; Pramatarova, A.; Biertho, L.; Gibson, M.; et al. Single-Cell Analysis of Human Adipose Tissue Identifies Depot- and Disease-Specific Cell Types. Nat. Metab. 2020, 2, 97–109. [Google Scholar] [CrossRef]
- Song, A.; Dai, W.; Jang, M.J.; Medrano, L.; Li, Z.; Zhao, H.; Shao, M.; Tan, J.; Li, A.; Ning, T.; et al. Low- and High-Thermogenic Brown Adipocyte Subpopulations Coexist in Murine Adipose Tissue. J. Clin. Investig. 2019, 130, 247–257. [Google Scholar] [CrossRef] [PubMed]
- Rajbhandari, P.; Arneson, D.; Hart, S.K.; Ahn, I.S.; Diamante, G.; Santos, L.C.; Zaghari, N.; Feng, A.-C.; Thomas, B.J.; Vergnes, L.; et al. Single Cell Analysis Reveals Immune Cell–Adipocyte Crosstalk Regulating the Transcription of Thermogenic Adipocytes. eLife 2019, 8, e49501. [Google Scholar] [CrossRef] [PubMed]
- Sárvári, A.K.; Van Hauwaert, E.L.; Markussen, L.K.; Gammelmark, E.; Marcher, A.-B.; Ebbesen, M.F.; Nielsen, R.; Brewer, J.R.; Madsen, J.G.S.; Mandrup, S. Plasticity of Epididymal Adipose Tissue in Response to Diet-Induced Obesity at Single-Nucleus Resolution. Cell Metab. 2021, 33, 437–453.e5. [Google Scholar] [CrossRef]
- Nguyen, Q.H.; Pervolarakis, N.; Nee, K.; Kessenbrock, K. Experimental Considerations for Single-Cell RNA Sequencing Approaches. Front. Cell Dev. Biol. 2018, 6, 108. [Google Scholar] [CrossRef]
- Sun, W.; Dong, H.; Balaz, M.; Slyper, M.; Drokhlyansky, E.; Colleluori, G.; Giordano, A.; Kovanicova, Z.; Stefanicka, P.; Balazova, L.; et al. SnRNA-Seq Reveals a Subpopulation of Adipocytes That Regulates Thermogenesis. Nature 2020, 587, 98–102. [Google Scholar] [CrossRef]
- Butler, A.; Hoffman, P.; Smibert, P.; Papalexi, E.; Satija, R. Integrating Single-Cell Transcriptomic Data across Different Conditions, Technologies, and Species. Nat. Biotechnol. 2018, 36, 411–420. [Google Scholar] [CrossRef]
- Stuart, T.; Butler, A.; Hoffman, P.; Hafemeister, C.; Papalexi, E.; Mauck, W.M.; Hao, Y.; Stoeckius, M.; Smibert, P.; Satija, R. Comprehensive Integration of Single-Cell Data. Cell 2019, 177, 1888–1902.e21. [Google Scholar] [CrossRef]
- Tirosh, I.; Izar, B.; Prakadan, S.M.; Wadsworth, M.H.; Treacy, D.; Trombetta, J.J.; Rotem, A.; Rodman, C.; Lian, C.; Murphy, G.; et al. Dissecting the Multicellular Ecosystem of Metastatic Melanoma by Single-Cell RNA-Seq. Science 2016, 352, 189–196. [Google Scholar] [CrossRef]
- Hafemeister, C.; Satija, R. Normalization and Variance Stabilization of Single-Cell RNA-Seq Data Using Regularized Negative Binomial Regression. Genome Biol. 2019, 20, 296. [Google Scholar] [CrossRef] [PubMed]
- Baran, Y.; Bercovich, A.; Sebe-Pedros, A.; Lubling, Y.; Giladi, A.; Chomsky, E.; Meir, Z.; Hoichman, M.; Lifshitz, A.; Tanay, A. MetaCell: Analysis of Single-Cell RNA-Seq Data Using K-Nn Graph Partitions. Genome Biol. 2019, 20, 206. [Google Scholar] [CrossRef]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma, A. Enrichr: Interactive and Collaborative HTML5 Gene List Enrichment Analysis Tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A Comprehensive Gene Set Enrichment Analysis Web Server 2016 Update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef]
- Linderman, G.C.; Zhao, J.; Kluger, Y. Zero-Preserving Imputation of ScRNA-Seq Data Using Low-Rank Approximation. BioRxiv 2018. [Google Scholar] [CrossRef]
- Alquicira-Hernandez, J.; Powell, J.E. Nebulosa recovers single-cell gene expression signals by kernel density estimation. Bioinformatics 2021, 37, 2485–2487. [Google Scholar] [CrossRef]
- The UniProt Consortium. UniProt: A Worldwide Hub of Protein Knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef] [PubMed]
- The Gene Ontology Consortium. Gene Ontology Consortium: Going Forward. Nucleic Acids Res. 2015, 43, D1049–D1056. [Google Scholar] [CrossRef] [PubMed]
- Roh, H.C.; Tsai, L.T.Y.; Shao, M.; Tenen, D.; Shen, Y.; Kumari, M.; Lyubetskaya, A.; Jacobs, C.; Dawes, B.; Gupta, R.K.; et al. Warming Induces Significant Reprogramming of Beige, but Not Brown, Adipocyte Cellular Identity. Cell Metab. 2018, 27, 1121–1137.e5. [Google Scholar] [CrossRef]
- Bendtsen, J.D.; Jensen, L.J.; Blom, N.; von Heijne, G.; Brunak, S. Feature-Based Prediction of Non-Classical and Leaderless Protein Secretion. Protein Eng. Des. Sel. 2004, 17, 349–356. [Google Scholar] [CrossRef]
- Armenteros, J.J.A. SignalP 5.0 Improves Signal Peptide Predictions Using Deep Neural Networks. Nat. Biotechnol. 2019, 37, 8. [Google Scholar] [CrossRef]
- Starruß, J.; de Back, W.; Brusch, L.; Deutsch, A. Morpheus: A User-Friendly Modeling Environment for Multiscale and Multicellular Systems Biology. Bioinformatics 2014, 30, 1331–1332. [Google Scholar] [CrossRef]
- Simpson, R.J.; Kalra, H.; Mathivanan, S. ExoCarta as a Resource for Exosomal Research. J. Extracell. Vesicles 2012, 1, 18374. [Google Scholar] [CrossRef]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L.L. Predicting Transmembrane Protein Topology with a Hidden Markov Model: Application to Complete Genomes11Edited by F. Cohen. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef]
- Efremova, M.; Vento-Tormo, M.; Teichmann, S.A.; Vento-Tormo, R. CellPhoneDB: Inferring Cell–Cell Communication from Combined Expression of Multi-Subunit Ligand–Receptor Complexes. Nat. Protoc. 2020, 15, 1484–1506. [Google Scholar] [CrossRef]
- Vento-Tormo, R.; Efremova, M.; Botting, R.A.; Turco, M.Y.; Vento-Tormo, M.; Meyer, K.B.; Park, J.-E.; Stephenson, E.; Polański, K.; Goncalves, A.; et al. Single-Cell Reconstruction of the Early Maternal–Fetal Interface in Humans. Nature 2018, 563, 347–353. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.; Mao, Q.; Tang, Y.; Wang, L.; Chawla, R.; Pliner, H.A.; Trapnell, C. Reversed Graph Embedding Resolves Complex Single-Cell Trajectories. Nat. Methods 2017, 14, 979–982. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.; Hill, A.; Packer, J.; Lin, D.; Ma, Y.-A.; Trapnell, C. Single-Cell MRNA Quantification and Differential Analysis with Census. Nat. Methods 2017, 14, 309–315. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Cacchiarelli, D.; Grimsby, J.; Pokharel, P.; Li, S.; Morse, M.; Lennon, N.J.; Livak, K.J.; Mikkelsen, T.S.; Rinn, J.L. The Dynamics and Regulators of Cell Fate Decisions Are Revealed by Pseudotemporal Ordering of Single Cells. Nat. Biotechnol. 2014, 32, 381–386. [Google Scholar] [CrossRef]
- Chen, E.Y.; Xu, H.; Gordonov, S.; Lim, M.P.; Perkins, M.H.; Ma’ayan, A. Expression2Kinases: MRNA Profiling Linked to Multiple Upstream Regulatory Layers. Bioinformatics 2012, 28, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Clarke, D.J.B.; Kuleshov, M.V.; Schilder, B.M.; Torre, D.; Duffy, M.E.; Keenan, A.B.; Lachmann, A.; Feldmann, A.S.; Gundersen, G.W.; Silverstein, M.C.; et al. EXpression2Kinases (X2K) Web: Linking Expression Signatures to Upstream Cell Signaling Networks. Nucleic Acids Res. 2018, 46, W171–W179. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Tang, Y.; Li, M.; Wang, J.; Pan, Y.; Wu, F.X. CytoNCA: A Cytoscape Plugin for Centrality Analysis and Evaluation of Protein Interaction Networks. Biosystems 2015, 127, 67–72. [Google Scholar] [CrossRef]
- Bardou, P.; Mariette, J.; Escudié, F.; Djemiel, C.; Klopp, C. Jvenn: An Interactive Venn Diagram Viewer. BMC Bioinform. 2014, 15, 293. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Sebastian, B.M.; Axhemi, A.; Chen, X.; Hillian, A.D.; Jacobsen, D.W.; Nagy, L.E. Ethanol-Induced Oxidative Stress via the CYP2E1 Pathway Disrupts Adiponectin Secretion from Adipocytes: ETHANOL-INDUCED OXIDATIVE STRESS. Alcohol. Clin. Exp. Res. 2012, 36, 214–222. [Google Scholar] [CrossRef]
- Chen, Y.; Ikeda, K.; Yoneshiro, T.; Scaramozza, A.; Tajima, K.; Wang, Q.; Kim, K.; Shinoda, K.; Sponton, C.H.; Brown, Z.; et al. Thermal Stress Induces Glycolytic Beige Fat Formation via a Myogenic State. Nature 2019, 565, 180–185. [Google Scholar] [CrossRef]
- Rahbani, J.F.; Roesler, A.; Hussain, M.F.; Samborska, B.; Dykstra, C.B.; Tsai, L.; Jedrychowski, M.P.; Vergnes, L.; Reue, K.; Spiegelman, B.M.; et al. Creatine Kinase B Controls Futile Creatine Cycling in Thermogenic Fat. Nature 2021, 590, 480–485. [Google Scholar] [CrossRef]
- Sun, Y.; Rahbani, J.F.; Jedrychowski, M.P.; Riley, C.L.; Vidoni, S.; Bogoslavski, D.; Hu, B.; Dumesic, P.A.; Zeng, X.; Wang, A.B.; et al. Mitochondrial TNAP Controls Thermogenesis by Hydrolysis of Phosphocreatine. Nature 2021, 593, 580–585. [Google Scholar] [CrossRef] [PubMed]
- Rabhi, N.; Belkina, A.C.; Desevin, K.; Cortez, B.N.; Farmer, S.R. Shifts of Immune Cell Populations Differ in Response to Different Effectors of Beige Remodeling of Adipose Tissue. IScience 2020, 23, 101765. [Google Scholar] [CrossRef]
- Lee, Y.-H.; Kim, S.-N.; Kwon, H.-J.; Granneman, J.G. Metabolic Heterogeneity of Activated Beige/Brite Adipocytes in Inguinal Adipose Tissue. Sci. Rep. 2017, 7, 39794. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.-X.; Zhao, X.-Y.; Meng, Z.-X.; Kern, M.; Dietrich, A.; Chen, Z.; Cozacov, Z.; Zhou, D.; Okunade, A.L.; Su, X.; et al. The Brown Fat–Enriched Secreted Factor Nrg4 Preserves Metabolic Homeostasis through Attenuation of Hepatic Lipogenesis. Nat. Med. 2014, 20, 1436–1443. [Google Scholar] [CrossRef] [PubMed]
- Rosell, M.; Kaforou, M.; Frontini, A.; Okolo, A.; Chan, Y.-W.; Nikolopoulou, E.; Millership, S.; Fenech, M.E.; MacIntyre, D.; Turner, J.O.; et al. Brown and White Adipose Tissues: Intrinsic Differences in Gene Expression and Response to Cold Exposure in Mice. Am. J. Physiol.-Endocrinol. Metab. 2014, 306, E945–E964. [Google Scholar] [CrossRef]
- Hotamisligil, G.; Shargill, N.; Spiegelman, B. Adipose Expression of Tumor Necrosis Factor-Alpha: Direct Role in Obesity-Linked Insulin Resistance. Science 1993, 259, 87–91. [Google Scholar] [CrossRef] [PubMed]
- Huh, J.Y.; Park, Y.J.; Kim, J.B. Adipocyte CD1d Determines Adipose Inflammation and Insulin Resistance in Obesity. Adipocyte 2018, 7, 129–136. [Google Scholar] [CrossRef]
- Kazak, L.; Chouchani, E.T.; Jedrychowski, M.P.; Erickson, B.K.; Shinoda, K.; Cohen, P.; Vetrivelan, R.; Lu, G.Z.; Laznik-Bogoslavski, D.; Hasenfuss, S.C.; et al. A Creatine-Driven Substrate Cycle Enhances Energy Expenditure and Thermogenesis in Beige Fat. Cell 2015, 163, 643–655. [Google Scholar] [CrossRef]
- Kajimura, S.; Spiegelman, B.M.; Seale, P. Brown and Beige Fat: Physiological Roles beyond Heat Generation. Cell Metab. 2015, 22, 546–559. [Google Scholar] [CrossRef]
- Szabo, I.; Zoratti, M. Now UCP(Rotein), Now You Don’t: UCP1 Is Not Mandatory for Thermogenesis. Cell Metab. 2017, 25, 761–762. [Google Scholar] [CrossRef][Green Version]
- Kajimura, S.; Seale, P.; Spiegelman, B.M. Transcriptional Control of Brown Fat Development. Cell Metab. 2010, 11, 257–262. [Google Scholar] [CrossRef]
- Puigserver, P.; Wu, Z.; Park, C.W.; Graves, R.; Wright, M.; Spiegelman, B.M. A Cold-Inducible Coactivator of Nuclear Receptors Linked to Adaptive Thermogenesis. Cell 1998, 92, 829–839. [Google Scholar] [CrossRef]
- Rajakumari, S.; Wu, J.; Ishibashi, J.; Lim, H.-W.; Giang, A.-H.; Won, K.-J.; Reed, R.R.; Seale, P. EBF2 Determines and Maintains Brown Adipocyte Identity. Cell Metab. 2013, 17, 562–574. [Google Scholar] [CrossRef] [PubMed]
- Seale, P.; Kajimura, S.; Spiegelman, B.M. Transcriptional Control of Brown Adipocyte Development and Physiological Function—Of Mice and Men. Genes Dev. 2009, 23, 788–797. [Google Scholar] [CrossRef] [PubMed]
- Milet, C.; Bléher, M.; Allbright, K.; Orgeur, M.; Coulpier, F.; Duprez, D.; Havis, E. Egr1 Deficiency Induces Browning of Inguinal Subcutaneous White Adipose Tissue in Mice. Sci. Rep. 2017, 7, 16153. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Biagi, C.A.O., Jr.; Cury, S.S.; Alves, C.P.; Rabhi, N.; Silva, W.A., Jr.; Farmer, S.R.; Carvalho, R.F.; Batista, M.L., Jr. Multidimensional Single-Nuclei RNA-Seq Reconstruction of Adipose Tissue Reveals Adipocyte Plasticity Underlying Thermogenic Response. Cells 2021, 10, 3073. https://doi.org/10.3390/cells10113073
Biagi CAO Jr., Cury SS, Alves CP, Rabhi N, Silva WA Jr., Farmer SR, Carvalho RF, Batista ML Jr. Multidimensional Single-Nuclei RNA-Seq Reconstruction of Adipose Tissue Reveals Adipocyte Plasticity Underlying Thermogenic Response. Cells. 2021; 10(11):3073. https://doi.org/10.3390/cells10113073
Chicago/Turabian StyleBiagi, Carlos Alberto Oliveira, Jr., Sarah Santiloni Cury, Cleidson Pádua Alves, Nabil Rabhi, Wilson Araujo Silva, Jr., Stephen R. Farmer, Robson Francisco Carvalho, and Miguel Luiz Batista, Jr. 2021. "Multidimensional Single-Nuclei RNA-Seq Reconstruction of Adipose Tissue Reveals Adipocyte Plasticity Underlying Thermogenic Response" Cells 10, no. 11: 3073. https://doi.org/10.3390/cells10113073
APA StyleBiagi, C. A. O., Jr., Cury, S. S., Alves, C. P., Rabhi, N., Silva, W. A., Jr., Farmer, S. R., Carvalho, R. F., & Batista, M. L., Jr. (2021). Multidimensional Single-Nuclei RNA-Seq Reconstruction of Adipose Tissue Reveals Adipocyte Plasticity Underlying Thermogenic Response. Cells, 10(11), 3073. https://doi.org/10.3390/cells10113073







