Viral Membrane Fusion Proteins and RNA Sorting Mechanisms for the Molecular Delivery by Exosomes
Abstract
1. Introduction
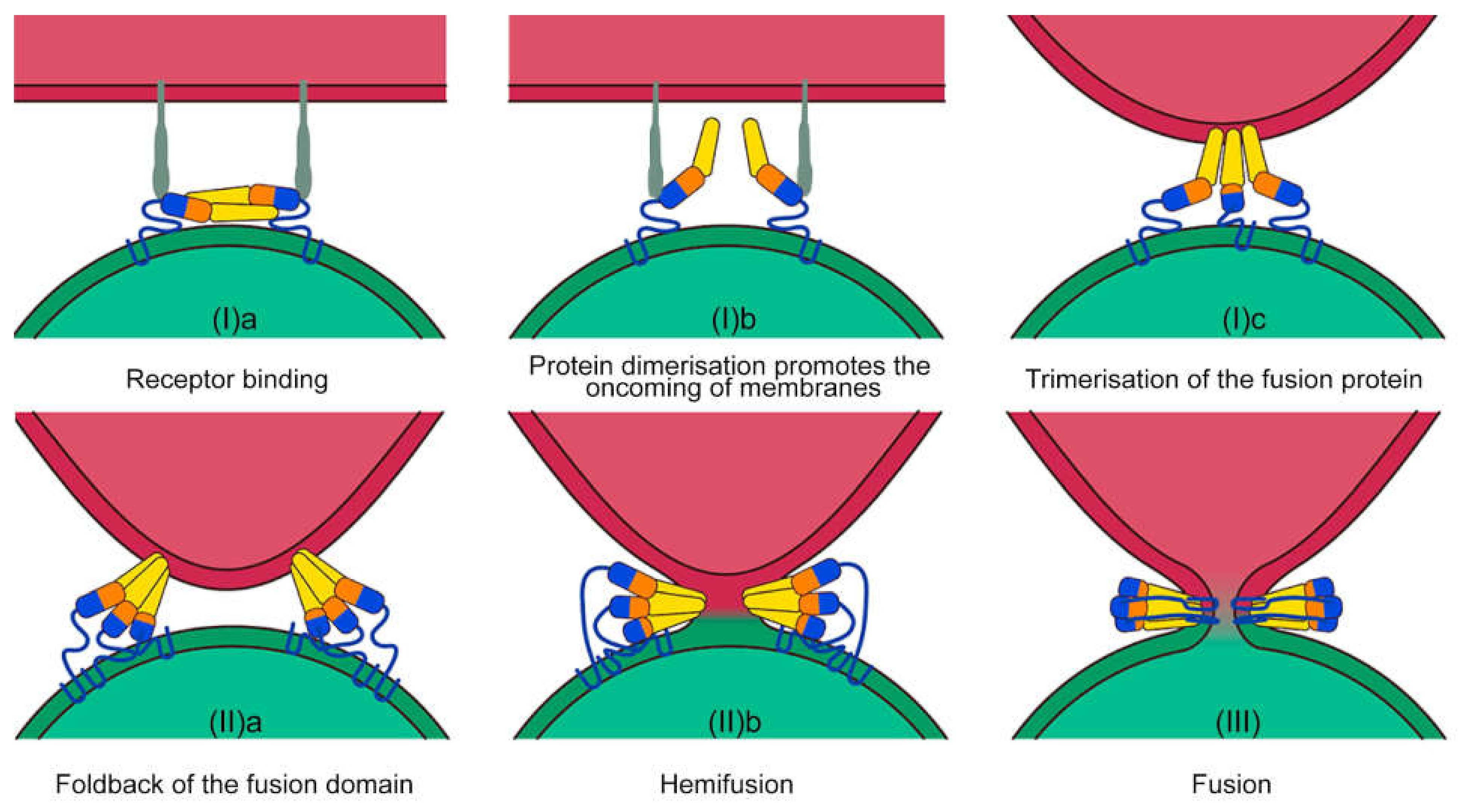
2. The Process of Membrane Fusion: Breaking the Energy Barrier
3. Eukaryotic Orthologues of Viral Fusion Proteins
4. Exploitation of the Host Mechanisms: EVs for Infection
5. Exploitation of Viral Sorting Mechanisms for RNA Loading into EVs
6. Conclusions and Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wilson, R.C.; Gilbert, L.A. The Promise and Challenge of in Vivo Delivery for Genome Therapeutics. ACS Chem. Biol. 2018, 13, 376–382. [Google Scholar] [CrossRef] [PubMed]
- Kimura, T.; Ferran, B.; Tsukahara, Y.; Shang, Q.; Desai, S.; Fedoce, A.; Pimentel, D.R.; Luptak, I.; Adachi, T.; Ido, Y.; et al. Production of adeno-associated virus vectors for in vitro and in vivo applications. Sci. Rep. 2019, 9, 13601. [Google Scholar] [CrossRef] [PubMed]
- Duan, D.; Yue, Y.; Yan, Z.; Yang, J.; Engelhardt, J.F. Endosomal processing limits gene transfer to polarized airway epithelia by adeno-associated virus. J. Clin. Invest. 2000, 105, 1573–1587. [Google Scholar] [CrossRef] [PubMed]
- Markusic, D.M.; Herzog, R.W.; Aslanidi, G.V.; Hoffman, B.E.; Li, B.; Li, M.; Jayandharan, G.R.; Ling, C.; Zolotukhin, I.; Ma, W.; et al. High-efficiency transduction and correction of murine hemophilia B using AAV2 vectors devoid of multiple surface-exposed tyrosines. Mol. Ther. 2010, 18, 2048–2056. [Google Scholar] [CrossRef] [PubMed]
- Zhong, L.; Li, B.; Jayandharan, G.; Mah, C.S.; Govindasamy, L.; Agbandje-McKenna, M.; Herzog, R.W.; Weigel-Van Aken, K.A.; Hobbs, J.A.; Zolotukhin, S.; et al. Tyrosine-phosphorylation of AAV2 vectors and its consequences on viral intracellular trafficking and transgene expression. Virology 2008, 381, 194–202. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Samulski, R.J. Engineering adeno-associated virus vectors for gene therapy. Nat. Rev. Genet. 2020, 21, 255–272. [Google Scholar] [CrossRef] [PubMed]
- Engin, A.B.; Hayes, A.W. The impact of immunotoxicity in evaluation of the nanomaterials safety. Toxicol. Res. Appl. 2018, 2, 239–244. [Google Scholar] [CrossRef]
- Li, X.; Liu, X.; Huang, J.; Fan, Y.; Cui, F.-Z. Biomedical investigation of CNT based coatings. Surf. Coatings Technol. 2011, 206, 759–766. [Google Scholar] [CrossRef]
- Meng, H.; Leong, W.; Leong, K.W.; Chen, C.; Zhao, Y. Walking the line: The fate of nanomaterials at biological barriers. Biomaterials 2018, 174, 41–53. [Google Scholar] [CrossRef]
- Al-Maawi, S.; Orlowska, A.; Sader, R.; James Kirkpatrick, C.; Ghanaati, S. In vivo cellular reactions to different biomaterials—Physiological and pathological aspects and their consequences. Semin. Immunol. 2017, 29, 49–61. [Google Scholar] [CrossRef]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [PubMed]
- Shurtleff, M.J.; Yao, J.; Qin, Y.; Nottingham, R.M.; Temoche-Diaz, M.M.; Schekman, R.; Lambowitz, A.M. Broad role for YBX1 in defining the small noncoding RNA composition of exosomes. Proc. Natl. Acad. Sci. USA 2017, 114, E8987–E8995. [Google Scholar] [CrossRef]
- Thakur, B.K.; Zhang, H.; Becker, A.; Matei, I.; Huang, Y.; Costa-Silva, B.; Zheng, Y.; Hoshino, A.; Brazier, H.; Xiang, J.; et al. Double-stranded DNA in exosomes: A novel biomarker in cancer detection. Cell Res. 2014, 24, 766–769. [Google Scholar] [CrossRef]
- Pegtel, D.M.; Cosmopoulos, K.; Thorley-Lawson, D.A.; Van Eijndhoven, M.A.J.; Hopmans, E.S.; Lindenberg, J.L.; De Gruijl, T.D.; Würdinger, T.; Middeldorp, J.M. Functional delivery of viral miRNAs via exosomes. Proc. Natl. Acad. Sci. USA 2010, 107, 6328–6333. [Google Scholar] [CrossRef]
- Sarko, D.K.; McKinney, C.E. Exosomes: Origins and therapeutic potential for neurodegenerative disease. Front. Neurosci. 2017, 11, 82–90. [Google Scholar] [CrossRef] [PubMed]
- Antimisiaris, S.G.; Mourtas, S.; Marazioti, A. Exosomes and exosome-inspired vesicles for targeted drug delivery. Pharmaceutics 2018, 10, 218–220. [Google Scholar] [CrossRef] [PubMed]
- Straubinger, R.M.; Hong, K.; Friend, D.S.; Papahadjopoulos, D. Endocytosis of liposomes and intracellular fate of encapsulated molecules: Encounter with a low pH compartment after internalization in coated vesicles. Cell 1983, 32, 1069–1079. [Google Scholar] [CrossRef]
- Harrison, S.C. Viral membrane fusion. Virology 2015, 479–480, 498–507. [Google Scholar] [CrossRef]
- Martens, S.; McMahon, H.T. Mechanisms of membrane fusion: Disparate players and common principles. Nat. Rev. Mol. Cell Biol. 2008, 9, 543–556. [Google Scholar] [CrossRef]
- Wilschut, J.; Hoekstra, D. Membrane fusion: From liposomes to biological membranes. Trends Biochem. Sci. 1984, 9, 479–483. [Google Scholar] [CrossRef]
- Kube, S.; Hersch, N.; Naumovska, E.; Gensch, T.; Hendriks, J.; Franzen, A.; Landvogt, L.; Siebrasse, J.P.; Kubitscheck, U.; Hoffmann, B.; et al. Fusogenic liposomes as nanocarriers for the delivery of intracellular proteins. Langmuir 2017, 33, 1051–1059. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Bahreman, A.; Daudey, G.; Bussmann, J.; Olsthoorn, R.C.L.; Kros, A. Drug delivery via cell membrane fusion using lipopeptide modified liposomes. ACS Cent. Sci. 2016, 2, 621–630. [Google Scholar] [CrossRef] [PubMed]
- Gurung, S.; Perocheau, D.; Touramanidou, L.; Baruteau, J. The exosome journey: From biogenesis to uptake and intracellular signalling. Cell Commun. Signal. 2021, 19, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, 64–78. [Google Scholar] [CrossRef]
- Pegtel, D.M.; Gould, S.J. Exosomes. Annu. Rev. Biochem. 2019, 88, 487–514. [Google Scholar] [CrossRef] [PubMed]
- Rand, R.P.; Parsegian, V.A. Physical force considerations in model and biological membranes. Can. J. Biochem. Cell Biol. 1984, 62, 752–759. [Google Scholar] [CrossRef] [PubMed]
- Parsegian, V.A.; Fuller, N.; Rand, R.P. Measured work of deformation and repulsion of lecithin bilayers. Proc. Natl. Acad. Sci. USA 1979, 76, 2750–2754. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.K. Architecture of a nascent viral fusion pore. EMBO J. 2010, 29, 1299–1311. [Google Scholar] [CrossRef]
- Rand, R.P.; Parsegian, V.A. Hydration forces between phospholipid bilayers. BBA Rev. Biomembr. 1989, 988, 351–376. [Google Scholar] [CrossRef]
- Lentz, B.R.; Malinin, V.; Haque, M.E.; Evans, K. Protein machines and lipid assemblies: Current views of cell membrane fusion. Curr. Opin. Struct. Biol. 2000, 10, 607–615. [Google Scholar] [CrossRef]
- Brukman, N.G.; Uygur, B.; Podbilewicz, B.; Chernomordik, L.V. How cells fuse. J. Cell Biol. 2019, 218, 1436–1451. [Google Scholar] [CrossRef]
- Mayer, A. Membrane fusion in eukaryotic cells. Annu. Rev. Cell Dev. Biol. 2002, 18, 109–116. [Google Scholar] [CrossRef]
- Kielian, M. Mechanisms of virus membrane fusion proteins. Annu. Rev. Virol. 2014, 1, 171–189. [Google Scholar] [CrossRef]
- Van Dongen, H.M.; Masoumi, N.; Witwer, K.W.; Pegtel, D.M. Extracellular Vesicles Exploit Viral Entry Routes for Cargo Delivery. Microbiol. Mol. Biol. Rev. 2016, 80, 369–386. [Google Scholar] [CrossRef]
- Skehel, J.J.; Wiley, D.C. Receptor binding and membrane fusion in virus entry: The influenza hemagglutinin. Annu. Rev. Biochem. 2000, 69, 531–569. [Google Scholar] [CrossRef]
- Stiasny, K.; Kössl, C.; Lepault, J.; Rey, F.A.; Heinz, F.X. Characterization of a structural intermediate of flavivirus membrane fusion. PLoS Pathog. 2007, 3, 0191–0199. [Google Scholar] [CrossRef]
- Ryham, R.J.; Klotz, T.S.; Yao, L.; Cohen, F.S. Calculating Transition Energy Barriers and Characterizing Activation States for Steps of Fusion. Biophys. J. 2016, 110, 1110–1124. [Google Scholar] [CrossRef]
- Efrat, A.; Chernomordik, L.V.; Kozlov, M.M. Point-like protrusion as a prestalk intermediate in membrane fusion pathway. Biophys. J. 2007, 92, L61. [Google Scholar] [CrossRef]
- Li, Y.; Han, X.; Lai, A.L.; Bushweller, J.H.; Cafiso, D.S.; Tamm, L.K. Membrane Structures of the Hemifusion-Inducing Fusion Peptide Mutant G1S and theFusion-Blocking Mutant G1V of Influenza Virus HemagglutininSuggest a Mechanism for Pore Opening in MembraneFusion. J. Virol. 2005, 79, 12065–12076. [Google Scholar] [CrossRef]
- Chao, L.H.; Klein, D.E.; Schmidt, A.G.; Peña, J.M.; Harrison, S.C. Sequential conformational rearrangements in flavivirus membrane fusion. Elife 2014, 3, 43–49. [Google Scholar] [CrossRef]
- Molotkovsky, R.J.; Galimzyanov, T.R.; Jiménez-Munguía, I.; Pavlov, K.V.; Batishchev, O.V.; Akimov, S.A. Switching between successful and dead-end intermediates in membrane fusion. Int. J. Mol. Sci. 2017, 18, 2598. [Google Scholar] [CrossRef]
- White, J.M.; Delos, S.E.; Brecher, M.; Schornberg, K. Structures and mechanisms of viral membrane fusion proteins: Multiple variations on a common theme. Crit. Rev. Biochem. Mol. Biol. 2008, 43, 189–219. [Google Scholar] [CrossRef]
- Kozlov, M.M.; McMahon, H.T.; Chernomordik, L.V. Protein-driven membrane stresses in fusion and fission. Trends Biochem. Sci. 2010, 35, 699–706. [Google Scholar] [CrossRef]
- Chernomordik, L.V.; Kozlov, M.M. Protein-Lipid Interplay in Fusion and Fission of Biological Membranes. Annu. Rev. Biochem. 2003, 72, 175–207. [Google Scholar] [CrossRef]
- Harrison, S.C. Mechanism of Membrane Fusion by Viral Envelope Proteins. Adv. Virus Res. 2005, 64, 231–261. [Google Scholar]
- Gaudin, Y.; Ruigrok, R.W.; Knossow, M.; Flamand, A. Low-pH conformational changes of rabies virus glycoprotein and their role in membrane fusion. J. Virol. 1993, 67, 1365–1372. [Google Scholar] [CrossRef]
- Pancera, M.; Zhou, T.; Druz, A.; Georgiev, I.S.; Soto, C.; Gorman, J.; Huang, J.; Acharya, P.; Chuang, G.Y.; Ofek, G.; et al. Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature 2014, 514, 455–461. [Google Scholar] [CrossRef]
- Shi, Y.; Barton, K.; De Maria, A.; Petrash, J.M.; Shiels, A.; Bassnett, S. The stratified syncytium of the vertebrate lens. J. Cell Sci. 2009, 122, 1607–1615. [Google Scholar] [CrossRef]
- Chen, E.H.; Pryce, B.A.; Tzeng, J.A.; Gonzalez, G.A.; Olson, E.N. Control of myoblast fusion by a guanine nucleotide exchange factor, loner, and its effector ARF6. Cell 2003, 114, 751–762. [Google Scholar] [CrossRef]
- Valansi, C.; Moi, D.; Leikina, E.; Matveev, E.; Graña, M.; Chernomordik, L.V.; Romero, H.; Aguilar, P.S.; Podbilewicz, B. Arabidopsis HAP2/GCS1 is a gamete fusion protein homologous to somatic and viral fusogens. J. Cell Biol. 2017, 216, 571–581. [Google Scholar] [CrossRef]
- Podbilewicz, B. Virus and cell fusion mechanisms. Annu. Rev. Cell Dev. Biol. 2014, 30, 111–139. [Google Scholar] [CrossRef]
- Vance, T.D.R.; Lee, J.E. Virus and eukaryote fusogen superfamilies. Curr. Biol. 2020, 30, R750–R754. [Google Scholar] [CrossRef]
- Soygur, B.; Sati, L. The role of syncytins in human reproduction and reproductive organ cancers. Reproduction 2016, 152, R167–R178. [Google Scholar] [CrossRef]
- Fu, B.; Ma, H.; Liu, D. Endogenous retroviruses function as gene expression regulatory elements during mammalian pre-implantation embryo development. Int. J. Mol. Sci. 2019, 20, 790. [Google Scholar] [CrossRef]
- Denner, J. Expression and function of endogenous retroviruses in the placenta. Apmis 2016, 124, 31–43. [Google Scholar] [CrossRef] [PubMed]
- Lokossou, A.G.; Toudic, C.; Nguyen, P.T.; Elisseeff, X.; Vargas, A.; Rassart, É.; Lafond, J.; Leduc, L.; Bourgault, S.; Gilbert, C.; et al. Endogenous retrovirus-encoded Syncytin-2 contributes to exosome-mediated immunosuppression of T cells. Biol. Reprod. 2020, 102, 185–198. [Google Scholar] [CrossRef] [PubMed]
- Blaise, S.; Ruggieri, A.; Dewannieux, M.; Cosset, F.-L.; Heidmann, T. Identification of an Envelope Protein from the FRD Family of Human Endogenous Retroviruses (HERV-FRD) Conferring Infectivity and Functional Conservation among Simians. J. Virol. 2004, 78, 1050–1054. [Google Scholar] [CrossRef]
- Badierah, R.A.; Uversky, V.N.; Redwan, E.M. Dancing with Trojan horses: An interplay between the extracellular vesicles and viruses. J. Biomol. Struct. Dyn. 2021, 39, 3034–3060. [Google Scholar] [CrossRef]
- Mause, S.F.; Weber, C. Microparticles: Protagonists of a novel communication network for intercellular information exchange. Circ. Res. 2010, 107, 1047–1057. [Google Scholar] [CrossRef]
- Zaborowski, M.P.; Balaj, L.; Breakefield, X.O.; Lai, C.P. Extracellular Vesicles: Composition, Biological Relevance, and Methods of Study. Bioscience 2015, 65, 783–797. [Google Scholar] [CrossRef]
- Hoen, E.N.; Cremer, T.; Gallo, R.C.; Margolis, L.B. Extracellular vesicles and viruses: Are they close relatives? Proc. Natl. Acad. Sci. USA 2016, 113, 9155–9161. [Google Scholar] [CrossRef] [PubMed]
- Wurdinger, T.; Gatson, N.N.; Balaj, L.; Kaur, B.; Breakefield, X.O.; Pegtel, D.M. Extracellular Vesicles and Their Convergence with Viral Pathways. Adv. Virol. 2012, 2012, 12. [Google Scholar] [CrossRef]
- Meckes, D.G. Exosomal Communication Goes Viral. J. Virol. 2015, 89, 5200–5203. [Google Scholar] [CrossRef] [PubMed]
- Ruiss, R.; Jochum, S.; Mocikat, R.; Hammerschmidt, W.; Zeidler, R. EBV-gp350 confers B-cell tropism to tailored exosomes is a neo-antigen in normal and malignant B cells-a new option for the treatment of B-CLL. PLoS ONE 2011, 6, e25294. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Woodson, M.; Sherman, M.B.; Neelakanta, G.; Sultana, H. Exosomes mediate Zika virus transmission through SMPD3 neutral Sphingomyelinase in cortical neurons. Emerg. Microbes Infect. 2019, 8, 307–326. [Google Scholar] [CrossRef]
- Kim, E.J.; Jacobs, M.W.; Ito-Cole, T.; Callaway, E.M. Improved Monosynaptic Neural Circuit Tracing Using Engineered Rabies Virus Glycoproteins. Cell Rep. 2016, 15, 692–699. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Woodson, M.; Neupane, B.; Bai, F.; Sherman, M.B.; Choi, K.H.; Neelakanta, G.; Sultana, H. Exosomes serve as novel modes of tick-borne flavivirus transmission from arthropod to human cells and facilitates dissemination of viral RNA and proteins to the vertebrate neuronal cells. PLoS Pathog. 2018, 14, e1006764. [Google Scholar] [CrossRef] [PubMed]
- Sanjuán, R. Collective Infectious Units in Viruses. Trends Microbiol. 2017, 25, 402–412. [Google Scholar] [CrossRef] [PubMed]
- Rozmyslowicz, T.; Majka, M.; Kijowski, J.; Murphy, S.L.; Conover, D.O.; Poncz, M.; Ratajczak, J.; Gaulton, G.N.; Ratajczak, M.Z. Platelet- and megakaryocyte-derived microparticles transfer CXCR4 receptor to CXCR4-null cells and make them susceptible to infection by X4-HIV. Aids 2003, 17, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Mack, M.; Kleinschmidt, A.; Brühl, H.; Klier, C.; Nelson, P.J.; Cihak, J.; Plachý, J.; Stangassinger, M.; Erfle, V.; Schlöndorff, D. Transfer of the chemokine receptor CCR5 between cells by membrane- derived microparticles: A mechanism for cellular human immunodeficiency virus 1 infection. Nat. Med. 2000, 6, 769–775. [Google Scholar] [CrossRef]
- Feng, Z.; Hensley, L.; McKnight, K.L.; Hu, F.; Madden, V.; Ping, L.; Jeong, S.H.; Walker, C.; Lanford, R.E.; Lemon, S.M. A pathogenic picornavirus acquires an envelope by hijacking cellular membranes. Nature 2013, 496, 367–371. [Google Scholar] [CrossRef] [PubMed]
- Yao, Z.; Qiao, Y.; Li, X.; Chen, J.; Ding, J.; Bai, L.; Shen, F.; Shi, B.; Liu, J.; Peng, L.; et al. Exosomes Exploit the Virus Entry Machinery and Pathway To Transmit Alpha Interferon-Induced Antiviral Activity. J. Virol. 2018, 92, e01578-18. [Google Scholar] [CrossRef]
- Yang, Y.; Han, Q.; Hou, Z.; Zhang, C.; Tian, Z.; Zhang, J. Exosomes mediate hepatitis B virus (HBV) transmission and NK-cell dysfunction. Cell. Mol. Immunol. 2017, 14, 465–475. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishnaiah, V.; Thumann, C.; Fofana, I.; Habersetzer, F.; Pan, Q.; De Ruiter, P.E.; Willemsen, R.; Demmers, J.A.A.; Raj, V.S.; Jenster, G.; et al. Exosome-mediated transmission of hepatitis C virus between human hepatoma Huh7.5 cells. Proc. Natl. Acad. Sci. USA 2013, 110, 13109–13113. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.E.; Rossignol, E.D.; Chang, D.; Zaia, J.; Forrester, I.; Raja, K.; Winbigler, H.; Nicastro, D.; Jackson, W.T.; Bullitt, E. Complexity and ultrastructure of infectious extracellular vesicles from cells infected by non-enveloped virus. Sci. Rep. 2020, 10, 7939. [Google Scholar] [CrossRef] [PubMed]
- Izquierdo-Useros, N.; Naranjo-Gómez, M.; Archer, J.; Hatch, S.C.; Erkizia, I.; Blanco, J.; Borràs, F.E.; Puertas, M.C.; Connor, J.H.; Fernádez-Figueras, M.T.; et al. Capture and transfer of HIV-1 particles by mature dendritic cells converges with the exosome-dissemination pathway. Blood 2009, 113, 2732–2741. [Google Scholar] [CrossRef]
- Kadiu, I.; Narayanasamy, P.; Dash, P.K.; Zhang, W.; Gendelman, H.E. Biochemical and Biologic Characterization of Exosomes and Microvesicles as Facilitators of HIV-1 Infection in Macrophages. J. Immunol. 2012, 189, 744–754. [Google Scholar] [CrossRef] [PubMed]
- Narayanan, A.; Iordanskiy, S.; Das, R.; Van Duyne, R.; Santos, S.; Jaworski, E.; Guendel, I.; Sampey, G.; Dalby, E.; Iglesias-Ussel, M.; et al. Exosomes derived from HIV-1-infected cells contain trans-activation response element RNA. J. Biol. Chem. 2013, 288, 20014–20033. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Wu, N.; Gan, X.; Yan, W.; Morrell, J.C.; Gould, S.J. Higher-order oligomerization targets plasma membrane proteins and HIV Gag to exosomes. PLoS Biol. 2007, 5, 1267–1283. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Schierer, S.; Blume, K.; Dindorf, J.; Wittki, S.; Xiang, W.; Ostalecki, C.; Koliha, N.; Wild, S.; Schuler, G.; et al. HIV-Nef and ADAM17-Containing Plasma Extracellular Vesicles Induce and Correlate with Immune Pathogenesis in Chronic HIV Infection. EBioMedicine 2016, 6, 103–113. [Google Scholar] [CrossRef]
- Raymond, A.D.; Campbell-Sims, T.C.; Khan, M.; Lang, M.; Huang, M.B.; Bond, V.C.; Powell, M.D. HIV type 1 Nef is released from infected cells in CD45+ microvesicles and is present in the plasma of HIV-infected individuals. AIDS Res. Hum. Retrovir. 2011, 27, 167–178. [Google Scholar] [CrossRef]
- Sims, B.; Farrow, A.L.; Williams, S.D.; Bansal, A.; Krendelchtchikov, A.; Matthews, Q.L. Tetraspanin blockage reduces exosome-mediated HIV-1 entry. Arch. Virol. 2018, 163, 1683. [Google Scholar] [CrossRef]
- Sims, B.; Farrow, A.L.; Williams, S.D.; Bansal, A.; Krendelchtchikov, A.; Gu, L.; Matthews, Q.L. Role of TIM-4 in exosome-dependent entry of HIV-1 into human immune cells. Int. J. Nanomedicine 2017, 12, 4823. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Ruiz, J.M.; Osuna-Ramos, J.F.; De Jesús-González, L.A.; Hurtado-Monzón, A.M.; Farfan-Morales, C.N.; Cervantes-Salazar, M.; Bolaños, J.; Cigarroa-Mayorga, O.E.; Martín-Martínez, E.S.; Medina, F.; et al. Isolation and characterization of exosomes released from mosquito cells infected with dengue virus. Virus Res. 2019, 266, 1–14. [Google Scholar] [CrossRef]
- Vora, A.; Zhou, W.; Londono-Renteria, B.; Woodson, M.; Sherman, M.B.; Colpitts, T.M.; Neelakanta, G.; Sultana, H. Arthropod EVs mediate dengue virus transmission through interaction with a tetraspanin domain containing glycoprotein Tsp29Fb. Proc. Natl. Acad. Sci. USA 2018, 115, E6604–E6613. [Google Scholar] [CrossRef] [PubMed]
- Pleet, M.L.; DeMarino, C.; Stonier, S.W.; Dye, J.M.; Jacobson, S.; Aman, M.J.; Kashanchi, F. Extracellular vesicles and ebola virus: A new mechanism of immune evasion. Viruses 2019, 11, 410. [Google Scholar] [CrossRef]
- Chahar, H.S.; Corsello, T.; Kudlicki, A.S.; Komaravelli, N.; Casola, A. Respiratory Syncytial Virus Infection Changes Cargo Composition of Exosome Released from Airway Epithelial Cells. Sci. Rep. 2018, 8, 387–401. [Google Scholar] [CrossRef]
- Sadeghipour, S.; Mathias, R.A. Herpesviruses hijack host exosomes for viral pathogenesis. Semin. Cell Dev. Biol. 2017, 67, 91–100. [Google Scholar] [CrossRef]
- Teow, S.Y.; Liew, K.; Khoo, A.S.B.; Peh, S.C. Pathogenic role of exosomes in epstein-barr virus (EBV)-associated cancers. Int. J. Biol. Sci. 2017, 13, 1276–1286. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Nanbo, A.; Sun, L.; Lin, Z. Extracellular vesicles in epstein-barr virus’ life cycle and pathogenesis. Microorganisms 2019, 7, 48. [Google Scholar] [CrossRef] [PubMed]
- Votteler, J.; Sundquist, W.I. Virus budding and the ESCRT pathway. Cell Host Microbe 2013, 14, 232–241. [Google Scholar] [CrossRef]
- Demirov, D.G.; Ono, A.; Orenstein, J.M.; Freed, E.O. Overexpression of the N-terminal domain of TSG101 inhibits HIV-1 budding by blocking late domain function. Proc. Natl. Acad. Sci. USA 2002, 99, 955–960. [Google Scholar] [CrossRef] [PubMed]
- Martin-Serrano, J.; Zang, T.; Bieniasz, P.D. Role of ESCRT-I in Retroviral Budding. J. Virol. 2003, 77, 4794–4804. [Google Scholar] [CrossRef] [PubMed]
- VerPlank, L.; Bouamr, F.; LaGrassa, T.J.; Agresta, B.; Kikonyogo, A.; Leis, J.; Carter, C.A. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55Gag. Proc. Natl. Acad. Sci. USA 2001, 98, 7724–7729. [Google Scholar] [CrossRef] [PubMed]
- Villarroya-Beltri, C.; Baixauli, F.; Mittelbrunn, M.; Fernández-Delgado, I.; Torralba, D.; Moreno-Gonzalo, O.; Baldanta, S.; Enrich, C.; Guerra, S.; Sánchez-Madrid, F. ISGylation controls exosome secretion by promoting lysosomal degradation of MVB proteins. Nat. Commun. 2016, 7, 13588. [Google Scholar] [CrossRef]
- Di Bonito, P.; Ridolfi, B.; Columba-Cabezas, S.; Giovannelli, A.; Chiozzini, C.; Manfredi, F.; Anticoli, S.; Arenaccio, C.; Federico, M. HPV-E7 delivered by engineered exosomes elicits a protective CD8+ T cell-mediated immune response. Viruses 2015, 7, 1079. [Google Scholar] [CrossRef]
- Manfredi, F.; Di Bonito, P.; Arenaccio, C.; Anticoli, S.; Federico, M. Incorporation of heterologous proteins in engineered exosomes. In Methods in Molecular Biology; Federico, M., Ed.; Humana Press: New York, NY, USA, 2016; Volume 1448, pp. 249–260. [Google Scholar]
- McNamara, R.P.; Costantini, L.M.; Myers, T.A.; Schouest, B.; Maness, N.J.; Griffith, J.D.; Damania, B.A.; Maclean, A.G.; Dittmer, D.P. Nef secretion into extracellular vesicles or exosomes is conserved across human and simian immunodeficiency viruses. MBio 2018, 9, e02344-17. [Google Scholar] [CrossRef] [PubMed]
- Anticoli, S.; Manfredi, F.; Chiozzini, C.; Arenaccio, C.; Olivetta, E.; Ferrantelli, F.; Capocefalo, A.; Falcone, E.; Ruggieri, A.; Federico, M. An Exosome-Based Vaccine Platform Imparts Cytotoxic T Lymphocyte Immunity Against Viral Antigens. Biotechnol. J. 2018, 13, e1700443. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, A. In vivo tissue-tropism of adeno-associated viral vectors. Curr. Opin. Virol. 2016, 21, 75–80. [Google Scholar] [CrossRef]
- Ayora-Talavera, G. Sialic acid receptors: Focus on their role in influenza infection. J. Receptor. Ligand Channel Res. 2018, 10, 1–11. [Google Scholar] [CrossRef]
- Kubota, M.; Matsuoka, R.; Suzuki, T.; Yonekura, K.; Yanagi, Y.; Hashiguchi, T. Molecular Mechanism of the Flexible Glycan Receptor Recognition by Mumps Virus. J. Virol. 2019, 93, e00344-19. [Google Scholar] [CrossRef] [PubMed]
- Marcink, T.C.; Wang, T.; des Georges, A.; Porotto, M.; Moscona, A. Human parainfluenza virus fusion complex glycoproteins imaged in action on authentic viral surfaces. PLoS Pathog. 2020, 16, e1008883. [Google Scholar] [CrossRef] [PubMed]
- Pearce, O.M.T.; Läubli, H. Sialic acids in cancer biology and immunity. Glycobiology 2016, 26, 111–128. [Google Scholar] [CrossRef]
- Li, W. The Hepatitis B Virus Receptor. Annu. Rev. Cell Dev. Biol. 2015, 31, 125–147. [Google Scholar] [CrossRef] [PubMed]
- Salhab, A.; Amer, J.; Lu, Y.; Safadi, R. Sodium+/taurocholate cotransporting polypeptide as target therapy for liver fibrosis. Gut 2021, 2020, 323345. [Google Scholar] [CrossRef]
- Cocquerel, L.; Voisset, C.; Dubuisson, J. Hepatitis C virus entry: Potential receptors and their biological functions. J. Gen. Virol. 2006, 87, 1075–1084. [Google Scholar] [CrossRef]
- Zhang, Y.; Qian, H.; Xu, A.; Yang, G. Increased expression of CD81 is associated with poor prognosis of prostate cancer and increases the progression of prostate cancer cells in vitro. Exp. Ther. Med. 2019, 19, 755–761. [Google Scholar] [CrossRef]
- Feng, H.; Wang, M.; Wu, C.; Yu, J.; Wang, D.; Ma, J.; Han, J. High scavenger receptor class B type i expression is related to tumor aggressiveness and poor prognosis in lung adenocarcinoma. Medicine 2018, 97, e0203. [Google Scholar] [CrossRef]
- Bates, P.; Young, J.A.T.; Varmus, H.E. A receptor for subgroup A Rous sarcoma virus is related to the low density lipoprotein receptor. Cell 1993, 74, 1043–1051. [Google Scholar] [CrossRef]
- Finkelshtein, D.; Werman, A.; Novick, D.; Barak, S.; Rubinstein, M. LDL receptor and its family members serve as the cellular receptors for vesicular stomatitis virus. Proc. Natl. Acad. Sci. USA 2013, 110, 7306–7311. [Google Scholar] [CrossRef]
- Migita, T.; Ruiz, S.; Fornari, A.; Fiorentino, M.; Priolo, C.; Zadra, G.; Inazuka, F.; Grisanzio, C.; Palescandolo, E.; Shin, E.; et al. Fatty Acid Synthase: A Metabolic Enzyme and Candidate Oncogene in Prostate Cancer. JNCI J. Natl. Cancer Inst. 2009, 101, 519–532. [Google Scholar] [CrossRef] [PubMed]
- Gallagher, E.J.; Zelenko, Z.; Neel, B.A.; Antoniou, I.M.; Rajan, L.; Kase, N.; LeRoith, D. Elevated tumor LDLR expression accelerates LDL cholesterol-mediated breast cancer growth in mouse models of hyperlipidemia. Oncogene 2017, 36, 6462–6471. [Google Scholar] [CrossRef] [PubMed]
- Yoon, V.; Fridkis-Hareli, M.; Munisamy, S.; Lee, J.; Anastasiades, D.; Stevceva, L. The GP120 Molecule of HIV-1 and its Interaction with T Cells. Curr. Med. Chem. 2010, 17, 741–749. [Google Scholar] [CrossRef] [PubMed]
- Hussaini, M.O.; Kreisel, F.H.; Hassan, A.; Nguyen, T.T.; Frater, J.L. CD4-positive diffuse large B-cell lymphoma: A variant with aggressive clinical potential. World J. Methodol. 2016, 6, 181. [Google Scholar] [CrossRef]
- Anderson, C.S.; Chu, C.Y.; Wang, Q.; Mereness, J.A.; Ren, Y.; Donlon, K.; Bhattacharya, S.; Misra, R.S.; Walsh, E.E.; Pryhuber, G.S.; et al. CX3CR1 as a respiratory syncytial virus receptor in pediatric human lung. Pediatr. Res. 2020, 87, 862–867. [Google Scholar] [CrossRef]
- Griffiths, C.D.; Bilawchuk, L.M.; McDonough, J.E.; Jamieson, K.C.; Elawar, F.; Cen, Y.; Duan, W.; Lin, C.; Song, H.; Casanova, J.L.; et al. IGF1R is an entry receptor for respiratory syncytial virus. Nature 2020, 583, 615–619. [Google Scholar] [CrossRef]
- Yuan, J.; Yin, Z.; Tao, K.; Wang, G.; Gao, J. Function of insulin-like growth factor 1 receptor in cancer resistance to chemotherapy (Review). Oncol. Lett. 2018, 15, 41–47. [Google Scholar] [CrossRef]
- Gonçalves, D.U.; Proietti, F.A.; Ribas, J.G.R.; Araújo, M.G.; Pinheiro, S.R.; Guedes, A.C.; Carneiro-Proietti, A.B.F. Epidemiology, treatment, and prevention of human T-cell leukemia virus type 1-associated diseases. Clin. Microbiol. Rev. 2010, 23, 577–589. [Google Scholar] [CrossRef]
- Carvalho, K.C.; Cunha, I.W.; Rocha, R.M.; Ayala, F.R.; Cajaiba, M.M.; Begnami, M.D.; Vilela, R.S.; Paiva, G.R.; Andrade, R.G.; Soares, F.A. GLUT1 expression in malignant tumors and its use as an immunodiagnostic marker. Clinics 2011, 66, 965–972. [Google Scholar] [CrossRef]
- Tatsuo, H.; Yanagi, Y. The morbillivirus receptor SLAM (CD150). Microbiol. Immunol. 2002, 46, 135–142. [Google Scholar] [CrossRef]
- Romanets-Korbut, O.; Najakshin, A.M.; Yurchenko, M.; Malysheva, T.A.; Kovalevska, L.; Shlapatska, L.M.; Zozulya, Y.A.; Taranin, A.V.; Horvat, B.; Sidorenko, S.P. Expression of CD150 in tumors of the central nervous system: Identification of a novel isoform. PLoS ONE 2015, 10, e0118302. [Google Scholar] [CrossRef]
- Bonaparte, M.I.; Dimitrov, A.S.; Bossart, K.N.; Crameri, G.; Mungall, B.A.; Bishop, K.A.; Choudhry, V.; Dimitrov, D.S.; Wang, L.-F.; Eaton, B.T.; et al. Ephrin-B2 ligand is a functional receptor for Hendra virus and Nipah virus. Proc. Natl. Acad. Sci. USA 2005, 102, 10652–10657. [Google Scholar] [CrossRef] [PubMed]
- Alam, S.M.; Fujimoto, J.; Jahan, I.; Sato, E.; Tamaya, T. Overexpression of ephrinB2 and EphB4 in tumor advancement of uterine endometrial cancers. Ann. Oncol. 2007, 18, 485–490. [Google Scholar] [CrossRef]
- Jia, H.P.; Look, D.C.; Shi, L.; Hickey, M.; Pewe, L.; Netland, J.; Farzan, M.; Wohlford-Lenane, C.; Perlman, S.; McCray, P.B. ACE2 Receptor Expression and Severe Acute Respiratory Syndrome Coronavirus Infection Depend on Differentiation of Human Airway Epithelia. J. Virol. 2005, 79, 14614–14621. [Google Scholar] [CrossRef] [PubMed]
- Errarte, P.; Beitia, M.; Perez, I.; Manterola, L.; Lawrie, C.H.; Solano-Iturri, J.D.; Calvete-Candenas, J.; Unda, M.; López, J.I.; Larrinaga, G. Expression and activity of angiotensin-regulating enzymes is associated with prognostic outcome in clear cell renal cell carcinoma patients. PLoS ONE 2017, 12, e0181711. [Google Scholar] [CrossRef]
- Kuhn, J.H.; Radoshitzky, S.R.; Guth, A.C.; Warfield, K.L.; Li, W.; Vincent, M.J.; Towner, J.S.; Nichol, S.T.; Bavari, S.; Choe, H.; et al. Conserved receptor-binding domains of Lake Victoria marburgvirus and Zaire ebolavirus bind a common receptor. J. Biol. Chem. 2006, 281, 15951–15958. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Xu, K.; Chen, L.; Zhou, Y.; Jiang, J. Prognostic value of TIM-1 expression in human non-small-cell lung cancer. J. Transl. Med. 2019, 17, 1–12. [Google Scholar] [CrossRef]
- Cao, W.; Henry, M.D.; Borrow, P.; Yamada, H.; Elder, J.H.; Ravkov, E.V.; Nichol, S.T.; Compans, R.W.; Campbell, K.P.; Oldstone, M.B.A. Identification of α-dystroglycan as a receptor for lymphocytic choriomeningitis virus and Lassa fever virus. Science 1998, 282, 2079–2081. [Google Scholar] [CrossRef]
- Kunz, S.; Rojek, J.M.; Perez, M.; Spiropoulou, C.F.; Oldstone, M.B.A. Characterization of the Interaction of Lassa Fever Virus with Its Cellular Receptor α-Dystroglycan. J. Virol. 2005, 79, 5979–5987. [Google Scholar] [CrossRef]
- Sciandra, F.; Bigotti, M.G.; Giardina, B.; Bozzi, M.; Brancaccio, A. Genetic Engineering of Dystroglycan in Animal Models of Muscular Dystrophy. Biomed. Res. Int. 2015, 2015, 635792. [Google Scholar] [CrossRef]
- Cohen-Dvashi, H.; Kilimnik, I.; Diskin, R. Structural basis for receptor recognition by Lujo virus. Nat. Microbiol. 2018, 3, 1153–1160. [Google Scholar] [CrossRef] [PubMed]
- Yasuoka, H.; Kodama, R.; Tsujimoto, M.; Yoshidome, K.; Akamatsu, H.; Nakahara, M.; Inagaki, M.; Sanke, T.; Nakamura, Y. Neuropilin-2 expression in breast cancer: Correlation with lymph node metastasis, poor prognosis, and regulation of CXCR4 expression. BMC Cancer 2009, 9, 1–7. [Google Scholar] [CrossRef]
- Cong, H.; Jiang, Y.; Tien, P. Identification of the Myelin Oligodendrocyte Glycoprotein as a Cellular Receptor for Rubella Virus. J. Virol. 2011, 85, 11038–11047. [Google Scholar] [CrossRef] [PubMed]
- Shu, Y.; Long, Y.; Wang, S.; Hu, W.; Zhou, J.; Xu, H.; Chen, C.; Ou, Y.; Lu, Z.; Lau, A.Y.; et al. Brain histopathological study and prognosis in MOG antibody-associated demyelinating pseudotumor. Ann. Clin. Transl. Neurol. 2019, 6, 392–396. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.; Srivastava, R.; Nessler, S.; Grummel, V.; Sommer, N.; Brück, W.; Hartung, H.P.; Stadelmann, C.; Hemmer, B. Identification of a pathogenic antibody response to native myelin oligodendrocyte glycoprotein in multiple sclerosis. Proc. Natl. Acad. Sci. USA 2006, 103, 19057–19062. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Kim, A.S.; Kafai, N.M.; Earnest, J.T.; Shah, A.P.; Case, J.B.; Basore, K.; Gilliland, T.C.; Sun, C.; Nelson, C.A.; et al. LDLRAD3 is a receptor for Venezuelan equine encephalitis virus. Nature 2020, 588, 308–314. [Google Scholar] [CrossRef] [PubMed]
- Campion, O.; Al Khalifa, T.; Langlois, B.; Thevenard-Devy, J.; Salesse, S.; Savary, K.; Schneider, C.; Etique, N.; Dedieu, S.; Devy, J. Contribution of the Low-Density Lipoprotein Receptor Family to Breast Cancer Progression. Front. Oncol. 2020, 10, 882. [Google Scholar] [CrossRef] [PubMed]
- Lo, Y.-L.; Liou, G.-G.; Lyu, J.-H.; Hsiao, M.; Hsu, T.-L.; Wong, C.-H. Dengue Virus Infection Is through a Cooperative Interaction between a Mannose Receptor and CLEC5A on Macrophage as a Multivalent Hetero-Complex. PLoS ONE 2016, 11, e0166474. [Google Scholar] [CrossRef]
- Colpitts, T.M.; Conway, M.J.; Montgomery, R.R.; Fikrig, E. West Nile virus: Biology, transmission, and human infection. Clin. Microbiol. Rev. 2012, 25, 635–648. [Google Scholar] [CrossRef]
- Liu, D.R.; Guan, Q.L.; Gao, M.T.; Jiang, L.; Kang, H.X. Mannose receptor as a potential biomarker for gastric cancer: A pilot study. Int. J. Biol. Markers 2017, 32, e278–e283. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Sengupta, N.; Chaudhuri, A.; Akbar, I.; Singh, N.; Chakraborty, S.; Suryawanshi, A.R.; Bhattacharyya, A.; Basu, A. PLVAP and GKN3 Are Two Critical Host Cell Receptors Which Facilitate Japanese Encephalitis Virus Entry Into Neurons. Sci. Rep. 2018, 8, 1–16. [Google Scholar] [CrossRef]
- Guo, L.; Zhang, H.; Hou, Y.; Wei, T.; Liu, J. Plasmalemma vesicle-associated protein: A crucial component of vascular homeostasis (Review). Exp. Ther. Med. 2016, 12, 1639–1644. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Chen, J.; Yang, M. Glucagon-like peptide-2 receptor is a receptor for tick-borne encephalitis virus to infect nerve cells. Biol. Sci. 2021. in preprint. [Google Scholar] [CrossRef]
- Orhan, A.; Gögenur, I.; Kissow, H. The intestinotrophic effects of glucagon-like peptide-2 in relation to intestinal neoplasia. J. Clin. Endocrinol. Metab. 2018, 103, 2827–2837. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, Z.; Liu, R.; Shuai, L.; Wang, X.; Luo, J.; Wang, C.; Chen, W.; Wang, X.; Ge, J.; et al. Metabotropic glutamate receptor subtype 2 is a cellular receptor for rabies virus. PLoS Pathog. 2018, 14, e1007189. [Google Scholar] [CrossRef]
- Aleman, M.; Milbank, A.; Harsch, K.M.; Detore, N.; Klein, E.; Heston, W. Expression of metabotropic glutamate receptors in human prostate cancers and its potential signaling through prostate specific membrane antigen. Cancer Res. 2004, 64. Available online: https://cancerres.aacrjournals.org/content/64/7_Supplement/1196.2.short#page (accessed on 6 October 2021).
- Arcella, A.; Carpinelli, G.; Battaglia, G.; D’Onofrio, M.; Santoro, F.; Ngomba, R.T.; Bruno, V.; Casolini, P.; Giangaspero, F.; Nicoletti, F. Pharmacological blockade of group II metabotropic glutamate receptors reduces the growth of glioma cells in vivo. Neuro. Oncol. 2005, 7, 236–245. [Google Scholar] [CrossRef]
- Yan, Y.; Vasudevan, S.; Nguyen, H.; Merlin, D. Intestinal Epithelial CD98: An Oligomeric and Multifunctional Protein. Biochim. Biophys. Acta 2008, 1780, 1087. [Google Scholar] [CrossRef] [PubMed]
- Cantor, J.M.; Ginsberg, M.H. CD98 at the crossroads of adaptive immunity and cancer. J. Cell Sci. 2012, 125, 1373–1382. [Google Scholar] [CrossRef] [PubMed]
- Spear, P.G. Herpes simplex virus: Receptors and ligands for cell entry. Cell. Microbiol. 2004, 6, 401–410. [Google Scholar] [CrossRef] [PubMed]
- Vicente, C.M.; da Silva, D.A.; Sartorio, P.V.; Silva, T.D.; Saad, S.S.; Nader, H.B.; Forones, N.M.; Toma, L. Heparan sulfate proteoglycans in human colorectal cancer. Anal. Cell. Pathol. 2018, 2018, 8389595. [Google Scholar] [CrossRef] [PubMed]
- Ross, S.R.; Schofield, J.J.; Farr, C.J.; Bucan, M. Mouse transferrin receptor 1 is the cell entry receptor for mouse mammary tumor virus. Proc. Natl. Acad. Sci. USA 2002, 99, 12386–12390. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Li, X.; Dong, D.; Zhang, B.; Xue, Y.; Shang, P. Transferrin receptor 1 in cancer: A new sight for cancer therapy. Am. J. Cancer Res. 2018, 8, 916. [Google Scholar] [PubMed]
- Esnault, C.; Priet, S.; Ribet, D.; Vernochet, C.; Bruls, T.; Lavialle, C.; Weissenbach, J.; Heidmann, T. A placenta-specific receptor for the fusogenic, endogenous retrovirus-derived, human syncytin-2. Proc. Natl. Acad. Sci. USA 2008, 105, 17532–17537. [Google Scholar] [CrossRef]
- Toda, K.; Nishikawa, G.; Iwamoto, M.; Itatani, Y.; Takahashi, R.; Sakai, Y.; Kawada, K. Clinical role of ASCT2 (SLC1A5) in KRAS-mutated colorectal cancer. Int. J. Mol. Sci. 2017, 18, 1632. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Jiang, X.; Bao, J.; Wang, Y.; Liu, H.; Tang, L. Exosomes in pathogen infections: A bridge to deliver molecules and link functions. Front. Immunol. 2018, 9, 90. [Google Scholar] [CrossRef]
- Kim, G.B.; Nam, G.H.; Hong, Y.; Woo, J.; Cho, Y.; Kwon, I.C.; Yang, Y.; Kim, I.S. Xenogenization of tumor cells by fusogenic exosomes in tumor microenvironment ignites and propagates antitumor immunity. Sci. Adv. 2020, 6, eaaz2083. [Google Scholar] [CrossRef] [PubMed]
- Kima, I.S.; Jenni, S.; Stanifer, M.L.; Roth, E.; Whelan, S.P.J.; Van Oijen, A.M.; Harrison, S.C. Mechanism of membrane fusion induced by vesicular stomatitis virus G protein. Proc. Natl. Acad. Sci. USA 2017, 114, E28–E36. [Google Scholar] [CrossRef] [PubMed]
- Meyer, C.; Losacco, J.; Stickney, Z.; Li, L.; Marriott, G.; Lu, B. Pseudotyping exosomes for enhanced protein delivery in mammalian cells. Int. J. Nanomedicine 2017, 12, 3153–3170. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Hong, Y.; Nam, G.H.; Chung, J.H.; Koh, E.; Kim, I.S. Virus-Mimetic Fusogenic Exosomes for Direct Delivery of Integral Membrane Proteins to Target Cell Membranes. Adv. Mater. 2017, 29, 1605604. [Google Scholar] [CrossRef]
- Masciopinto, F.; Giovani, C.; Campagnoli, S.; Galli-Stampino, L.; Colombatto, P.; Brunetto, M.; Yen, T.S.B.; Houghton, M.; Pileri, P.; Abrignani, S. Association of hepatitis C virus envelope proteins with exosomes. Eur. J. Immunol. 2004, 34, 2834–2842. [Google Scholar] [CrossRef] [PubMed]
- Vallhov, H.; Gutzeit, C.; Johansson, S.M.; Nagy, N.; Paul, M.; Li, Q.; Friend, S.; George, T.C.; Klein, E.; Scheynius, A.; et al. Exosomes Containing Glycoprotein 350 Released by EBV-Transformed B Cells Selectively Target B Cells through CD21 and Block EBV Infection In Vitro. J. Immunol. 2011, 186, 73–82. [Google Scholar] [CrossRef]
- Alvarez-Erviti, L.; Seow, Y.; Yin, H.; Betts, C.; Lakhal, S.; Wood, M.J.A. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol. 2011, 29, 341–345. [Google Scholar] [CrossRef]
- Fu, Y.; Xiong, S. Tagged extracellular vesicles with the RBD of the viral spike protein for delivery of antiviral agents against SARS-COV-2 infection. J. Control. Release 2021, 335, 584–595. [Google Scholar] [CrossRef] [PubMed]
- Salomon, C.; Nuzhat, Z.; Dixon, C.L.; Menon, R. Placental Exosomes During Gestation: Liquid Biopsies Carrying Signals for the Regulation of Human Parturition. Curr. Pharm. Des. 2018, 24, 974–982. [Google Scholar] [CrossRef] [PubMed]
- Yue, B.; Yang, H.; Wang, J.; Ru, W.; Wu, J.; Huang, Y.; Lan, X.; Lei, C.; Chen, H. Exosome biogenesis, secretion and function of exosomal miRNAs in skeletal muscle myogenesis. Cell Prolif. 2020, 53, e12857. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, A.; Costa-Silva, B.; Shen, T.-L.; Rodrigues, G.; Hashimoto, A.; Tesic Mark, M.; Molina, H.; Kohsaka, S.; Di Giannatale, A.; Ceder, S.; et al. Tumour exosome integrins determine organotropic metastasis. Nature 2015, 527, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Wortzel, I.; Dror, S.; Kenific, C.M.; Lyden, D. Exosome-Mediated Metastasis: Communication from a Distance. Dev. Cell 2019, 49, 347–360. [Google Scholar] [CrossRef]
- Eckwahl, M.J.; Telesnitsky, A.; Wolin, S.L. Host RNA packaging by retroviruses: A newly synthesized story. MBio 2016, 7, e02025-15. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Li, Y.; Liu, Y.; Xu, W.; Zhu, X. Exosomal Non-coding RNAs-Mediated Crosstalk in the Tumor Microenvironment. Front. Cell Dev. Biol. 2021, 9, 646864. [Google Scholar] [CrossRef]
- Van Eijndhoven, M.A.J.; Baglio, S.R.; Pegtel, D.M. Packaging RNA drugs into extracellular vesicles. Nat. Biomed. Eng. 2020, 4, 6–8. [Google Scholar] [CrossRef] [PubMed]
- Villarroya-Beltri, C.; Gutiérrez-Vázquez, C.; Sánchez-Cabo, F.; Pérez-Hernández, D.; Vázquez, J.; Martin-Cofreces, N.; Martinez-Herrera, D.J.; Pascual-Montano, A.; Mittelbrunn, M.; Sánchez-Madrid, F. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat. Commun. 2013, 4, 2980. [Google Scholar] [CrossRef] [PubMed]
- McKenzie, A.J.; Hoshino, D.; Hong, N.H.; Cha, D.J.; Franklin, J.L.; Coffey, R.J.; Patton, J.G.; Weaver, A.M. KRAS-MEK Signaling Controls Ago2 Sorting into Exosomes. Cell Rep. 2016, 15, 978–987. [Google Scholar] [CrossRef]
- Iavello, A.; Frech, V.S.L.; Gai, C.; Deregibus, M.C.; Quesenberry, P.J.; Camussi, G. Role of Alix in miRNA packaging during extracellular vesicle biogenesis. Int. J. Mol. Med. 2016, 37, 958–966. [Google Scholar] [CrossRef] [PubMed]
- Hagiwara, K.; Katsuda, T.; Gailhouste, L.; Kosaka, N.; Ochiya, T. Commitment of Annexin A2 in recruitment of microRNAs into extracellular vesicles. FEBS Lett. 2015, 589, 4071–4078. [Google Scholar] [CrossRef] [PubMed]
- Statello, L.; Maugeri, M.; Garre, E.; Nawaz, M.; Wahlgren, J.; Papadimitriou, A.; Lundqvist, C.; Lindfors, L.; Collén, A.; Sunnerhagen, P.; et al. Identification of RNA-binding proteins in exosomes capable of interacting with different types of RNA: RBP-facilitated transport of RNAs into exosomes. PLoS ONE 2018, 13, e0195969. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, K.; Ghoshal, B.; Ghosh, S.; Chakrabarty, Y.; Shwetha, S.; Das, S.; Bhattacharyya, S.N. Reversible HuR-micro RNA binding controls extracellular export of miR-122 and augments stress response. EMBO Rep. 2016, 17, 1184–1203. [Google Scholar] [CrossRef]
- Wozniak, A.L.; Adams, A.; King, K.E.; Dunn, W.; Christenson, L.K.; Hung, W.T.; Weinman, S.A. The RNA binding protein FMR1 controls selective exosomal miRNA cargo loading during inflammation. J. Cell Biol. 2020, 219, e201912074. [Google Scholar] [CrossRef]
- Zietzer, A.; Hosen, M.R.; Wang, H.; Goody, P.R.; Sylvester, M.; Latz, E.; Nickenig, G.; Werner, N.; Jansen, F. The RNA-binding protein hnRNPU regulates the sorting of microRNA-30c-5p into large extracellular vesicles. J. Extracell. Vesicles 2020, 9, 1786967. [Google Scholar] [CrossRef] [PubMed]
- Fabbiano, F.; Corsi, J.; Gurrieri, E.; Trevisan, C.; Notarangelo, M.; D’Agostino, V.G. RNA packaging into extracellular vesicles: An orchestra of RNA-binding proteins? J. Extracell. Vesicles 2020, 10, e12043. [Google Scholar] [CrossRef] [PubMed]
- Stoltzfus, C.M. Synthesis and Processing of Avian Sarcoma Retrovirus RNA. Adv. Virus Res. 1988, 35, 1–38. [Google Scholar] [CrossRef] [PubMed]
- Kuzembayeva, M.; Dilley, K.; Sardo, L.; Hu, W.S. Life of psi: How full-length HIV-1 RNAs become packaged genomes in the viral particles. Virology 2014, 454–455, 362–370. [Google Scholar] [CrossRef]
- Lu, K.; Heng, X.; Summers, M.F. Structural Determinants and Mechanism of HIV-1 Genome Packaging. J. Mol. Biol. 2011, 410, 609–633. [Google Scholar] [CrossRef] [PubMed]
- Aldovini, A.; Young, R.A. Mutations of RNA and protein sequences involved in human immunodeficiency virus type 1 packaging result in production of noninfectious virus. J. Virol. 1990, 64, 1920–1926. [Google Scholar] [CrossRef]
- Clever, J.; Sassetti, C.; Parslow, T.G. RNA secondary structure and binding sites for gag gene products in the 5’ packaging signal of human immunodeficiency virus type 1. J. Virol. 1995, 69, 2101–2109. [Google Scholar] [CrossRef]
- McBride, M.; Schwartz, M.D.; Panganiban, A. Efficient encapsidation of human immunodeficiency virus type 1 vectors and further characterization of cis elements required for encapsidation. J. Virol. 1997, 71, 4544–4554. [Google Scholar] [CrossRef]
- Heng, X.; Kharytonchyk, K.; Garcia, E.L.; Lu, K.; Divakaruni, S.S.; LaCotti, C.; Edme, K.; Telesnitsky, A.; Summers, M.F. Identification of a minimal region of the HIV-1 5’-leader required for RNA dimerization, NC binding, and packaging. J. Mol. Biol. 2012, 417, 224–239. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Jun, H.J.; Jang, S.I.; You, J.C. The Determination of Importance of Sequences Neighboring the Psi Sequence in Lentiviral Vector Transduction and Packaging Efficiency. PLoS ONE 2012, 7, e50148. [Google Scholar] [CrossRef]
- Sakuragi, J.; Shioda, T.; Panganiban, A.T. Duplication of the Primary Encapsidation and Dimer Linkage Region of Human Immunodeficiency Virus Type 1 RNA Results in the Appearance of Monomeric RNA in Virions. J. Virol. 2001, 75, 2557–2565. [Google Scholar] [CrossRef]
- Mailler, E.; Bernacchi, S.; Marquet, R.; Paillart, J.-C.; Vivet-Boudou, V.; Smyth, R.P. The Life-Cycle of the HIV-1 Gag–RNA Complex. Viruses 2016, 8, 248. [Google Scholar] [CrossRef]
- Summers, M.F.; Henderson, L.E.; Chance, M.R.; Bess, J.W., Jr.; South, T.L.; Blake, P.R.; Sagi, I.; Perez-Alvarado, G.; Sowder, R.C.; Arthur, L.O.; et al. Nucleocapsid zinc fingers detected in retroviruses: EXAFS studies of intact viruses and the solution-state structure of the nucleocapsid protein from HIV-1. Protein Sci. 1992, 1, 563. [Google Scholar] [CrossRef]
- Cimarelli, A.; Sandin, S.; Höglund, S.; Luban, J. Basic Residues in Human Immunodeficiency Virus Type 1 Nucleocapsid Promote Virion Assembly via Interaction with RNA. J. Virol. 2000, 74, 3046–3057. [Google Scholar] [CrossRef]
- Campbell, S.; Rein, A. In vitro assembly properties of human immunodeficiency virus type 1 Gag protein lacking the p6 domain. J. Virol. 1999, 73, 2270–2279. [Google Scholar] [CrossRef]
- Nikolaitchik, O.A.; Dilley, K.A.; Fu, W.; Gorelick, R.J.; Tai, S.-H.S.; Soheilian, F.; Ptak, R.G.; Nagashima, K.; Pathak, V.K.; Hu, W.-S. Dimeric RNA Recognition Regulates HIV-1 Genome Packaging. PLoS Pathog. 2013, 9, e1003249. [Google Scholar] [CrossRef]
- Miyazaki, Y.; Garcia, E.L.; King, S.R.; Lyalla, K.; Loeliger, K.; Starck, P.; Syed, S.; Telesnitsky, A.; Summers, M.F. An RNA structural switch regulates diploid genome packaging by Moloney murine leukemia virus. J. Mol. Biol. 2010, 396, 141–152. [Google Scholar] [CrossRef]
- Shen, B.; Wu, N.; Yang, M.; Gould, S.J. Protein targeting to exosomes/microvesicles by plasma membrane anchors. J. Biol. Chem. 2011, 286, 14383–14395. [Google Scholar] [CrossRef]
- Nakagawa, S.; Takahashi, M.U. gEVE: A genome-based endogenous viral element database provides comprehensive viral protein-coding sequences in mammalian genomes. Database 2016, 2016, baw087. [Google Scholar] [CrossRef]
- Ashley, J.; Cordy, B.; Lucia, D.; Fradkin, L.G.; Budnik, V.; Thomson, T. Retrovirus-like Gag Protein Arc1 Binds RNA and Traffics across Synaptic Boutons. Cell 2018, 172, 262–274. [Google Scholar] [CrossRef]
- Abed, M.; Verschueren, E.; Budayeva, H.; Liu, P.; Kirkpatrick, D.S.; Reja, R.; Kummerfeld, S.K.; Webster, J.D.; Gierke, S.; Reichelt, M.; et al. The Gag protein PEG10 binds to RNA and regulates trophoblast stem cell lineage specification. PLoS ONE 2019, 14, e0214110. [Google Scholar] [CrossRef]
- Segel, M.; Lash, B.; Song, J.; Ladha, A.; Liu, C.C.; Jin, X.; Mekhedov, S.L.; Macrae, R.K.; Koonin, E.V.; Zhang, F. Mammalian retrovirus-like protein PEG10 packages its own mRNA and can be pseudotyped for mRNA delivery. Science 2021, 373, 882–889. [Google Scholar] [CrossRef]
- Gee, P.; Lung, M.S.Y.; Okuzaki, Y.; Sasakawa, N.; Iguchi, T.; Makita, Y.; Hozumi, H.; Miura, Y.; Yang, L.F.; Iwasaki, M.; et al. Extracellular nanovesicles for packaging of CRISPR-Cas9 protein and sgRNA to induce therapeutic exon skipping. Nat. Commun. 2020, 11, 1–18. [Google Scholar] [CrossRef]
- Koppers-Lalic, D.; Hogenboom, M.M.; Middeldorp, J.M.; Pegtel, D.M. Virus-modified exosomes for targeted RNA delivery; A new approach in nanomedicine. Adv. Drug Deliv. Rev. 2013, 65, 348–356. [Google Scholar] [CrossRef]

| Viruses | Exosome Content | References |
|---|---|---|
| Non-enveloped virus (Exosomes-like vesicles): | ||
| Hepatitis A, B, C | viral particles, viral RNA, proteins | [71,72,73,74] |
| Poliovirus (PV) | virions, viral RNA and replication proteins | [75] |
| Enveloped virus: | ||
| Human immunodeficiency virus (HIV) | virus and viral constituents (such as viral microRNA (miRNA), viral proteins Gag and Nef | [76,77,78,79,80,81,82,83] |
| dengue virus (DENV) | complete RNA genome and proteins of DENV | [84,85] |
| Ebola virus (EBOV) | proteins (VP40, GP, NP) and RNA | [86] |
| respiratory syncytial virus (RSV) | different mRNA species, small non-coding RNAs, nucleocapsid protein N, attachment protein G, and fusion protein F | [87] |
| alpha (Herpes Simplex Virus 1), beta (Human Cytomegalovirus, and Human Herpesvirus 6), and gamma (Epstein–Barr Virus, and Kaposi Sarcoma-associated Herpesvirus) herpesviruses | viral DNA, mRNAs, miRNAs, and some EBV proteins: EBV nuclear antigen-1 (EBNA-1) and latent membrane proteins 1 and 2 (LMP-1 and LMP-2) | [88,89,90] |
| Virus | Receptor of Fusion Protein | Localization in Normal Cells | Potential Application for Treatment of Human Pathologies with Upregulation of Viral Receptor |
|---|---|---|---|
| Influensa Mumps virus Human parainfluenza viruses | Sialic acid receptors | Ciliated epithelial cells [101,102,103] | Alteration in sialic acid processing that leads to an upregulation of sialylated glycans and its receptors in many tumors [104] |
| Hepatitis B virus | Sodium Taurocholate Cotransporting Polypeptide (NTCP) | Hepatocytes [105] | Use in target therapy for liver fibrosis and cancer [106] |
| Hepatitis C virus | CD81 tetraspanin, scavenger receptor class B type I (SR-B1) | Hepatocytes [107] | CD81 increases the progression of prostate cancer [108] High SR-B1 expression is observe in lung adenocarcinoma [109] |
| Rous sarcoma virus Vesicular stomatitis virus | Low density lipoprotein receptor (LDLR) | Bronchial epithelial cells [110] Epithelial cells [111] | Increased LDLR expression in Prostate cancer [112] and breast cancer [113] |
| Human immunodeficiency viruses (HIV) | CD209, CD4 | T cells [114] | Targeting to HIV infected cells. B and T cell lymphoma [115] |
| Respiratory syncytial virus | IGF1R, CX3CR1 | Bronchial epithelial cells [116,117] | Broad types and a range of cancers [118] |
| Human T-lymphotropic virus | glucose transporter-1 (GLUT-1) | T cells [119] | Broad types and a range of cancers [120] |
| Measles morbillivirus | CD150 | Immune cells [121] | Tumors of the Central Nervous System [122] |
| Nipah virus Hendra virus | EphrinB2 | Endothelial and smooth muscle cells in arterial vessels [123] | Uterine endometrial cancers [124] |
| Coronavirus | ACE2 | Small intestine, testis, kidneys, lungs. [125] | Expression of ACE2 was highest in renal cell carcinoma [126] |
| Zaire ebolavirus Marburg virus | T-cell immunoglobulin and mucin domain 1 (TIM-1) | Kidney & urinary bladder, intestine [127] | TIM-1 overexpression in human non-small-cell lung cancer [128] |
| Lymphocytic choriomeningitis virus Lassa virus | α-dystroglycan | Female and Muscle tissues [129,130] | Muscular diseases treatment [131] |
| Lujo mammarenavirus | neuropilin-2 (NRP2) | Female and Male tissues [132] | Overexpression in breast cancer [133] |
| Rubella virus | Myelin oligodendrocyte glycoprotein (MOG) | Oligodendrocyte [134] | Glioma [135] Multiple sclerosis [136] |
| Venezuelan Equine encephalitis virus | Low-density lipoprotein receptor class A domain-containing 3 (LDLRAD3) | Neuronal cells [137] | Breast Cancer [138] |
| Dengue virus West Nile virus | Mannose-binding receptor (MR), CD209 | Dendritic cells, Macrophage [139,140] | Gastric cancer [141] |
| Japanese Encephalitis Virus | PLVAP and GKN3 | Dendritic cells, Macrophage [142] | PLVAP was upregulated in tumors of the brain, lungs, breasts, stomach, liver, pancreas, colon, small intestine, kidneys, ovaries, prostate, uterus, skin and lymph nodes [143] |
| Tick-borne encephalitis virus | Glucagon-like peptide-2 receptor (GLP2R) | Nerve cells [144] | Gastrointestinal Tumors [145] |
| Rabies virus | Metabotropic glutamate receptor subtype 2 (mGluR2) | Nerve cells [146] | Prostate Cancer [147] Glioma [148] |
| Variola virus | CD98 | Epithelial cells [149] | B cell lymphomas [150] |
| Herpes simplex virus | Heparan sulfate (HS) | Epithelial cells [151] | Colorectal Cancer [152] |
| Mouse mammary tumor virus | Transferrin receptor 1 | Mammary epithelial cells [153] | TFR1 is abundantly expressed in liver, breast, lung and colon cancer cells [154] |
| Syncytin-1 | Na-dependent amino acid transporter 2 (ASCT2) | ASCT2 expression increases in highly proliferative cells such as inflammatory and stem cells [155] | Colorectal Cancer [156] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zubarev, I.; Vladimirtsev, D.; Vorontsova, M.; Blatov, I.; Shevchenko, K.; Zvereva, S.; Lunev, E.A.; Faizuloev, E.; Barlev, N. Viral Membrane Fusion Proteins and RNA Sorting Mechanisms for the Molecular Delivery by Exosomes. Cells 2021, 10, 3043. https://doi.org/10.3390/cells10113043
Zubarev I, Vladimirtsev D, Vorontsova M, Blatov I, Shevchenko K, Zvereva S, Lunev EA, Faizuloev E, Barlev N. Viral Membrane Fusion Proteins and RNA Sorting Mechanisms for the Molecular Delivery by Exosomes. Cells. 2021; 10(11):3043. https://doi.org/10.3390/cells10113043
Chicago/Turabian StyleZubarev, Ilya, Dmitry Vladimirtsev, Maria Vorontsova, Igor Blatov, Konstantin Shevchenko, Svetlana Zvereva, Evgenii A. Lunev, Evgeny Faizuloev, and Nikolay Barlev. 2021. "Viral Membrane Fusion Proteins and RNA Sorting Mechanisms for the Molecular Delivery by Exosomes" Cells 10, no. 11: 3043. https://doi.org/10.3390/cells10113043
APA StyleZubarev, I., Vladimirtsev, D., Vorontsova, M., Blatov, I., Shevchenko, K., Zvereva, S., Lunev, E. A., Faizuloev, E., & Barlev, N. (2021). Viral Membrane Fusion Proteins and RNA Sorting Mechanisms for the Molecular Delivery by Exosomes. Cells, 10(11), 3043. https://doi.org/10.3390/cells10113043







