Impacts of p97 on Proteome Changes in Human Cells during Coronaviral Replication
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines
2.2. Chemical Inhibitors
2.3. Generation of p97-Knockdown Ccell Line
2.4. HCoV Stock Production and Viral Titer Titration
2.5. HCoV Infection
2.6. Detection of Viral RNA by Real-Time PCR
2.7. Western Blotting
2.8. p97 Inhibitor Treatment
2.9. Cellular Viability
2.10. Stage-Limited Inhibition Assay
2.11. TMT Label Proteomics
2.12. Proteomic Data Processing
3. Results
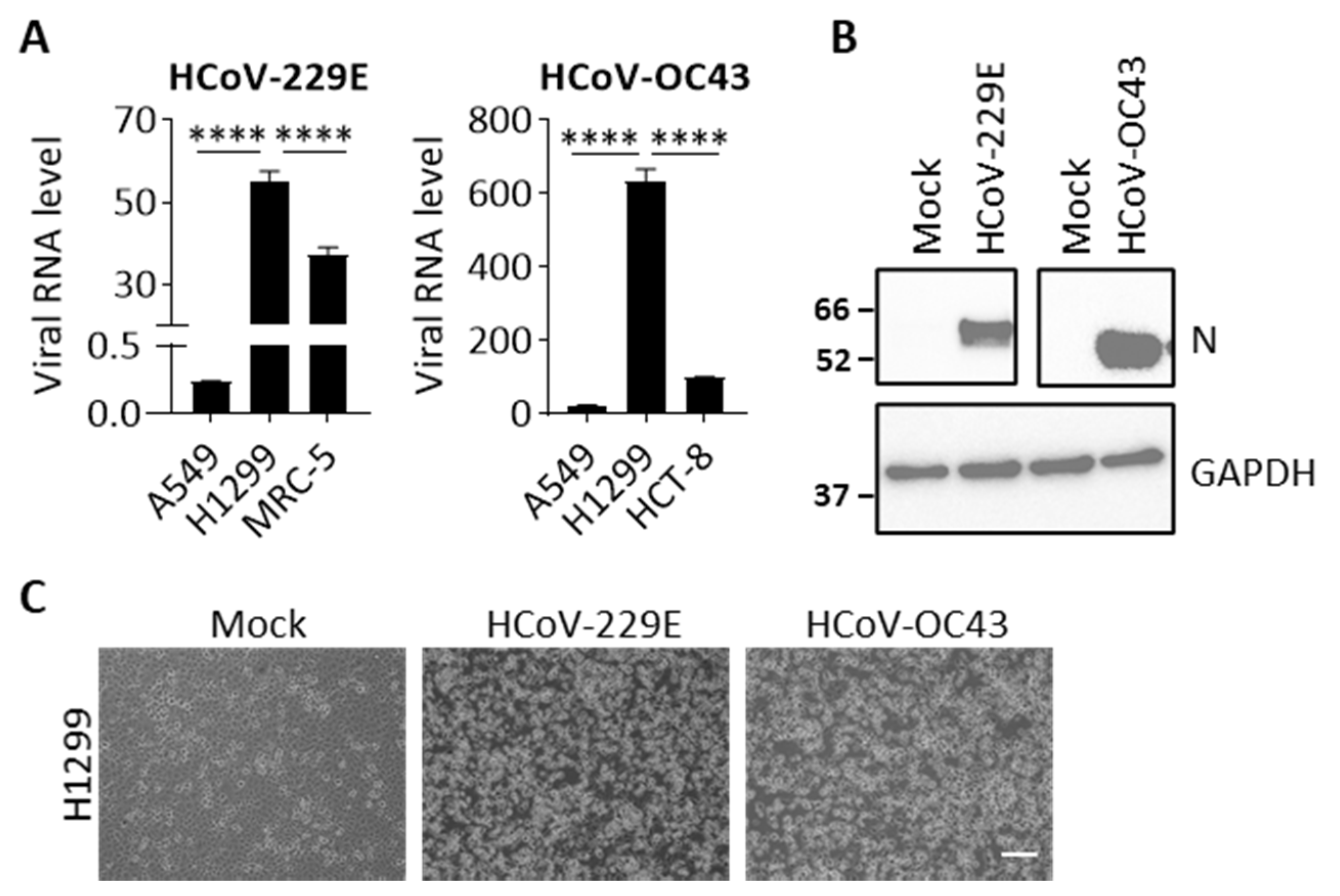
3.1. H1299 Is Susceptible to Both HCoV-229E and HCoV-OC43 Infection
3.2. p97 Is a Potential Therapeutic Target for HCoV Infection
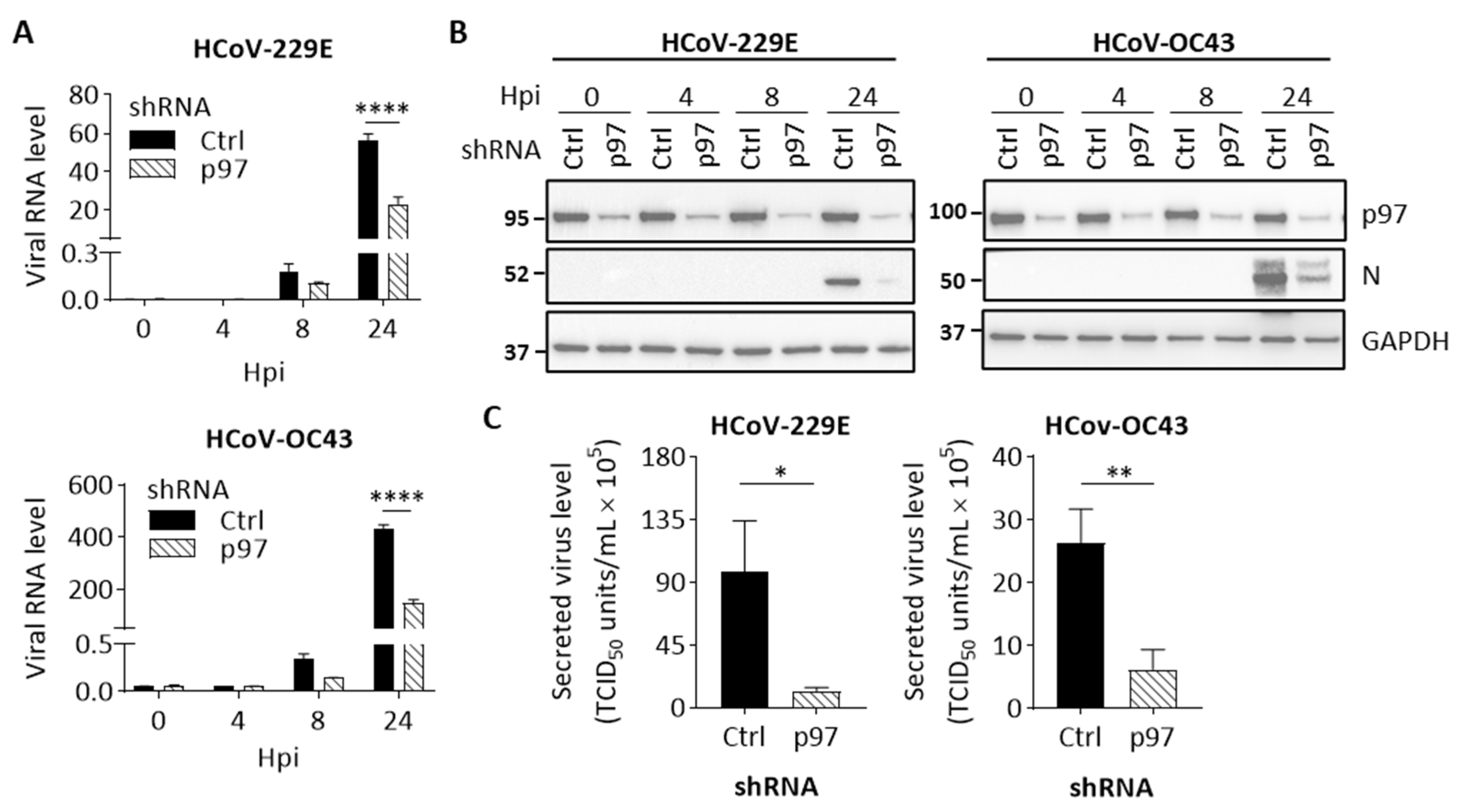
3.3. p97 Is Required for Early Stages of HCoV Replication
3.4. Effects of p97 on Multiple Pathways after HCoV Infection
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- V’kovski, P.; Kratzel, A.; Steiner, S.; Stalder, H.; Thiel, V. Coronavirus Biology and Replication: Implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021, 19, 155–170. [Google Scholar] [CrossRef]
- Liu, D.X.; Liang, J.Q.; Fung, T.S. Human Coronavirus-229E, -OC43, -NL63, and -HKU1 (Coronaviridae). Encycl. Virol. 2021, 2, 428–440. [Google Scholar] [CrossRef]
- Hu, B.; Guo, H.; Zhou, P.; Shi, Z.-L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021, 19, 141–154. [Google Scholar] [CrossRef]
- Rubin, D.; Chan-Tack, K.; Farley, J.; Sherwat, A. FDA Approval of Redmesivir—A Step in the Right Direction. N. Engl. J. Med. 2020, 383, 2598–2600. [Google Scholar] [CrossRef]
- Kokic, G.; Hillen, H.S.; Tegunov, D.; Dienemann, C.; Seitz, F.; Schmitzova, J.; Farnung, L.; Siewert, A.; Höbartner, C.; Cramer, P. Mechanism of SARS-CoV-2 Polymerase Stalling by Remdesivir. Nat. Commun. 2021, 12, 279. [Google Scholar] [CrossRef] [PubMed]
- Yeager, C.L.; Ashmun, R.A.; Williams, R.K.; Cardellichio, C.B.; Shapiro, L.H.; Look, A.T.; Holmes, K.V. Human Aminopeptidase N Is a Receptor for Human Coronavirus 229E. Nature 1992, 357, 420–422. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, H.; Pyrc, K.; van der Hoek, L.; Geier, M.; Berkhout, B.; Pöhlmann, S. Human Coronavirus NL63 Employs the Severe Acute Respiratory Syndrome Coronavirus Receptor for Cellular Entry. Proc. Natl. Acad. Sci. USA 2005, 102, 7988–7993. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Moore, M.J.; Vasilieva, N.; Sui, J.; Wong, S.K.; Berne, M.A.; Somasundaran, M.; Sullivan, J.L.; Luzuriaga, K.; Greenough, T.C.; et al. Angiotensin-Converting Enzyme 2 Is a Functional Receptor for the SARS Coronavirus. Nature 2003, 426, 450–454. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef]
- Raj, V.S.; Mou, H.; Smits, S.L.; Dekkers, D.H.W.; Müller, M.A.; Dijkman, R.; Muth, D.; Demmers, J.A.A.; Zaki, A.; Fouchier, R.A.M.; et al. Dipeptidyl Peptidase 4 Is a Functional Receptor for the Emerging Human Coronavirus-EMC. Nature 2013, 495, 251–254. [Google Scholar] [CrossRef] [PubMed]
- Shang, J.; Wan, Y.; Luo, C.; Ye, G.; Geng, Q.; Auerbach, A.; Li, F. Cell Entry Mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2020, 117, 11727–11734. [Google Scholar] [CrossRef]
- Yang, N.; Shen, H.-M. Targeting the Endocytic Pathway and Autophagy Process as a Novel Therapeutic Strategy in COVID-19. Int. J. Biol. Sci. 2020, 16, 1724–1731. [Google Scholar] [CrossRef] [PubMed]
- Van den Boom, J.; Meyer, H. VCP/P97-Mediated Unfolding as a Principle in Protein Homeostasis and Signaling. Mol. Cell 2018, 69, 182–194. [Google Scholar] [CrossRef] [PubMed]
- Ballar, P.; Fang, S. Regulation of ER-Associated Degradation via P97/VCP-Interacting Motif. Biochem. Soc. Trans. 2008, 36, 818–822. [Google Scholar] [CrossRef][Green Version]
- Parisi, E.; Yahya, G.; Flores, A.; Aldea, M. Cdc48/P97 Segregase Is Modulated by Cyclin-Dependent Kinase to Determine Cyclin Fate during G1 Progression. EMBO J. 2018, 37, e98724. [Google Scholar] [CrossRef] [PubMed]
- Papadopoulos, C.; Kirchner, P.; Bug, M.; Grum, D.; Koerver, L.; Schulze, N.; Poehler, R.; Dressler, A.; Fengler, S.; Arhzaouy, K.; et al. VCP/P97 Cooperates with YOD1, UBXD1 and PLAA to Drive Clearance of Ruptured Lysosomes by Autophagy. EMBO J. 2017, 36, 135–150. [Google Scholar] [CrossRef]
- Bouhaddou, M.; Memon, D.; Meyer, B.; White, K.M.; Rezelj, V.V.; Correa Marrero, M.; Polacco, B.J.; Melnyk, J.E.; Ulferts, S.; Kaake, R.M.; et al. The Global Phosphorylation Landscape of SARS-CoV-2 Infection. Cell 2020, 182, 685–712.e19. [Google Scholar] [CrossRef] [PubMed]
- Samavarchi-Tehrani, P.; Abdouni, H.; Knight, J.D.R.; Astori, A.; Samson, R.; Lin, Z.-Y.; Kim, D.-K.; Knapp, J.J.; St-Germain, J.; Go, C.D.; et al. A SARS-CoV-2–Host Proximity Interactome. bioRxiv 2020. Preprint. [Google Scholar] [CrossRef]
- Stukalov, A.; Girault, V.; Grass, V.; Bergant, V.; Karayel, O.; Urban, C.; Haas, D.A.; Huang, Y.; Oubraham, L.; Wang, A.; et al. Multi-Level Proteomics Reveals Host-Perturbation Strategies of SARS-CoV-2 and SARS-CoV. Nature 2020, 594, 246–252. [Google Scholar] [CrossRef]
- Wu, M.; Chen, Y.; Xia, H.; Wang, C.; Tan, C.Y.; Cai, X.; Liu, Y.; Ji, F.; Xiong, P.; Liu, R.; et al. Transcriptional and Proteomic Insights into the Host Response in Fatal COVID-19 Cases. Proc. Natl. Acad. Sci. USA 2020, 117, 28336–28343. [Google Scholar] [CrossRef] [PubMed]
- Radhakrishnan, S.K.; den Besten, W.; Deshaies, R.J. P97-Dependent Retrotranslocation and Proteolytic Processing Govern Formation of Active Nrf1 upon Proteasome Inhibition. eLife 2014, 3, e01856. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape Provides a Biologist-Oriented Resource for the Analysis of Systems-Level Datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Fonseka, P.; Pathan, M.; Chitti, S.V.; Kang, T.; Mathivanan, S. FunRich Enables Enrichment Analysis of OMICs Datasets. J. Mol. Biol. 2021, 433, 166747. [Google Scholar] [CrossRef] [PubMed]
- Pathan, M.; Keerthikumar, S.; Ang, C.-S.; Gangoda, L.; Quek, C.Y.J.; Williamson, N.A.; Mouradov, D.; Sieber, O.M.; Simpson, R.J.; Salim, A.; et al. FunRich: An Open Access Standalone Functional Enrichment and Interaction Network Analysis Tool. Proteomics 2015, 15, 2597–2601. [Google Scholar] [CrossRef]
- Pathan, M.; Keerthikumar, S.; Chisanga, D.; Alessandro, R.; Ang, C.-S.; Askenase, P.; Batagov, A.O.; Benito-Martin, A.; Camussi, G.; Clayton, A.; et al. A Novel Community Driven Software for Functional Enrichment Analysis of Extracellular Vesicles Data. J. Extracell. Vesicles 2017, 6, 1321455. [Google Scholar] [CrossRef] [PubMed]
- Jassal, B.; Matthews, L.; Viteri, G.; Gong, C.; Lorente, P.; Fabregat, A.; Sidiropoulos, K.; Cook, J.; Gillespie, M.; Haw, R.; et al. The Reactome Pathway Knowledgebase. Nucleic Acids Res. 2020, 48, D498–D503. [Google Scholar] [CrossRef] [PubMed]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. G:Profiler: A Web Server for Functional Enrichment Analysis and Conversions of Gene Lists (2019 Update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. Ggplot2; Springer New York: New York, NY, USA, 2009; ISBN 978-0-387-98140-6. [Google Scholar]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING V11: Protein-Protein Association Networks with Increased Coverage, Supporting Functional Discovery in Genome-Wide Experimental Datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Zhou, H.-J.; Wang, J.; Yao, B.; Wong, S.; Djakovic, S.; Kumar, B.; Rice, J.; Valle, E.; Soriano, F.; Menon, M.-K.; et al. Discovery of a First-in-Class, Potent, Selective, and Orally Bioavailable Inhibitor of the P97 AAA ATPase (CB-5083). J. Med. Chem. 2015, 58, 9480–9497. [Google Scholar] [CrossRef]
- Bojkova, D.; Klann, K.; Koch, B.; Widera, M.; Krause, D.; Ciesek, S.; Cinatl, J.; Münch, C. Proteomics of SARS-CoV-2-Infected Host Cells Reveals Therapy Targets. Nature 2020, 583, 469–472. [Google Scholar] [CrossRef]
- Wong, H.H.; Kumar, P.; Tay, F.P.L.; Moreau, D.; Liu, D.X.; Bard, F. Genome-Wide Screen Reveals Valosin-Containing Protein Requirement for Coronavirus Exit from Endosomes. J. Virol. 2015, 89, 11116–11128. [Google Scholar] [CrossRef] [PubMed]
- Dominguez Andres, A.; Feng, Y.; Campos, A.R.; Yin, J.; Yang, C.-C.; James, B.; Murad, R.; Kim, H.; Deshpande, A.J.; Gordon, D.E.; et al. SARS-CoV-2 ORF9c Is a Membrane-Associated Protein That Suppresses Antiviral Responses in Cells. bioRxiv 2020. Preprint. [Google Scholar] [CrossRef]
- Friedman, N.; Jacob-Hirsch, J.; Drori, Y.; Eran, E.; Kol, N.; Nayshool, O.; Mendelson, E.; Rechavi, G.; Mandelboim, M. Transcriptomic Profiling and Genomic Mutational Analysis of Human Coronavirus (HCoV)-229E -Infected Human Cells. PLoS ONE 2021, 16, e0247128. [Google Scholar] [CrossRef] [PubMed]
- Meyer, H.; Weihl, C.C. The VCP/P97 System at a Glance: Connecting Cellular Function to Disease Pathogenesis. J. Cell Sci. 2014, 127, 3877–3883. [Google Scholar] [CrossRef]
- Panda, D.; Rose, P.P.; Hanna, S.L.; Gold, B.; Hopkins, K.C.; Lyde, R.B.; Marks, M.S.; Cherry, S. Genome-Wide RNAi Screen Identifies SEC61A and VCP as Conserved Regulators of Sindbis Virus Entry. Cell Rep. 2013, 5, 1737–1748. [Google Scholar] [CrossRef]
- Ramanathan, H.N.; Zhang, S.; Douam, F.; Mar, K.B.; Chang, J.; Yang, P.L.; Schoggins, J.W.; Ploss, A.; Lindenbach, B.D. A Sensitive Yellow Fever Virus Entry Reporter Identifies Valosin-Containing Protein (VCP/P97) as an Essential Host Factor for Flavivirus Uncoating. mBio 2020, 11, e00467-20. [Google Scholar] [CrossRef] [PubMed]
- Phongphaew, W.; Kobayashi, S.; Sasaki, M.; Carr, M.; Hall, W.W.; Orba, Y.; Sawa, H. Valosin-Containing Protein (VCP/P97) Plays a Role in the Replication of West Nile Virus. Virus Res. 2017, 228, 114–123. [Google Scholar] [CrossRef]
- Carissimo, G.; Chan, Y.-H.; Utt, A.; Chua, T.-K.; Bakar, F.A.; Merits, A.; Ng, L.F.P. VCP/P97 Is a Proviral Host Factor for Replication of Chikungunya Virus and Other Alphaviruses. Front. Microbiol. 2019, 10, 2236. [Google Scholar] [CrossRef]
- Anderson, D.J.; Le Moigne, R.; Djakovic, S.; Kumar, B.; Rice, J.; Wong, S.; Wang, J.; Yao, B.; Valle, E.; Kiss von Soly, S.; et al. Targeting the AAA ATPase P97 as an Approach to Treat Cancer through Disruption of Protein Homeostasis. Cancer Cell 2015, 28, 653–665. [Google Scholar] [CrossRef] [PubMed]
- Sehrawat, S.; Khasa, R.; Deb, A.; Prajapat, S.K.; Mallick, S.; Basu, A.; Surjit, M.; Kalia, M.; Vrati, S. Valosin-Containing Protein/P97 Plays Critical Roles in the Japanese Encephalitis Virus Life Cycle. J. Virol. 2021, 95, e02336-20. [Google Scholar] [CrossRef]
- Zhang, J.; Hu, Y.; Hau, R.; Musharrafieh, R.; Ma, C.; Zhou, X.; Chen, Y.; Wang, J. Identification of NMS-873, an Allosteric and Specific P97 Inhibitor, as a Broad Antiviral against Both Influenza A and B Viruses. Eur. J. Pharm. Sci. 2019, 133, 86–94. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Soto-Acosta, R.; Ding, R.; Chen, L.; Geraghty, R.J. Anti-HCMV Activity by an Irreversible P97 Inhibitor LC-1310. Med. Chem. Res. 2021, 30, 440–448. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, K.-W.; Li, S.; Wang, F.; Ruiz-Lopez, N.M.; Houerbi, N.; Chou, T.-F. Impacts of p97 on Proteome Changes in Human Cells during Coronaviral Replication. Cells 2021, 10, 2953. https://doi.org/10.3390/cells10112953
Cheng K-W, Li S, Wang F, Ruiz-Lopez NM, Houerbi N, Chou T-F. Impacts of p97 on Proteome Changes in Human Cells during Coronaviral Replication. Cells. 2021; 10(11):2953. https://doi.org/10.3390/cells10112953
Chicago/Turabian StyleCheng, Kai-Wen, Shan Li, Feng Wang, Nallely M. Ruiz-Lopez, Nadia Houerbi, and Tsui-Fen Chou. 2021. "Impacts of p97 on Proteome Changes in Human Cells during Coronaviral Replication" Cells 10, no. 11: 2953. https://doi.org/10.3390/cells10112953
APA StyleCheng, K.-W., Li, S., Wang, F., Ruiz-Lopez, N. M., Houerbi, N., & Chou, T.-F. (2021). Impacts of p97 on Proteome Changes in Human Cells during Coronaviral Replication. Cells, 10(11), 2953. https://doi.org/10.3390/cells10112953





