Milk Exosome-Derived MicroRNA-2478 Suppresses Melanogenesis through the Akt-GSK3β Pathway
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
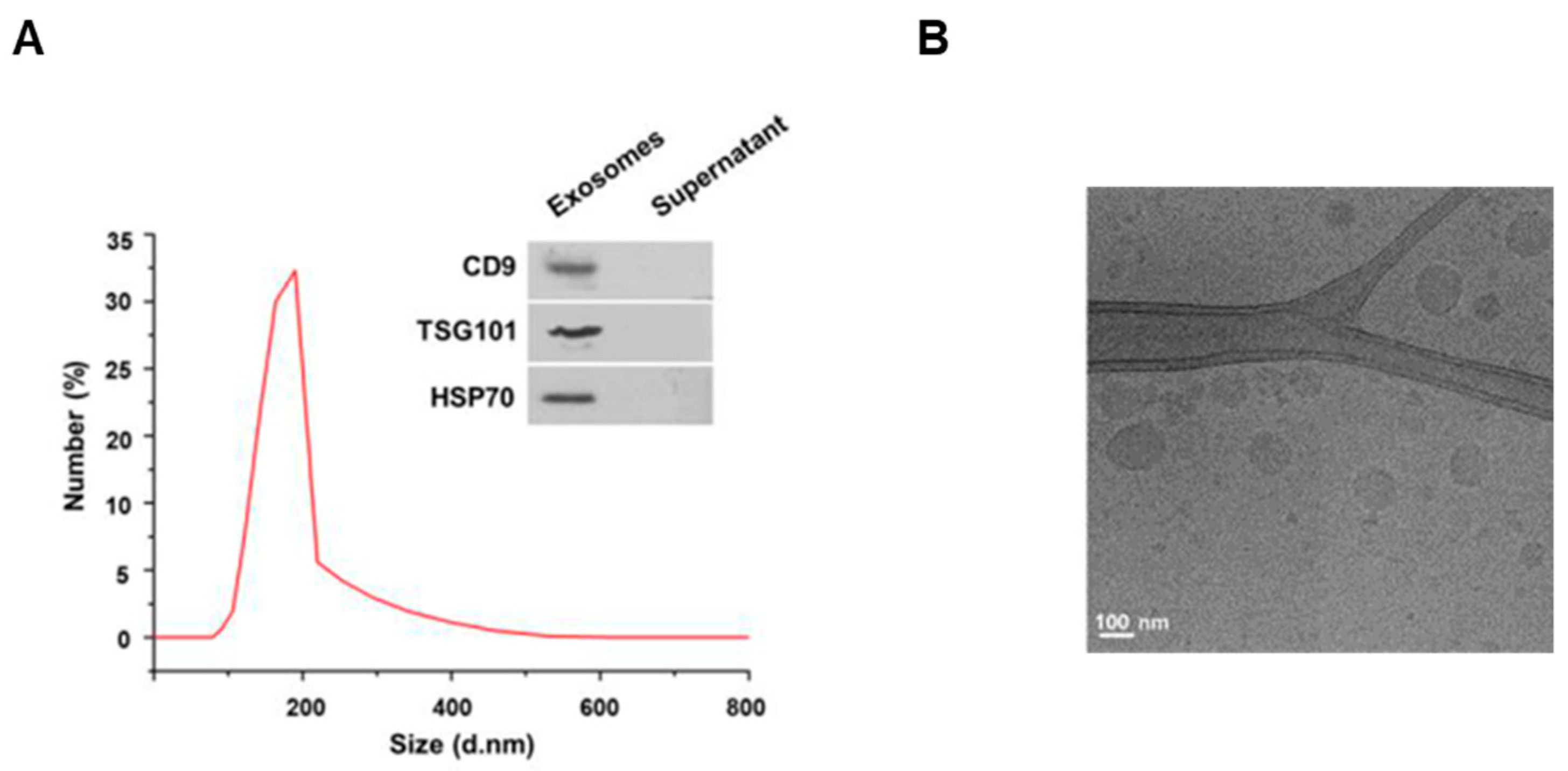
2.2. Exosomes Purification
2.3. Cryo-Electron Microscopy
2.4. Cell Viability Assay (WST Assay)
2.5. Melanin Content Measurement
2.6. Tyrosinase Activity Assay
2.7. Quantitative rReverse-Transcription Polymerase Chain Reaction (qRT-PCR)
2.8. Transfection and Luciferase Reporter Assay
2.9. Western Blotting
2.10. Human Skin Tissues
2.11. Histological Analysis
2.12. Statistical Analysis
3. Results
3.1. Characterization of the Exosomes Isolated from Milk
3.2. Tyrosinase Activity and Melanin Production Suppressed by Milk Exosomes in B16F10 Cells
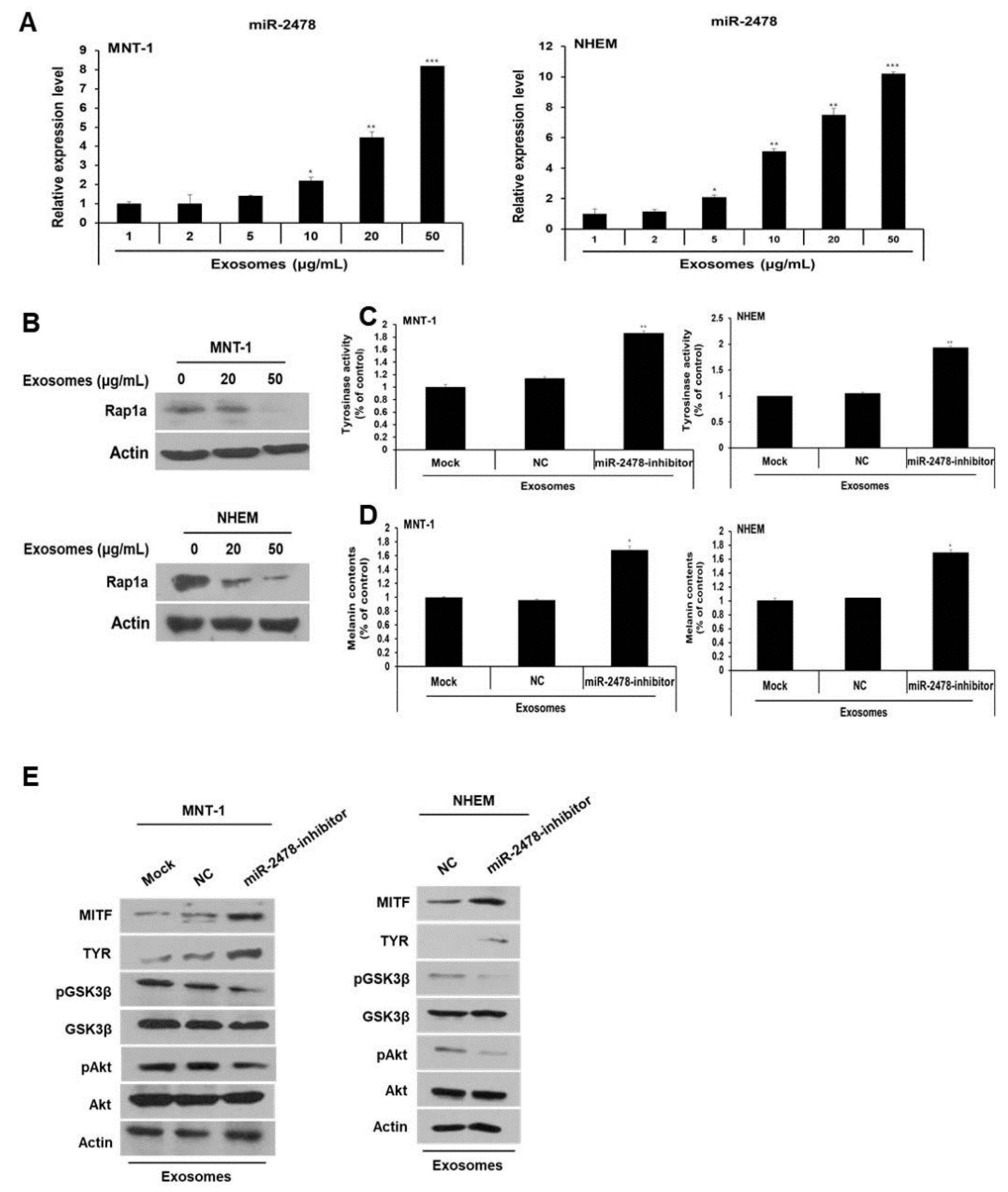
3.3. Enrichment of Bovine-Specific miR-2478 in Milk Exosomes
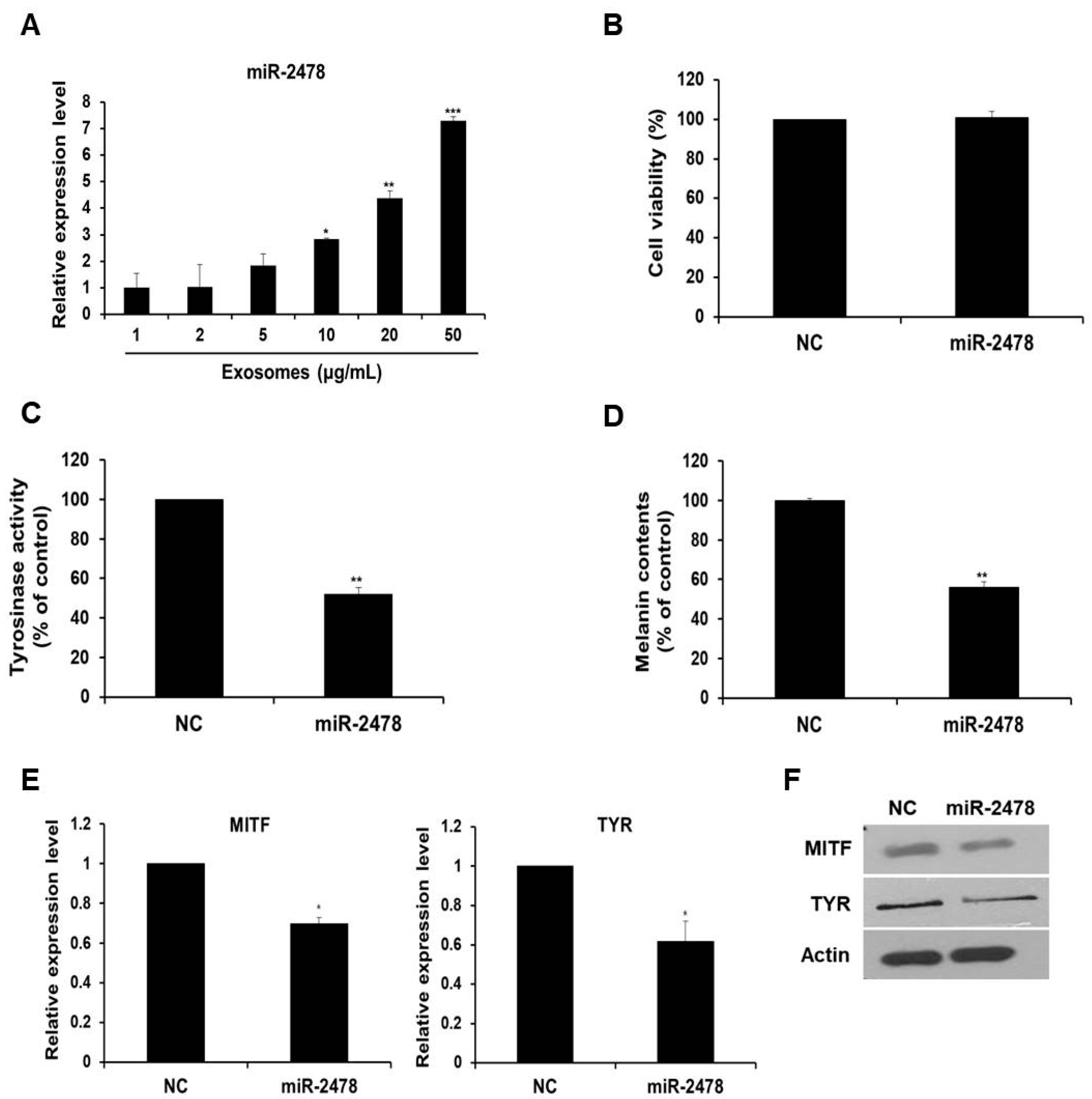
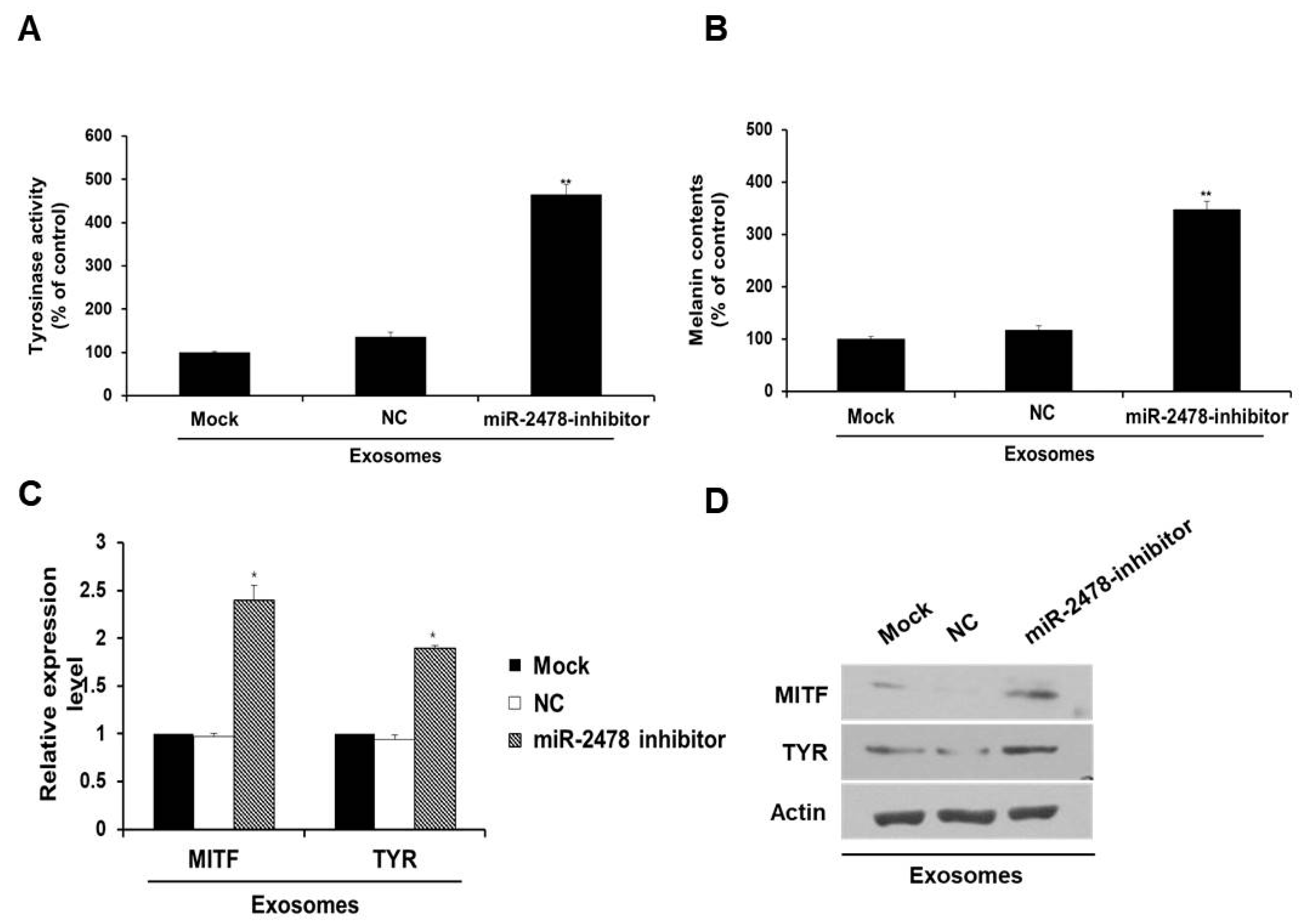
3.4. Bovine-Specific miR-2478 from Milk Exosomes Suppresses Melanogenesis
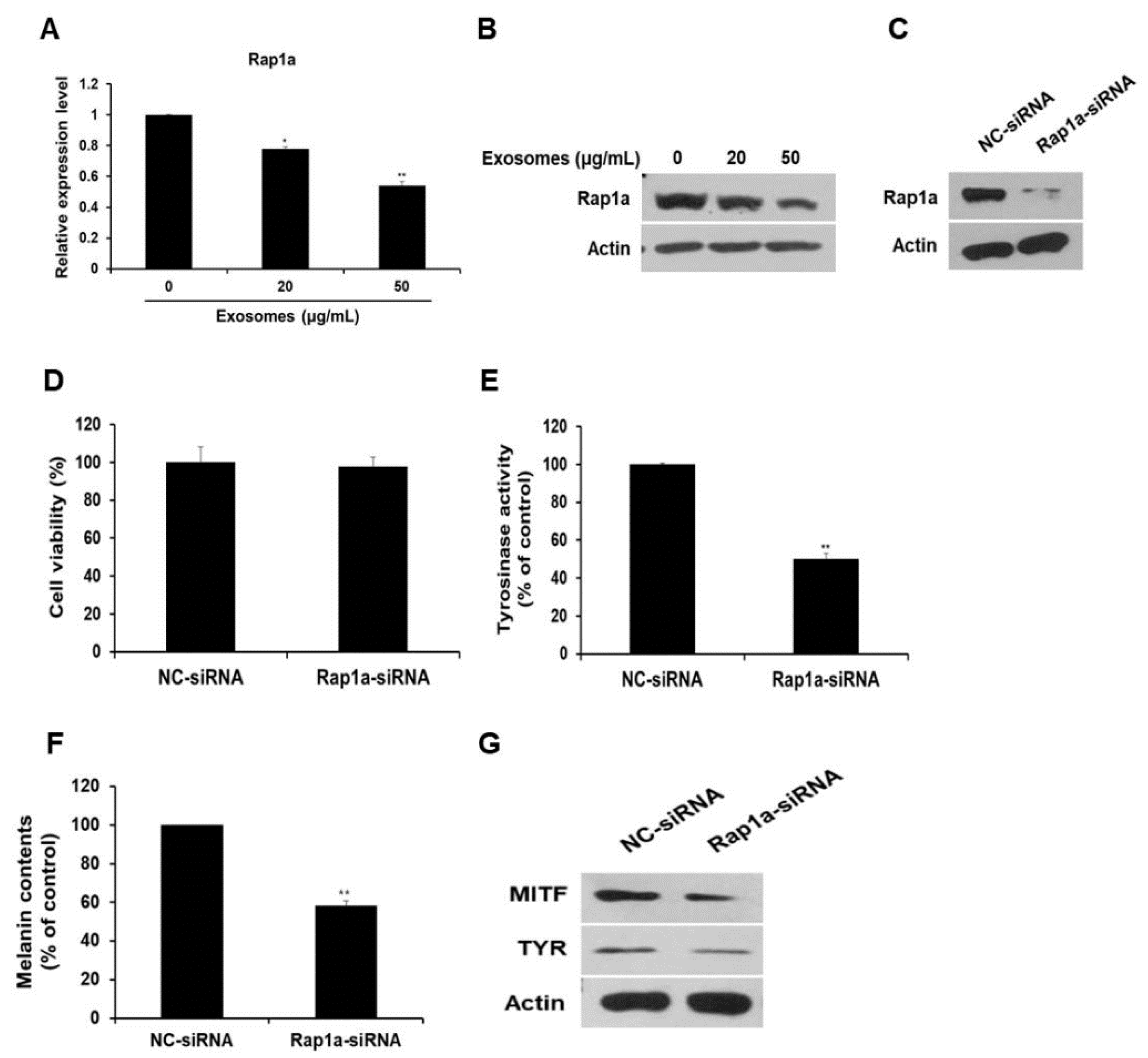
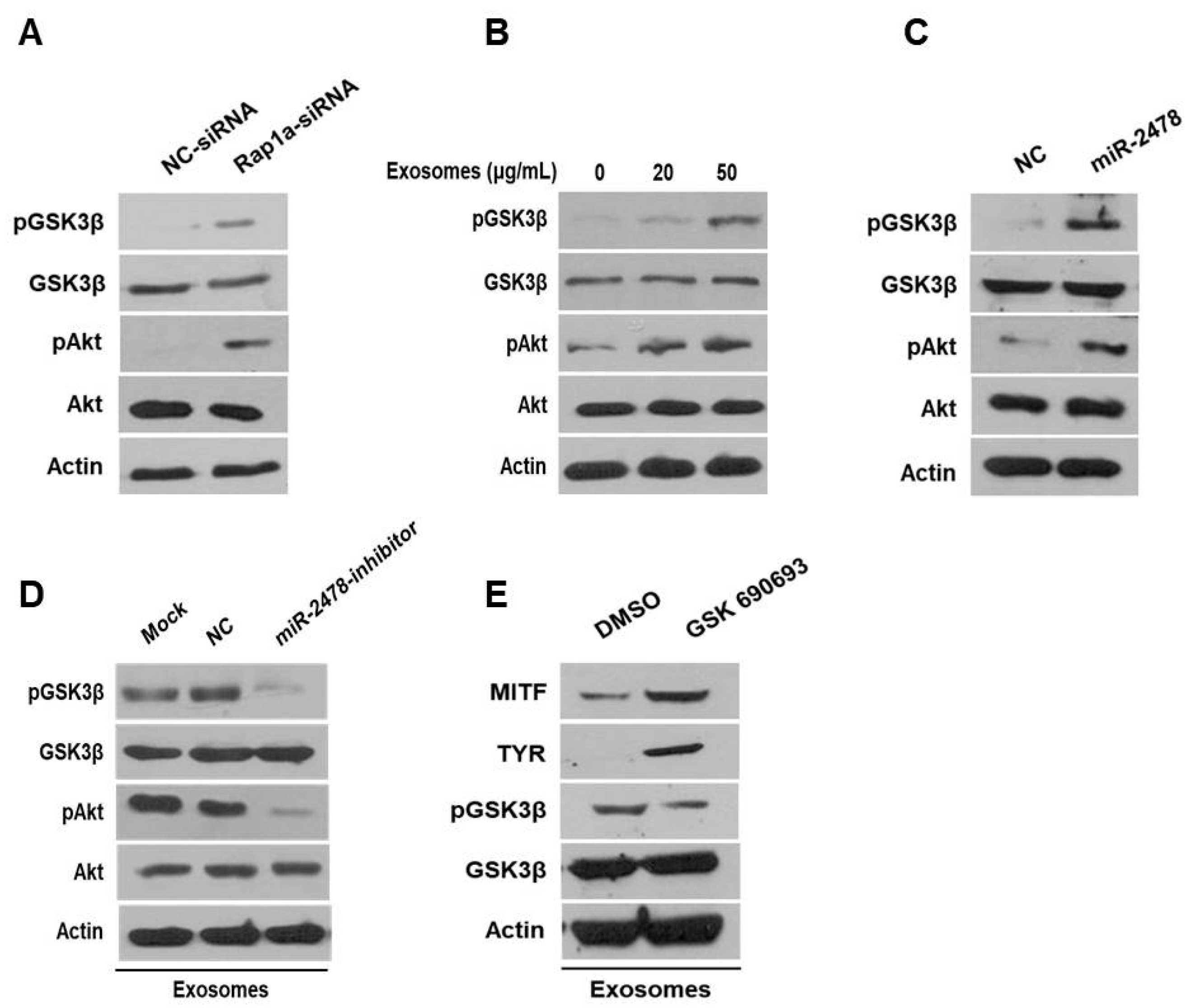
3.5. Rap1a Is Directly Targeted by miR-2478
3.6. Rap1a Promotes Melanogenesis
3.7. Milk Exosomes Suppress Melanogenesis through the Akt-GSK3β Pathway
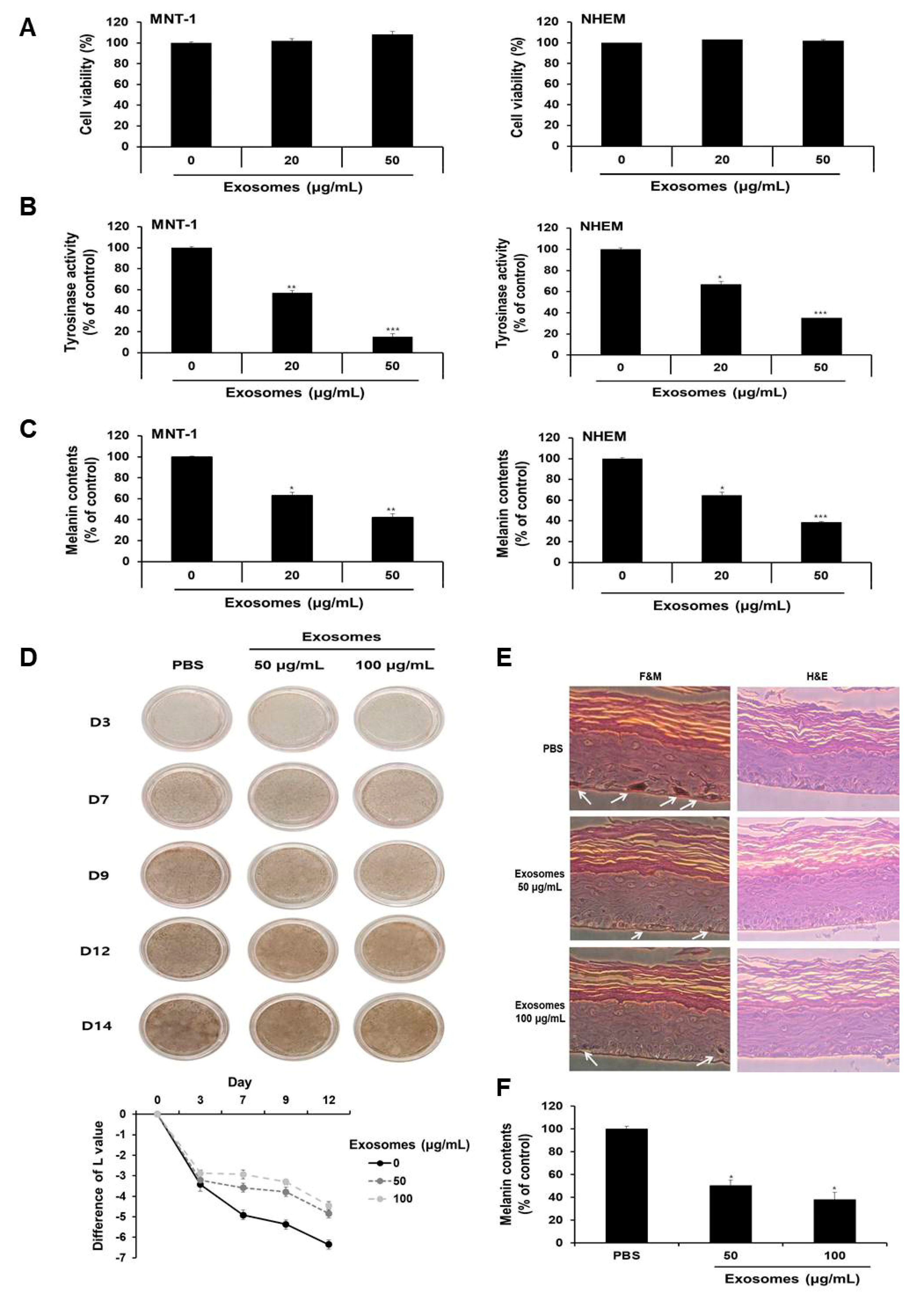
3.8. Milk Exosomes Inhibited Melanogenesis in Human Melanoma Cells, Melanocytes and MelanoDerm Tissue
3.9. miR-2478 Inhibited Melanogenesis in Human MNT-1 Melanoma Cells and NHEM
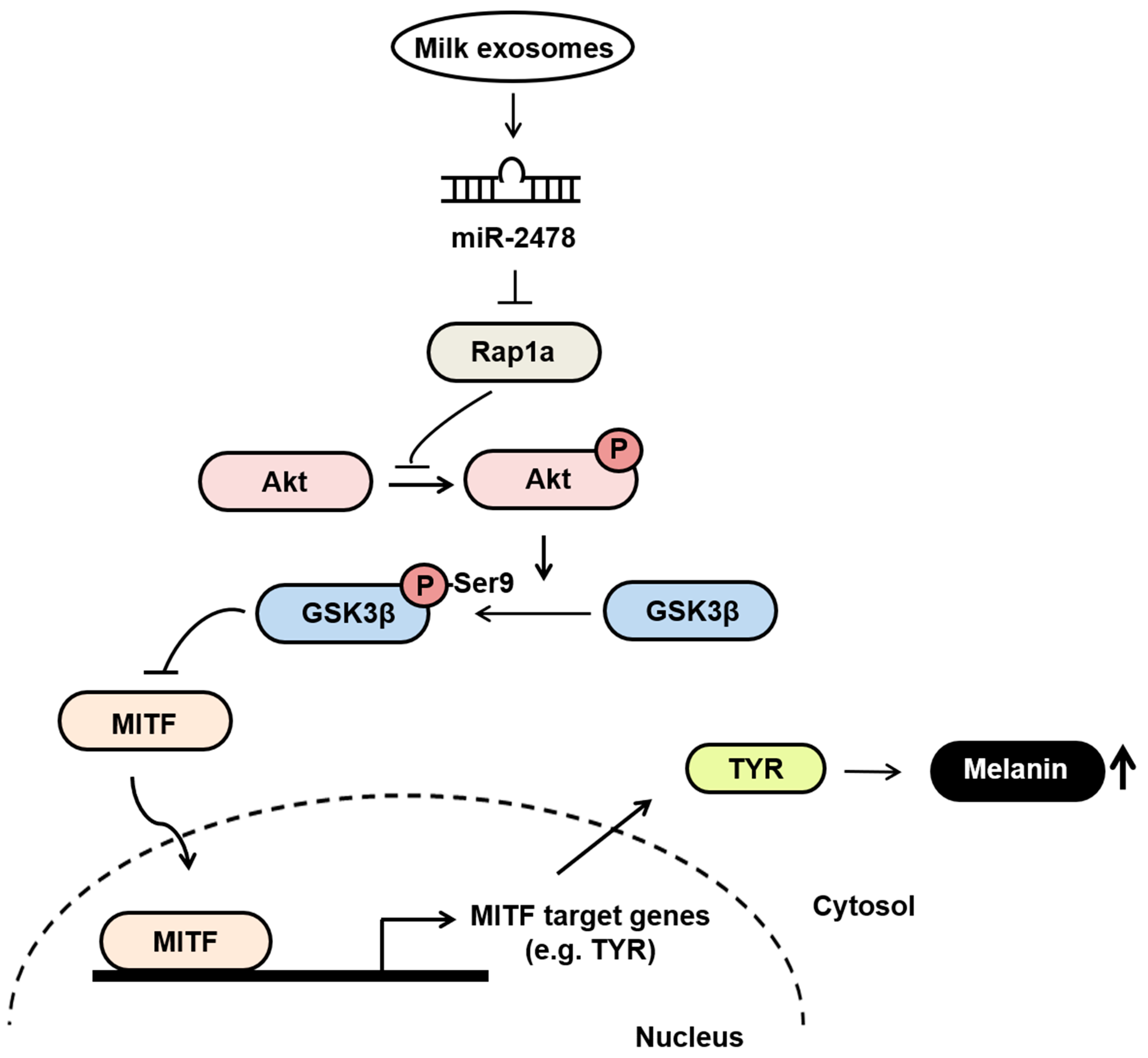
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Draelos, Z.D. Cosmeceuticals: What’s real, What’s not. Dermatol. Clin. 2019, 37, 107–115. [Google Scholar] [CrossRef]
- Searle, T.; Al-Niaimi, F.; Ali, F.R. The top 10 cosmeceuticals for facial hyperpigmentation. Dermatol. Ther. 2020, 33, e14095. [Google Scholar] [CrossRef]
- Slominski, R.M.; Zmijewski, M.A.; Slominski, A.T. The role of melanin pigment in melanoma. Exp. Dermatol. 2015, 24, 258–259. [Google Scholar] [CrossRef] [Green Version]
- Praetorius, C.; Sturm, R.A.; Steingrimsson, E. Sun-induced freckling: Ephelides and solar lentigines. Pigment Cell Melanoma Res. 2014, 27, 339–350. [Google Scholar] [CrossRef]
- Serre, C.; Busuttil, V.; Botto, J.M. Intrinsic and extrinsic regulation of human skin melanogenesis and pigmentation. Int. J. Cosmet. Sci. 2018, 40, 328–347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gebert, L.F.R.; MacRae, I.J. Regulation of microRNA function in animals. Nat. Rev. Mol. Cell Biol. 2019, 20, 21–37. [Google Scholar] [CrossRef]
- Ji, K.; Zhang, P.; Zhang, J.; Fan, R.; Liu, Y.; Yang, S.; Hu, S.; Liu, X.; Dong, C. MicroRNA 143-5p regulates alpaca melanocyte migration, proliferation and melanogenesis. Exp. Dermatol. 2018, 27, 166–171. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.H.; Lee, T.R.; Cho, E.G. SH3BP4, a novel pigmentation gene, is inversely regulated by miR-125b and MITF. Exp. Mol. Med. 2017, 49, e367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, N.H.; Choi, S.H.; Kim, C.H.; Lee, C.H.; Lee, T.R.; Lee, A.Y. Reduced MiR-675 in exosome in H19 RNA-related melanogenesis via MITF as a direct target. J. Investig. Dermatol. 2014, 134, 1075–1082. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Guo, J.; Tian, F.; Yang, N.; Yan, F.; Ding, Y.; Wei, J.; Hu, G.; Nie, G.; Sun, J. Field-Free Isolation of Exosomes from Extracellular Vesicles by Microfluidic Viscoelastic Flows. ACS Nano 2017, 11, 6968–6976. [Google Scholar] [CrossRef] [Green Version]
- Ha, D.; Yang, N.; Nadithe, V. Exosomes as therapeutic drug carriers and delivery vehicles across biological membranes: Current perspectives and future challenges. Acta Pharm. Sin. B. 2016, 6, 287–296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zempleni, J. Milk exosomes: Beyond dietary microRNAs. Genes Nutr. 2017, 12, 12. [Google Scholar] [CrossRef] [PubMed]
- Izumi, H.; Tsuda, M.; Sato, Y.; Kosaka, N.; Ochiya, T.; Iwamoto, H.; Namba, K.; Takeda, Y. Bovine milk exosomes contain microRNA and mRNA and are taken up by human macrophages. J. Dairy Sci. 2015, 98, 2920–2933. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manca, S.; Upadhyaya, B.; Mutai, E.; Desaulniers, A.T.; Cederberg, R.A.; White, B.R.; Zempleni, J. Milk exosomes are bioavailable and distinct microRNA cargos have unique tissue distribution patterns. Sci. Rep. 2018, 8, 11321. [Google Scholar] [CrossRef] [Green Version]
- Reinhardt, T.A.; Lippolis, J.D.; Nonnecke, B.J.; Sacco, R.E. Bovine milk exosome proteome. J. Proteom. 2012, 75, 1486–1492. [Google Scholar] [CrossRef]
- Izumi, H.; Kosaka, N.; Shimizu, T.; Sekine, K.; Ochiya, T.; Takase, M. Bovine milk contains microRNA and messenger RNA that are stable under degradative conditions. J. Dairy Sci. 2012, 95, 4831–4841. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakajima, M.; Shinoda, I.; Samejima, Y.; Miyauchi, H.; Fukuwatari, Y.; Hayasawa, H. Kappa-casein suppresses melanogenesis in cultured pigment cells. Pigment Cell Res. 1996, 9, 235–239. [Google Scholar] [CrossRef]
- Nakajima, M.; Shinoda, I.; Mikogami, T.; Iwamoto, H.; Hashimoto, S.; Miyauchi, H.; Fukuwatari, Y.; Hayasawa, H. Beta-lactoglobulin suppresses melanogenesis in cultured human melanocytes. Pigment Cell Res. 1997, 10, 410–413. [Google Scholar] [CrossRef] [PubMed]
- Rong, J.; Shan, C.; Liu, S.; Zheng, H.; Liu, C.; Liu, M.; Jin, F.; Wang, L. Skin resistance to UVB-induced oxidative stress and hyperpigmentation by the topical use of Lactobacillus helveticus NS8-fermented milk supernatant. J. Appl. Microbiol. 2017, 123, 511–523. [Google Scholar] [CrossRef]
- Park, J.Y.; Juhnn, Y.S. cAMP signaling increases histone deacetylase 8 expression via the Epac2-Rap1A-Akt pathway in H1299 lung cancer cells. Exp. Mol. Med. 2017, 49, e297. [Google Scholar] [CrossRef] [Green Version]
- Hwang, G.Y.; Choung, S.Y. Anti-melanogenic effects of Aster spathulifolius extract in UVB-exposed C57BL/6J mice and B16F10 melanoma cells through the regulation of MAPK/ERK and AKT/GSK3beta signaling. J. Pharm. Pharmacol. 2016, 68, 503–513. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.Y.; You, Y.J.; Liu, Y.J.; Hou, C.W.; Wu, C.S.; Wen, K.C.; Lin, C.Y.; Chiang, H.M. Sesamol Inhibited Melanogenesis by Regulating Melanin-Related Signal Transduction in B16F10 Cells. Int. J. Mol. Sci. 2018, 19, 1108. [Google Scholar] [CrossRef] [Green Version]
- Takano, K.; Hachiya, A.; Murase, D.; Tanabe, H.; Kasamatsu, S.; Takahashi, Y.; Moriwaki, S.; Hase, T. Quantitative changes in the secretion of exosomes from keratinocytes homeostatically regulate skin pigmentation in a paracrine manner. J. Dermatol. 2020, 47, 265–276. [Google Scholar] [CrossRef]
- Liu, Y.; Xue, L.; Gao, H.; Chang, L.; Yu, X.; Zhu, Z.; He, X.; Geng, J.; Dong, Y.; Li, H.; et al. Exosomal miRNA derived from keratinocytes regulates pigmentation in melanocytes. J. Dermatol. Sci. 2019, 93, 159–167. [Google Scholar] [CrossRef] [Green Version]
- Zhao, C.; Wang, D.; Wang, X.; Mao, Y.; Xu, Z.; Sun, Y.; Mei, X.; Song, J.; Shi, W. Down-regulation of exosomal miR-200c derived from keratinocytes in vitiligo lesions suppresses melanogenesis. J. Cell Mol. Med. 2020, 24, 12164–12175. [Google Scholar] [CrossRef] [PubMed]
- Cho, B.S.; Lee, J.; Won, Y.; Duncan, D.I.; Jin, R.C.; Lee, J.; Kwon, H.H.; Park, C.H.; Yang, S.H.; Park, B.C.; et al. Skin brightening efficacy of exosomes derived from human adipose tissue-derived stem/stromal cells: A prospective, split-face, randomized placebo-controlled study. Cosmetics 2020, 7, 90. [Google Scholar] [CrossRef]
- Lee, R.; Ko, H.J.; Kim, K.; Sohn, Y.; Min, S.Y.; Kim, J.A.; Na, D.; Yeon, J.H. Anti-melanogenic effects of extracellular vesicles derived from plant leaves and stems in mouse melanoma cells and human healthy skin. J. Extracell Vesicles 2019, 9, 1703480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adriano, B.; Cotto, N.M.; Chauhan, N.; Jaggi, M.; Chauhan, S.C.; Yallapu, M.M. Milk exosomes: Nature’s abundant nanoplatform for theranostic applications. Bioact. Mater. 2021, 6, 2479–2490. [Google Scholar] [CrossRef]
- Doyle, L.M.; Wang, M.Z. Overview of extracellular vesicles, their origin, composition, purpose, and methods for exosome isolation and analysis. Cells 2019, 8, 727. [Google Scholar] [CrossRef] [Green Version]
- Reif, S.; Shiff, Y.E.; Gerstl, R.G. Milk-derived exosomes (MDEs) have a different biological effect on normal fetal colon epithelial cells compared to colon tumor cells in a miRNA-dependent manner. J. Transl. Med. 2019, 17, 325. [Google Scholar] [CrossRef]
- Relif, S.; Shiff, Y.E.; Koroukhov, N.; Shilo, I.; Musseri, M.; Gerstl, R.G. Cow and human milk-derived exosomes ameliorate colitis in DSS murine model. Nutrients 2020, 12, 2589. [Google Scholar] [CrossRef]
- Yun, B.; Maburutse, B.E.; Kang, M.; Park, M.R.; Park, D.J.; Kim, Y.; Oh, S. Short communication: Dietary bovine milk-derived exosomes improve bone health in an osteoporosis-induced mouse model. J. Dairy Sci. 2020, 103, 7752–7760. [Google Scholar] [CrossRef] [PubMed]
- Arntz, O.J.; Pieters, B.C.; Oliveira, M.C.; Broeren, M.G.; Bennink, M.B.; de Vries, M.; van Lent, P.L.; Koenders, M.I.; van den Berg, W.B.; van der Kraan, P.M.; et al. Oral administration of bovine milk derived extracellular vesicles attenuates arthritis in two mouse models. Mol. Nutr. Food Res. 2015, 59, 1701–1712. [Google Scholar] [CrossRef]
- Samuel, M.; Chisanga, D.; Liem, M.; Keerthikumar, S.; Anand, S.; Ang, C.S.; Adda, C.G.; Versteegen, E.; Jois, M.; Mathivanan, S. Bovine milk-derived exosomes from colostrum are enriched with proteins implicated in immune response and growth. Sci. Rep. 2017, 7, 5933. [Google Scholar] [CrossRef] [PubMed]
- Benmoussa, A.; Gotti, C.; Bourassa, S.; Gilbert, C.; Provost, P. Identification of protein markers for extracellular vesicle (EV) subsets in cow’s milk. J. Proteom. 2019, 192, 78–88. [Google Scholar] [CrossRef]
- Zhao, Y.; Wang, P.; Meng, J.; Ji, Y.; Xu, D.; Chen, T.; Fan, R.; Yu, X.; Yao, J.; Dong, C. MicroRNA-27a-3p inhibits melanogenesis in mouse skin melanocytes by targeting Wnt3a. Int. J. Mol. Sci. 2015, 16, 10921–10933. [Google Scholar] [CrossRef] [Green Version]
- Guo, J.; Zhang, J.; Wang, W.; Cheung, F.W.; Lu, Y.; Ng, C.; Kung, H.; Liu, W. MicroRNA-218 inhibits melanogenesis by directly suppressing microphthalmia-associated transcription factor expression. RNA Biol. 2014, 11, 732–741. [Google Scholar] [CrossRef] [Green Version]
- Noguchi, S.; Kumazaki, M.; Yasui, Y.; Mori, T.; Yamada, N.; Akao, Y. MicroRNA-203 regulates melanosome transport and tyrosinase expression in melanoma cells by targeting kinesin superfamily protein 5b. J. Investig. Dermatol. 2014, 134, 461–469. [Google Scholar] [CrossRef] [PubMed] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bae, I.-S.; Kim, S.H. Milk Exosome-Derived MicroRNA-2478 Suppresses Melanogenesis through the Akt-GSK3β Pathway. Cells 2021, 10, 2848. https://doi.org/10.3390/cells10112848
Bae I-S, Kim SH. Milk Exosome-Derived MicroRNA-2478 Suppresses Melanogenesis through the Akt-GSK3β Pathway. Cells. 2021; 10(11):2848. https://doi.org/10.3390/cells10112848
Chicago/Turabian StyleBae, In-Seon, and Sang Hoon Kim. 2021. "Milk Exosome-Derived MicroRNA-2478 Suppresses Melanogenesis through the Akt-GSK3β Pathway" Cells 10, no. 11: 2848. https://doi.org/10.3390/cells10112848
APA StyleBae, I.-S., & Kim, S. H. (2021). Milk Exosome-Derived MicroRNA-2478 Suppresses Melanogenesis through the Akt-GSK3β Pathway. Cells, 10(11), 2848. https://doi.org/10.3390/cells10112848





