Macrophage-Derived Immunoglobulin M Inhibits Inflammatory Responses via Modulating Endoplasmic Reticulum Stress
Abstract
:1. Introduction
2. Materials and Methods
2.1. Mice
2.2. Isolation of Primary Macrophages
2.3. Cell Culture and Transfection
2.4. Protein Extraction and Western Blot Analysis
2.5. Immunofluorescence
2.6. Flow Cytometry Analysis
2.7. RT-PCR and Repertoire Analysis of IgM
2.8. Next-Generation Sequencing and Bioinformatic Analysis
2.9. Transwell Assay
2.10. ELISA of Cytokines
2.11. Glutathione S-Transferase (GST) Pull-Down Assay
2.12. Statistical Analysis
3. Results
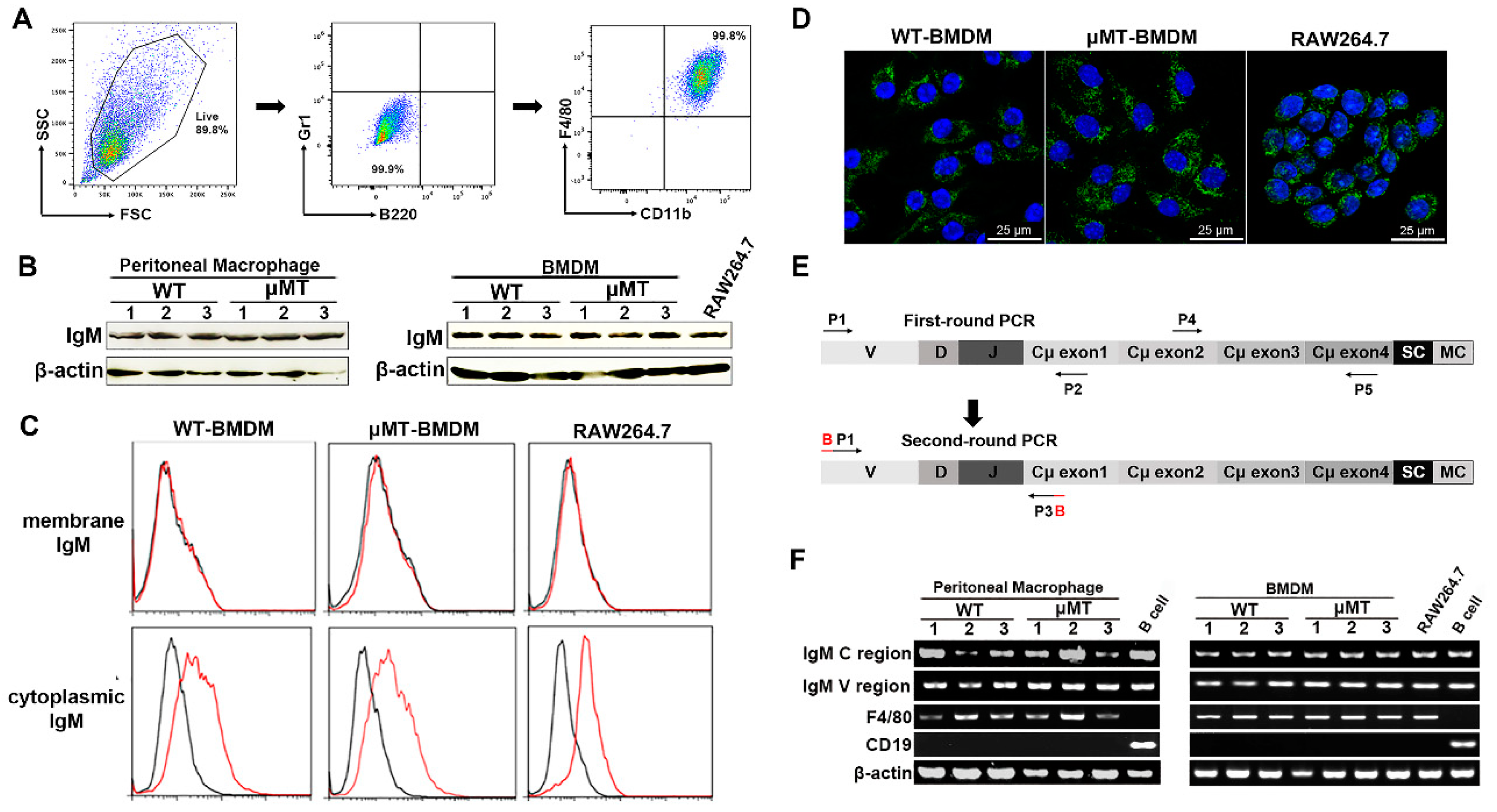
3.1. IgM Expression in Primary Macrophages of Adult Mice and Macrophage Cell Line-RAW264.7
3.2. The Repertoire of IgM Heavy Chain Variable Region Sequences Displays Restricted Diversity in Macrophages
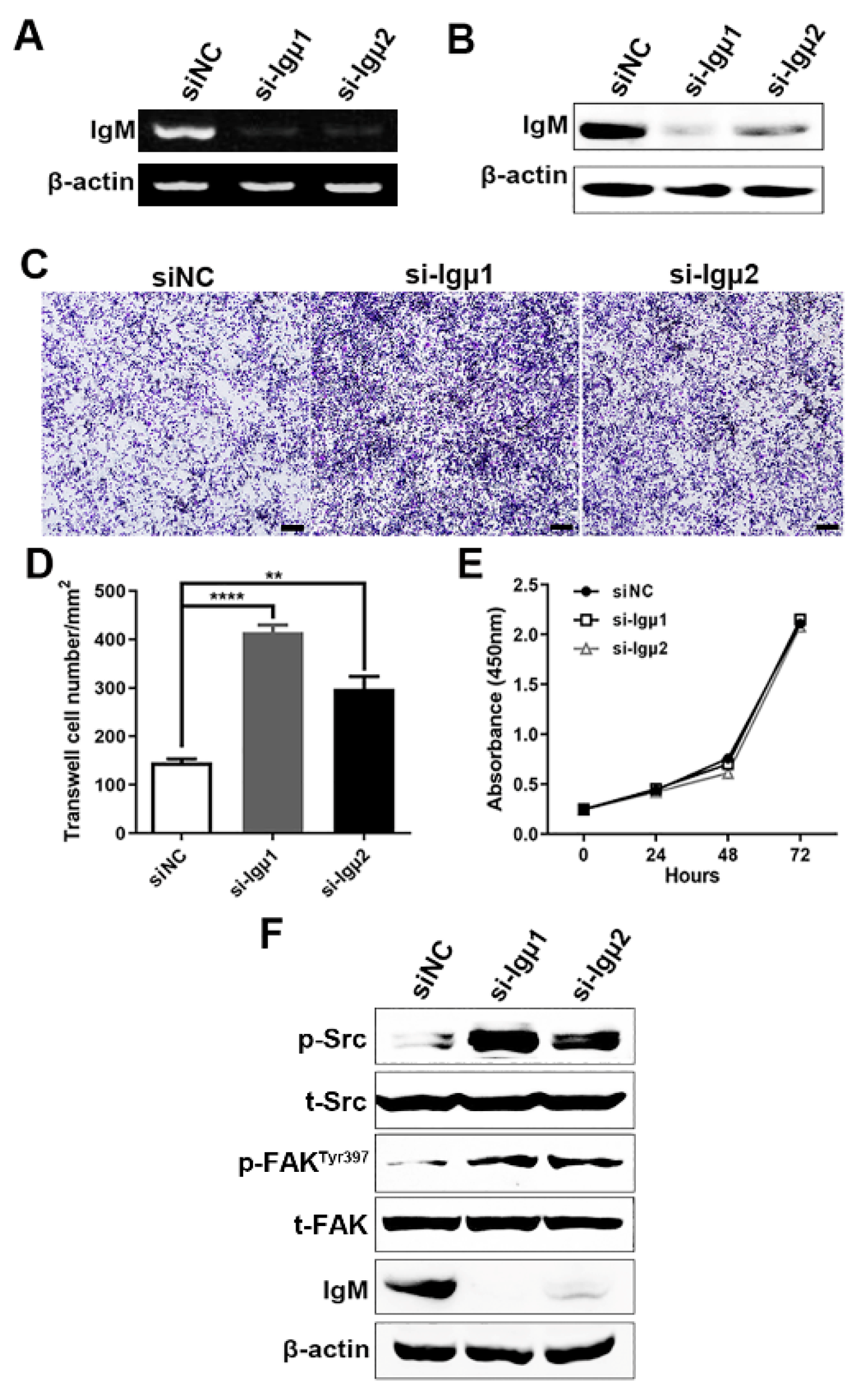
3.3. Knockdown of IgM Enhanced the Migratory Ability of Macrophages through Activating Src/FAK Axis
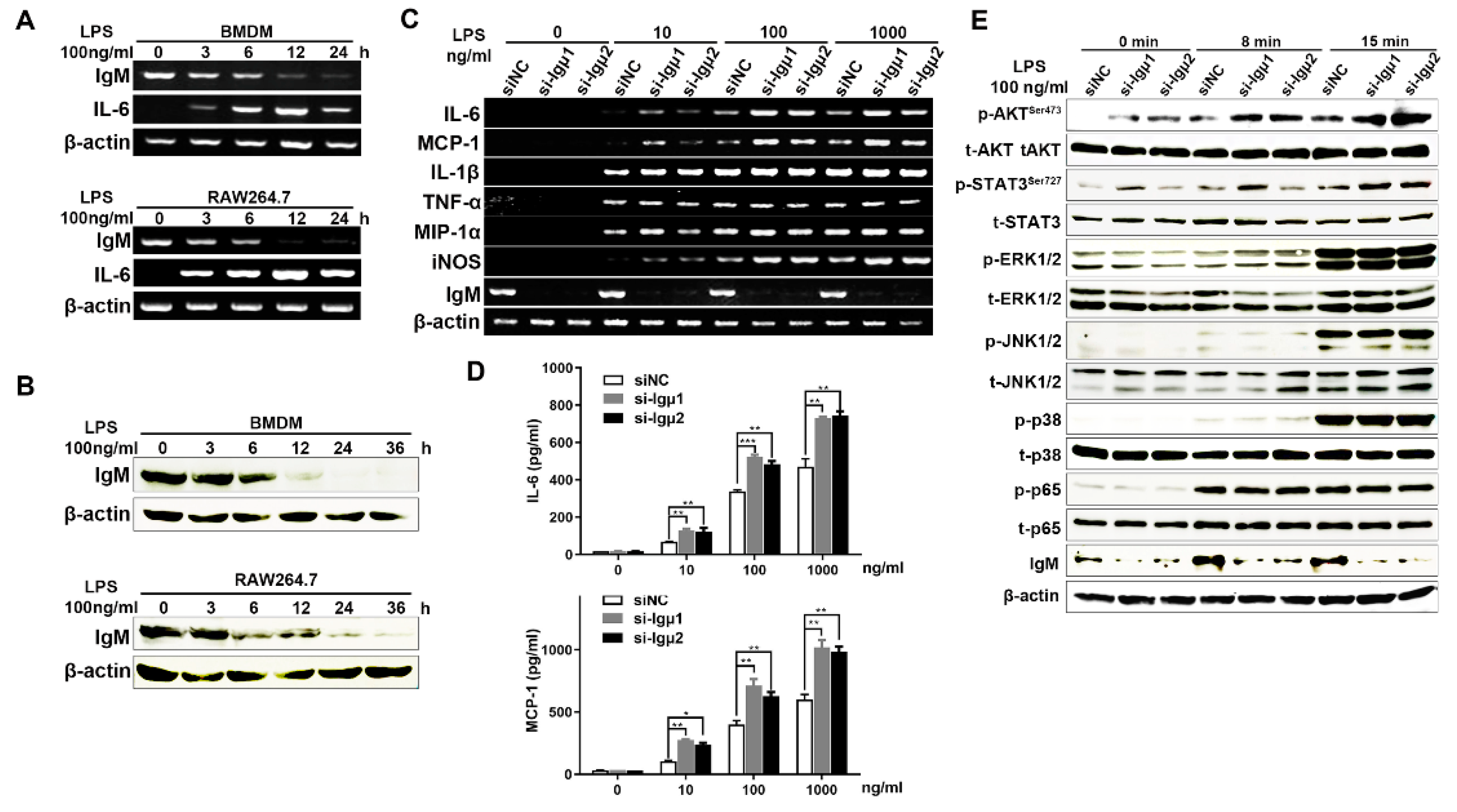
3.4. Knockdown of IgM Promoted Pro-Inflammatory Response of Macrophages through Secreting Pro-Inflammatory Mediators
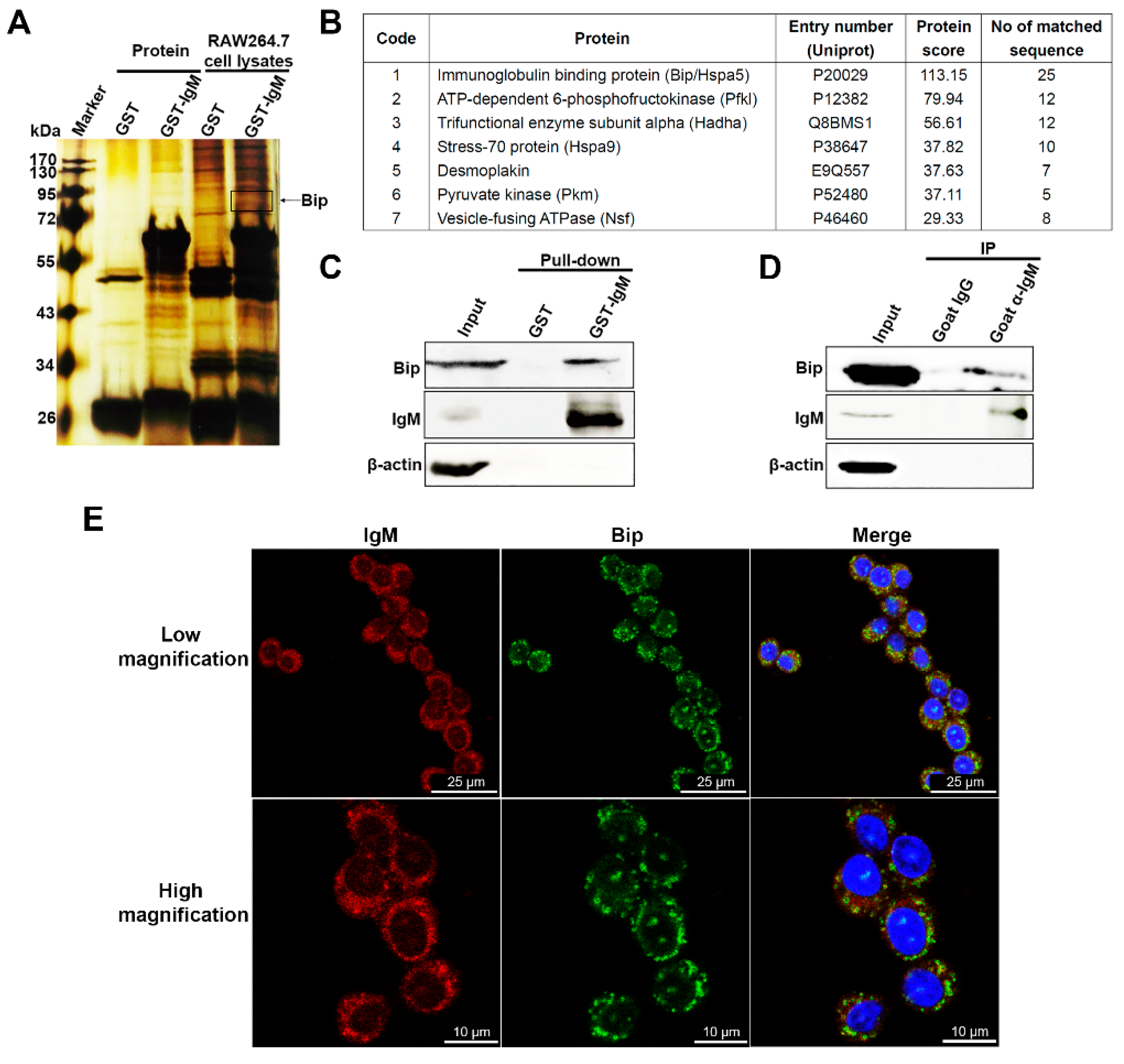
3.5. IgM Located in ER and Interacts with Bip in Macrophage
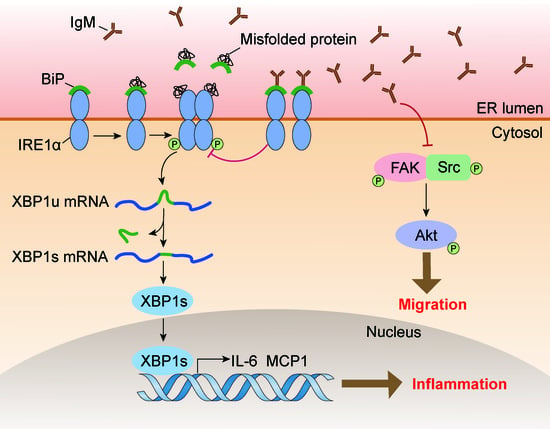
3.6. IgM Regulating Bip Expression via IRE1α-Mediated XBP1 mRNA Splicing Is Necessary for ER-Stress-Induced Macrophage Activation
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Blandino, R.; Baumgarth, N. Secreted IgM: New tricks for an old molecule. J. Leukoc. Biol. 2019, 106, 1021–1034. [Google Scholar] [CrossRef]
- Sathe, A.; Cusick, J.K. Biochemistry, Immunoglobulin M. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2021. [Google Scholar]
- Boes, M. Role of natural and immune IgM antibodies in immune responses. Mol. Immunol. 2000, 37, 1141–1149. [Google Scholar] [CrossRef]
- Haas, I.G.; Wabl, M. Immunoglobulin heavy chain binding protein. Nature 1983, 306, 387–389. [Google Scholar] [CrossRef]
- Anelli, T.; Ceppi, S.; Bergamelli, L.; Cortini, M.; Masciarelli, S.; Valetti, C.; Sitia, R. Sequential steps and checkpoints in the early exocytic compartment during secretory IgM biogenesis. EMBO J. 2007, 26, 4177–4188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colucci, M.; Stockmann, H.; Butera, A.; Masotti, A.; Baldassarre, A.; Giorda, E.; Petrini, S.; Rudd, P.M.; Sitia, R.; Emma, F.; et al. Sialylation of N-linked glycans influences the immunomodulatory effects of IgM on T cells. J. Immunol. 2015, 194, 151–157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiu, X.; Zhu, X.; Zhang, L.; Mao, Y.; Zhang, J.; Hao, P.; Li, G.; Lv, P.; Li, Z.; Sun, X.; et al. Human epithelial cancers secrete immunoglobulin g with unidentified specificity to promote growth and survival of tumor cells. Cancer Res. 2003, 63, 6488–6495. [Google Scholar] [PubMed]
- Babbage, G.; Ottensmeier, C.H.; Blaydes, J.; Stevenson, F.K.; Sahota, S.S. Immunoglobulin heavy chain locus events and expression of activation-induced cytidine deaminase in epithelial breast cancer cell lines. Cancer Res. 2006, 66, 3996–4000. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, F.; Zhang, L.; Zheng, J.; Zhao, L.; Huang, J.; Shao, W.; Liao, Q.; Ma, T.; Geng, L.; Yin, C.C.; et al. Spontaneous production of immunoglobulin M in human epithelial cancer cells. PLoS ONE 2012, 7, e51423. [Google Scholar] [CrossRef]
- Shao, W.; Hu, F.; Ma, J.; Zhang, C.; Liao, Q.; Zhu, Z.; Liu, E.; Qiu, X. Epithelial cells are a source of natural IgM that contribute to innate immune responses. Int. J. Biochem. Cell Biol. 2016, 73, 19–29. [Google Scholar] [CrossRef]
- Nourshargh, S.; Alon, R. Leukocyte migration into inflamed tissues. Immunity 2014, 41, 694–707. [Google Scholar] [CrossRef] [Green Version]
- Wynn, T.A.; Chawla, A.; Pollard, J.W. Macrophage biology in development, homeostasis and disease. Nature 2013, 496, 445–455. [Google Scholar] [CrossRef]
- Wu, Y.; Hirschi, K.K. Tissue-Resident Macrophage Development and Function. Front. Cell Dev. Biol. 2020, 8, 617879. [Google Scholar] [CrossRef]
- Locati, M.; Curtale, G.; Mantovani, A. Diversity, Mechanisms, and Significance of Macrophage Plasticity. Annu. Rev. Pathol. 2020, 15, 123–147. [Google Scholar] [CrossRef] [Green Version]
- Netea, M.G.; Balkwill, F.; Chonchol, M.; Cominelli, F.; Donath, M.Y.; Giamarellos-Bourboulis, E.J.; Golenbock, D.; Gresnigt, M.S.; Heneka, M.T.; Hoffman, H.M.; et al. A guiding map for inflammation. Nat. Immunol. 2017, 18, 826–831. [Google Scholar] [CrossRef] [Green Version]
- Fuchs, T.; Hahn, M.; Ries, L.; Giesler, S.; Busch, S.; Wang, C.; Han, J.; Schulze, T.J.; Puellmann, K.; Beham, A.W.; et al. Expression of combinatorial immunoglobulins in macrophages in the tumor microenvironment. PLoS ONE 2018, 13, e0204108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Busch, S.; Talamini, M.; Brenner, S.; Abdulazim, A.; Hanggi, D.; Neumaier, M.; Seiz-Rosenhagen, M.; Fuchs, T. Circulating monocytes and tumor-associated macrophages express recombined immunoglobulins in glioblastoma patients. Clin. Transl. Med. 2019, 8, 18. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Sun, X.; Gong, X.; He, Z.; Chen, L.; Qiu, X.; Yin, C.C. Rearrangement and expression of the immunoglobulin mu-chain gene in human myeloid cells. Cell. Mol. Immunol. 2014, 11, 94–104. [Google Scholar] [CrossRef] [Green Version]
- Qiu, X.; Sun, X.; He, Z.; Huang, J.; Hu, F.; Chen, L.; Lin, P.; You, M.J.; Medeiros, L.J.; Yin, C.C. Immunoglobulin gamma heavy chain gene with somatic hypermutation is frequently expressed in acute myeloid leukemia. Leukemia 2013, 27, 92–99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shao, W.; Zhang, C.; Liu, E.; Zhang, L.; Ma, J.; Zhu, Z.; Gong, X.; Qin, Z.; Qiu, X. Identification of Liver Epithelial Cell-derived Ig Expression in mu chain-deficient mice. Sci. Rep. 2016, 6, 23669. [Google Scholar] [CrossRef] [Green Version]
- Brezinschek, H.P.; Foster, S.J.; Brezinschek, R.I.; Dorner, T.; Domiati-Saad, R.; Lipsky, P.E. Analysis of the human VH gene repertoire. Differential effects of selection and somatic hypermutation on human peripheral CD5(+)/IgM+ and CD5(-)/IgM+ B cells. J. Clin. Investig. 1997, 99, 2488–2501. [Google Scholar] [CrossRef] [Green Version]
- Ward, C.; Rettig, T.A.; Hlavacek, S.; Bye, B.A.; Pecaut, M.J.; Chapes, S.K. Effects of spaceflight on the immunoglobulin repertoire of unimmunized C57BL/6 mice. Life Sci. Space Res. 2018, 16, 63–75. [Google Scholar] [CrossRef]
- Owen, K.A.; Pixley, F.J.; Thomas, K.S.; Vicente-Manzanares, M.; Ray, B.J.; Horwitz, A.F.; Parsons, J.T.; Beggs, H.E.; Stanley, E.R.; Bouton, A.H. Regulation of lamellipodial persistence, adhesion turnover, and motility in macrophages by focal adhesion kinase. J. Cell Biol. 2007, 179, 1275–1287. [Google Scholar] [CrossRef] [Green Version]
- Preissler, S.; Ron, D. Early Events in the Endoplasmic Reticulum Unfolded Protein Response. Cold Spring Harb. Perspect. Biol. 2019, 11, a033894. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Todd, D.J.; Lee, A.H.; Glimcher, L.H. The endoplasmic reticulum stress response in immunity and autoimmunity. Nat. Rev. Immunol. 2008, 8, 663–674. [Google Scholar] [CrossRef]
- Kopp, M.C.; Larburu, N.; Durairaj, V.; Adams, C.J.; Ali, M.M.U. UPR proteins IRE1 and PERK switch BiP from chaperone to ER stress sensor. Nat. Struct. Mol. Biol. 2019, 26, 1053–1062. [Google Scholar] [CrossRef] [PubMed]
- Bornemann, K.D.; Brewer, J.W.; Beck-Engeser, G.B.; Corley, R.B.; Haas, I.G.; Jack, H.M. Roles of heavy and light chains in IgM polymerization. Proc. Natl. Acad. Sci. USA 1995, 92, 4912–4916. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malhotra, J.D.; Kaufman, R.J. Endoplasmic reticulum stress and oxidative stress: A vicious cycle or a double-edged sword? Antioxid. Redox Signal. 2007, 9, 2277–2293. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Xia, M.; Sun, X.; He, Z.; Hu, F.; Chen, L.; Bueso-Ramos, C.E.; Qiu, X.; Yin, C.C. IGK with conserved IGKappaV/IGKappaJ repertoire is expressed in acute myeloid leukemia and promotes leukemic cell migration. Oncotarget 2015, 6, 39062–39072. [Google Scholar] [CrossRef] [Green Version]
- Mandric, I.; Rotman, J.; Yang, H.T.; Strauli, N.; Montoya, D.J.; Van Der Wey, W.; Ronas, J.R.; Statz, B.; Yao, D.; Petrova, V.; et al. Profiling immunoglobulin repertoires across multiple human tissues using RNA sequencing. Nat. Commun. 2020, 11, 3126. [Google Scholar] [CrossRef]
- Jiang, D.; Ge, J.; Liao, Q.; Ma, J.; Liu, Y.; Huang, J.; Wang, C.; Xu, W.; Zheng, J.; Shao, W.; et al. IgG and IgA with potential microbial-binding activity are expressed by normal human skin epidermal cells. Int. J. Mol. Sci. 2015, 16, 2574–2590. [Google Scholar] [CrossRef] [Green Version]
- Tang, J.; Zhang, J.; Liu, Y.; Liao, Q.; Huang, J.; Geng, Z.; Xu, W.; Sheng, Z.; Lee, G.; Zhang, Y.; et al. Lung squamous cell carcinoma cells express non-canonically glycosylated IgG that activates integrin-FAK signaling. Cancer Lett. 2018, 430, 148–159. [Google Scholar] [CrossRef]
- Zhang, C.; Xiao, L.; Huang, Y.; Zhang, L.; Jiang, D.; Shao, W.; Zheng, J.; Hu, F.; Chu, M.; Huang, J.; et al. NBIGV-DB: A dedicated database of non-B cell derived immunoglobulin variable region. Gene 2021, 772, 145378. [Google Scholar] [CrossRef] [PubMed]
- Maa, M.C.; Chang, M.Y.; Chen, Y.J.; Lin, C.H.; Yu, C.J.; Yang, Y.L.; Li, J.; Chen, P.R.; Tang, C.H.; Lei, H.Y.; et al. Requirement of inducible nitric-oxide synthase in lipopolysaccharide-mediated Src induction and macrophage migration. J. Biol. Chem. 2008, 283, 31408–31416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitra, S.K.; Hanson, D.A.; Schlaepfer, D.D. Focal adhesion kinase: In command and control of cell motility. Nat. Rev. Mol. Cell Biol. 2005, 6, 56–68. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Zhang, F.; Qin, K.; Wu, M.; Li, H.; Zhu, H.; Ning, Q.; Lei, P.; Shen, G. Glucose-Regulated Protein 78-Induced Myeloid Antigen-Presenting Cells Maintained Tolerogenic Signature upon LPS Stimulation. Front. Immunol. 2016, 7, 552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feige, M.J.; Groscurth, S.; Marcinowski, M.; Shimizu, Y.; Kessler, H.; Hendershot, L.M.; Buchner, J. An unfolded CH1 domain controls the assembly and secretion of IgG antibodies. Mol. Cell 2009, 34, 569–579. [Google Scholar] [CrossRef] [Green Version]
- Hetz, C.; Glimcher, L.H. Fine-tuning of the unfolded protein response: Assembling the IRE1alpha interactome. Mol. Cell 2009, 35, 551–561. [Google Scholar] [CrossRef] [Green Version]
- Hetz, C.; Papa, F.R. The Unfolded Protein Response and Cell Fate Control. Mol. Cell 2018, 69, 169–181. [Google Scholar] [CrossRef] [Green Version]
- Adams, C.J.; Kopp, M.C.; Larburu, N.; Nowak, P.R.; Ali, M.M.U. Structure and Molecular Mechanism of ER Stress Signaling by the Unfolded Protein Response Signal Activator IRE1. Front. Mol. Biosci. 2019, 6, 11. [Google Scholar] [CrossRef] [Green Version]
- Martinon, F.; Chen, X.; Lee, A.H.; Glimcher, L.H. TLR activation of the transcription factor XBP1 regulates innate immune responses in macrophages. Nat. Immunol. 2010, 11, 411–418. [Google Scholar] [CrossRef]


| Gene Name | Primer Sequence 5′-3′ | |
|---|---|---|
| IgM variable region | sense primer (P1) | CTTCCGGAATTCSARGTNMAGCTGSAGSAGTCWGG |
| external antisense primer (P2) | CTTCAAGAAGGTGAGACCC | |
| internal antisense primer (P3) | GACATTTGGGAAGGACTGACTCTC | |
| IgM constant region | sense primer (P4) | GCTAAAGGATGGGAAGCT |
| antisense primer (P5) | GGATGCTGTGGGTAAAGT | |
| F4/80 | sense primer | AAGTGACTCACCTTGTGGTCC |
| antisense primer | TAGATGCAAAGCCAGGGTGG | |
| CD19 | sense primer | AAGGAAGCGAATGACTGACCC |
| antisense primer | GAAGTCCATCATCCTGCCA | |
| β-actin | sense primer | CAGGGTGTGATGGTGGGAAT |
| antisense primer | GGAAAAGAGCCTCAGGGCAT | |
| IL-6 | sense primer | GCCTTCTTGGGACTGATGCT |
| antisense primer | GCCACTCCTTCTGTGACTCC | |
| MCP-1 | sense primer | GGCTCAGCCAGATGCAGTTA |
| antisense primer | GTGCTTGAGGTGGTTGTGGA | |
| IL-1β | sense primer | TGCCACCTTTTGACAGTGAT |
| antisense primer | TGGGTGTGCCGTCTTTCATT | |
| TNF-α | sense primer | ACGTGGAACTGGCAGAAGAG |
| antisense primer | TGAGGGTCTGGGCCATAGAA | |
| MIP-1α | sense primer | ACTGCCCTTGCTGTTCTTCT |
| antisense primer | GTCTCTTTGGAGTCAGCGCA | |
| iNOS | sense primer | GTGGTGACAAGCACATTTGG |
| antisense primer | GGCTGGACTTTTCACTCTGC | |
| XBP-1 | sense primer | GAACCAGGAGTTAAGAACACG |
| antisense primer | AGGCAACAGTGTCAGAGTCC |
| Sample | Total Sequences | Productive Sequences | Productive Percentage (%) |
|---|---|---|---|
| No.1 WT-BMDM | 3843 | 3704 | 96.38 |
| No.2 WT-BMDM | 2380 | 2320 | 97.48 |
| No.3 WT-BMDM | 2688 | 2596 | 96.58 |
| No.1 μMT-BMDM | 842 | 816 | 96.91 |
| No.2 μMT-BMDM | 929 | 845 | 90.96 |
| No.3 μMT-BMDM | 1553 | 1514 | 97.49 |
| RAW264.7 | 8295 | 7946 | 95.79 |
| No.1 WT-B | 1930 | 1773 | 91.97 |
| No.2 WT-B | 2022 | 1827 | 90.36 |
| No.3 WT-B | 2085 | 1943 | 93.19 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gong, X.; Yan, H.; Ma, J.; Zhu, Z.; Zhang, S.; Xu, W.; Huang, J.; Qiu, X. Macrophage-Derived Immunoglobulin M Inhibits Inflammatory Responses via Modulating Endoplasmic Reticulum Stress. Cells 2021, 10, 2812. https://doi.org/10.3390/cells10112812
Gong X, Yan H, Ma J, Zhu Z, Zhang S, Xu W, Huang J, Qiu X. Macrophage-Derived Immunoglobulin M Inhibits Inflammatory Responses via Modulating Endoplasmic Reticulum Stress. Cells. 2021; 10(11):2812. https://doi.org/10.3390/cells10112812
Chicago/Turabian StyleGong, Xiaoting, Huige Yan, Junfan Ma, Zhu Zhu, Shenghua Zhang, Weiyan Xu, Jing Huang, and Xiaoyan Qiu. 2021. "Macrophage-Derived Immunoglobulin M Inhibits Inflammatory Responses via Modulating Endoplasmic Reticulum Stress" Cells 10, no. 11: 2812. https://doi.org/10.3390/cells10112812
APA StyleGong, X., Yan, H., Ma, J., Zhu, Z., Zhang, S., Xu, W., Huang, J., & Qiu, X. (2021). Macrophage-Derived Immunoglobulin M Inhibits Inflammatory Responses via Modulating Endoplasmic Reticulum Stress. Cells, 10(11), 2812. https://doi.org/10.3390/cells10112812