The CAM Model for CIC-DUX4 Sarcoma and Its Potential Use for Precision Medicine
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cells and Media
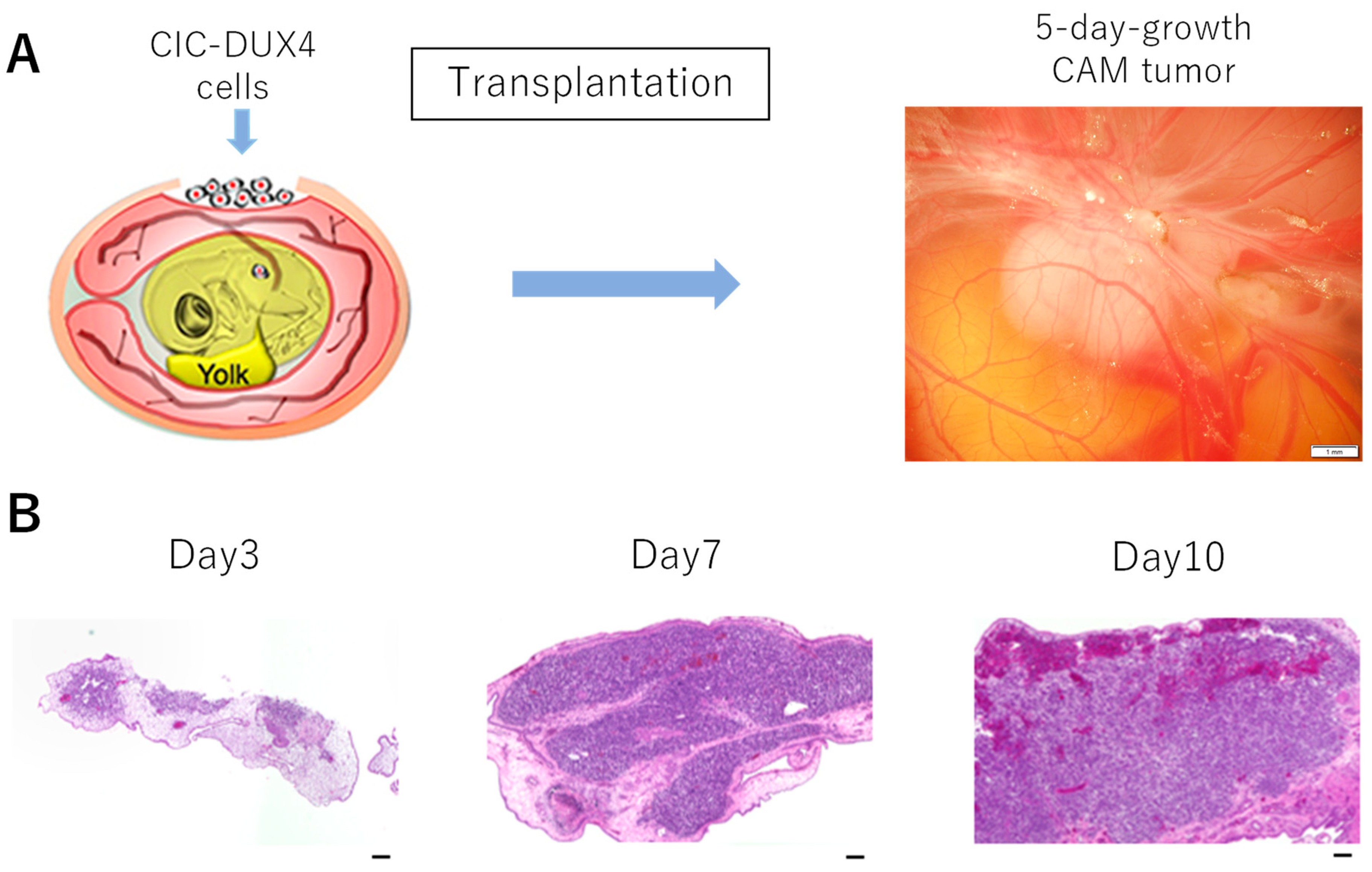
2.2. CAM Assay
2.3. Immunohistochemistry
2.4. Immunoblotting
2.5. RNA Isolation and RT-PCR
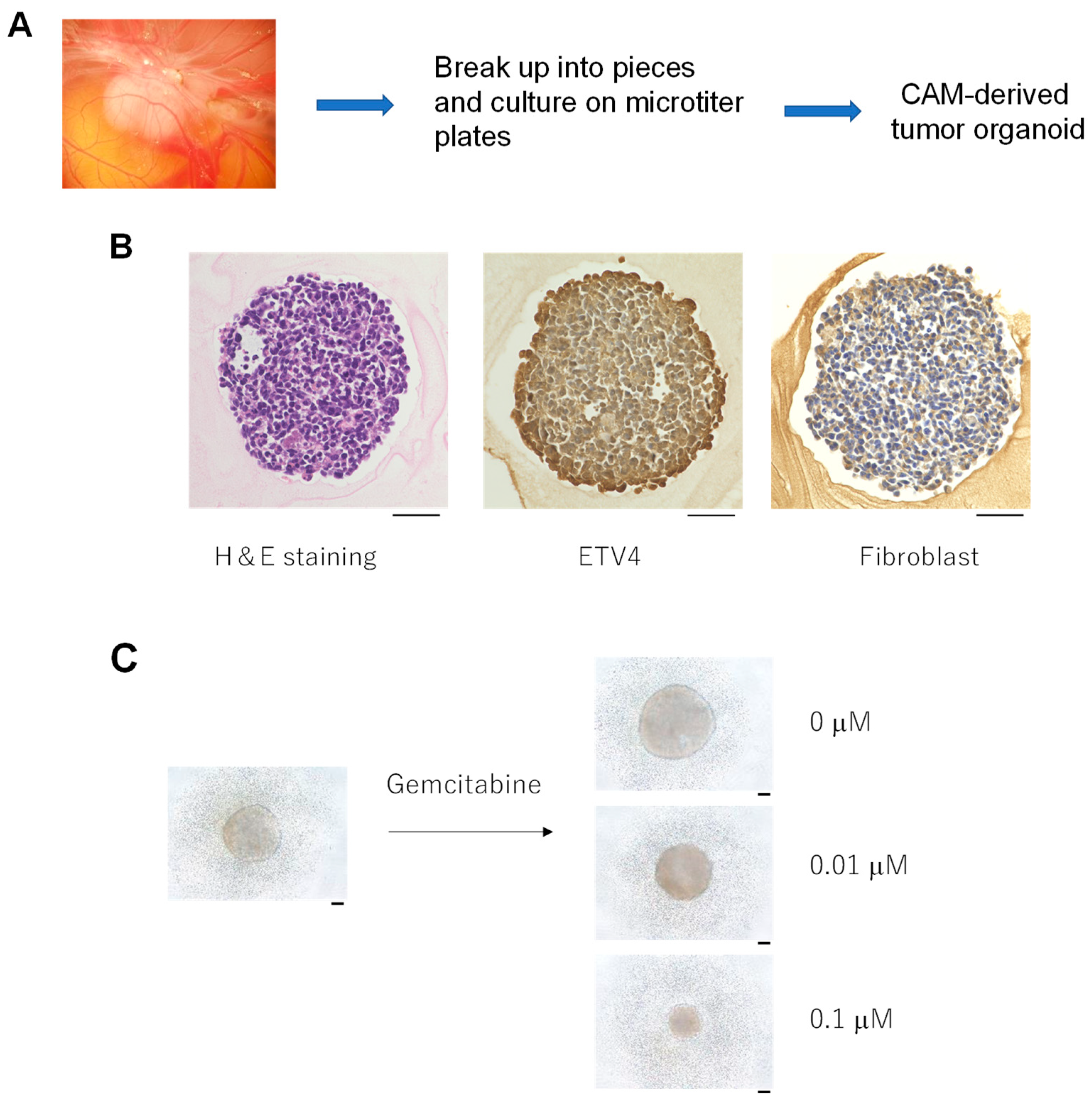
2.6. Formation of Tumor Organoids from CAM Tumor
3. Results
3.1. Formation of CIC-DUX4 Tumor on the CAM Membrane
3.2. Molecular Features and Tumor Microenvironment
3.3. CAM Tumor Can Be Transferred to a Fresh CAM
3.4. Formation of Tumor Organoids from the CAM Tumor
3.5. The CIC-DUX4 Gene Is Retained in the CAM, Passaged CAM and CAM-Derived Tumor Organoids
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Italiano, A.; Sung, Y.S.; Zhang, L.; Singer, S.; Maki, R.G.; Coindre, J.M.; Antonescu, C.R. High prevalence of CIC fusion with double homeobox (DUX4) transcription factors in EWSR1-negative undifferentiated small blue round cell sarcomas. Genes Chromosomes Cancer 2012, 51, 207–218. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, A.; Goto, K.; Kodaira, M.; Kobayashi, E.; Kawamoto, H.; Mori, T.; Yoshimoto, S.; Endo, O.; Kodama, N.; Kushima, R.; et al. Cic-rearranged sarcomas: A Study of 20 cases and comparisons with Ewing sarcomas. Am. J. Surg Pathol. 2016, 40, 313–323. [Google Scholar] [CrossRef]
- Specht, K.; Sung, Y.S.; Zhang, L.; Richter, G.H.; Fletcher, C.D.; Antonescu, C.R. Distinct transcriptional signature and immunoprofile of CIC-DUX4 fusion-positive round cell tumors compared to EWSR1-rearranged Ewing sarcomas: Further evidence toward distinct pathologic entities. Genes Chromosomes Cancer 2014, 53, 622–633. [Google Scholar] [CrossRef] [Green Version]
- Kawamura-Saito, M.; Yamazaki, Y.; Kaneko, K.; Kawaguchi, N.; Kanda, H.; Mukai, H.; Gotoh, T.; Motoi, T.; Fukayama, M.; Aburatani, H.; et al. Fusion between CIC and DUX4 up-regulates PEA3 family genes in Ewing-like sarcomas with t(4;19) (q35;q13) translocation. Hum. Mol. Genet. 2006, 15, 2125–2137. [Google Scholar] [CrossRef]
- Antonescu, C.R.; Owosho, A.A.; Zhang, L.; Chen, S.; Deniz, K.; Huryn, J.M.; Kao, Y.C.; Huang, S.C.; Singer, S.; Tap, W.; et al. Sarcomas with CIC-rearrangements are a distinct pathologic entity with aggressive outcome: A clinicopathologic and molecular study of 115 Cases. Am. J. Surg. Pathol. 2017, 41, 941–949. [Google Scholar] [CrossRef]
- Delattre, O.; Zucman, J.; Plougastel, B.; Desmaze, C.; Melot, T.; Peter, M.; Kovar, H.; Joubert, I.; de Jong, P.; Rouleau, G.; et al. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature 1992, 359, 162–165. [Google Scholar] [CrossRef] [PubMed]
- Sorensen, P.H.; Lessnick, S.L.; Lopez-Terrada, D.; Liu, X.F.; Triche, T.J.; Denny, C.T. A second Ewing’s sarcoma translocation, t(21;22), fuses the EWS gene to another ETS-family transcription factor, ERG. Nat. Genet. 1994, 6, 146–151. [Google Scholar] [CrossRef] [PubMed]
- Ajuria, L.; Nieva, C.; Winkler, C.; Kuo, D.; Samper, N.; Andreu, M.J.; Helman, A.; González-Crespo, S.; Paroush, Z.; Courey, A.; et al. Capicua DNA-binding sites are general response elements for RTK signaling in Drosophila. Development 2011, 138, 915–924. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tseng, A.S.; Tapon, N.; Kanda, H.; Cigizoglu, S.; Edelmann, L.; Pellock, B.; White, K.; Hariharan, I.K. Capicua regulates cell proliferation downstream of the receptor tyrosine kinase/ras signaling pathway. Curr. Biol. 2007, 17, 728–733. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, Y.; Ha, N.; Forés, M.; Xiang, J.; Gläßer, C.; Maldera, J.; Jiménez, G.; Edgar, B.A. EGFR/Ras Signaling Controls Drosophila Intestinal Stem Cell Proliferation via Capicua-Regulated Genes. PLoS Genet. 2015, 11, e1005634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dixit, M.; Ansseau, E.; Tassin, A.; Winokur, S.; Shi, R.; Qian, H.; Sauvage, S.; Mattéotti, C.; van Acker, A.M.; Leo, O.; et al. DUX4, a candidate gene of facioscapulohumeral muscular dystrophy, encodes a transcriptional activator of PITX1. Proc. Natl. Acad. Sci. USA 2007, 104, 18157–18162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, H.; Wang, Z.; Jin, S.; Hao, H.; Zheng, L.; Zhou, B.; Zhang, W.; Lv, H.; Yuan, Y. Dux4 induces cell cycle arrest at G1 phase through upregulation of p21 expression. Biochem. Biophys. Res. Commun. 2014, 446, 235–240. [Google Scholar] [CrossRef] [PubMed]
- Young, J.M.; Whiddon, J.L.; Yao, Z.; Kasinathan, B.; Snider, L.; Geng, L.N.; Balog, J.; Tawil, R.; van der Maarel, S.M.; Tapscott, S.J. DUX4 binding to retroelements creates promoters that are active in FSHD muscle and testis. PLoS Genet. 2013, 9, e1003947. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oyama, R.; Takahashi, M.; Yoshida, A.; Sakumoto, M.; Takai, Y.; Kito, F.; Shiozawa, K.; Qiao, Z.; Arai, Y.; Shibata, T.; et al. Generation of novel patient-derived CIC-DUX4 sarcoma xenografts and cell lines. Sci. Rep. 2017, 7, 4712. [Google Scholar] [CrossRef] [Green Version]
- Yoshimatsu, Y.; Noguchi, R.; Tsuchiya, R.; Kito, F.; Sei, A.; Sugaya, J.; Nakagawa, M.; Yoshida, A.; Iwata, S.; Kawai, A.; et al. Establishment and characterization of NCC-CD52-C1: A novel patient-derived cell line of CIC-DUX4 sarcoma. Hum. Cell 2020, 33, 427–436. [Google Scholar] [CrossRef]
- Okimoto, R.A.; Wu, W.; Nanjo, S.; Olivas, V.; Lin, Y.K.; Ponce, R.K.; Oyama, R.; Kondo, T.; Bivona, T.G. CIC-DUX4 oncoprotein drives sarcoma metastasis and tumorigenesis via distinct regulatory programs. J. Clin. Oncol. 2019, 129, 3401–3406. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.K.; Wu, W.; Ponce, R.K.; Kim, J.W.; Okimoto, R.A. Negative MAPK-ERK regulation sustains CIC-DUX4 oncoprotein expression in undifferentiated sarcoma. Proc. Nat. Acad. Sci. USA 2020, 117, 20776–20784. [Google Scholar] [CrossRef]
- Ren, Y.; Ouyang, Z.; Hou, Z.; Yan, Y.; Zhi, Z.; Shi, M.; Du, M.; Liu, H.; Wen, Y.; Shao, Y. CIC Is a Mediator of the ERK1/2-DUSP6 Negative Feedback Loop. iScience 2020, 23, 101635. [Google Scholar] [CrossRef]
- Vu, B.T.; Shahin, S.A.; Croissant, J.; Fatieiev, Y.; Matsumoto, K.; Le-Hoang Doan, T.; Yik, T.; Simargi, S.; Ratliff, L.; Mauriello, C.; et al. Chick chorioallantoic membrane assay as an in vivo model to study the effect of nanoparticle-based anticancer drugs in ovarian cancer. Sci. Rep. 2018, 8, 8524. [Google Scholar] [CrossRef]
- Nowak-Sliwinska, P.; Segura, T.; Iruela-Arispe, M.L. The chicken chorioallantoic membrane model in biology, medicine and bioengineering. Angiogenesis 2014, 17, 779–804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ribatti, D.; Alessandri, G.; Baronio, M.; Raffaghello, L.; Cosimo, E.; Marimpietri, D.; Montaldo, P.G.; De Falco, G.; Caruso, A.; Vacca, A.; et al. Inhibition of neuroblastoma-induced angiogenesis by fenretinide. Int. J. Cancer 2001, 94, 314–321. [Google Scholar] [CrossRef]
- Herrmann, A.; Moss, D.; See, V. The chorioallantoic membrane pf the chick embryo to assess tumor formation and metastasis. Methods Mol. Biol. 2016, 1464, 97–105. [Google Scholar]
- Klingenberg, M.; Becker, J.; Eberth, S.; Kube, D.; Wilting, J. The chick chorioallantoic membrane as an in vivo xenograft model for Burkitt lymphoma. BMC Cancer 2014, 14, 339. [Google Scholar] [CrossRef] [Green Version]
- Komatsu, A.; Matsumoto, K.; Saito, T.; Muto, M.; Tamanoi, F. Patient derived chicken egg tumor model (PDcE model): Current status and critical issues. Cells 2019, 8, 440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mapanao, A.K.; Che, P.P.; Sarogni, P.; Siminia, P.; Giovannetti, E.; Voliani, V. Tumor grafted—chick chorioallantoic membrane as an alternative model for biological cancer research and conventional/nanomaterial-based theranostics evaluation. Expert Opin. Drug Metab. Toxicol. 2021, 17, 947–968. [Google Scholar] [CrossRef] [PubMed]
- Sys, G.; Bockstal, M.V.; Forsyth, R.; Blake, M.; Poffyn, B.; Uyttendaele, D.; Bracke, M.; De Wever, O. Tumor grafts derived from sarcoma patients retain tumor morphology, viability and invasion potential and indicate disease outcomes in the chick chorioallantoic membrane model. Cancer Lett. 2012, 326, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Eckrich, J.; Kugler, P.; Buhr, C.R.; Ernst, B.P.; Mendler, S.; Baumgart, J.; Brieger, J.; Wiesmann, N. Monitoring of tumor growth and vascularization with repetitive ultrasonography in the chicken chorioallantoic-membrane-assay. Sci Rep. 2020, 10, 18585. [Google Scholar] [CrossRef] [PubMed]
- DeBord, L.C.; Pathak, R.R.; Villaneuva, M.; Liu, H.; Harrington, D.A.; Yu, W.; Lewis, M.T.; Sikora, A.G. The chick chorioallantoic membrane (CAM) as a versatile patient-derived xenograft (PDX) platform for precision medicine and preclinical research. Am. J. Cancer Res. 2018, 8, 1642–1660. [Google Scholar]
- Hu, J.; Ishihara, M.; Chin, A.I.; Wu, L. Establishment of xenografts of urological cancers on chicken chorioallantoic membrane (CAM) to study metastasis. Precis. Clin. Med. 2019, 2, 140–151. [Google Scholar] [CrossRef] [Green Version]
- Choi, E.K.; Thomas, D.G.; McHugh, J.B.; Patel, R.M.; Roulston, D.; Schuetze, S.M.; Chugh, R.; Biermann, J.S.; Lucas, D.R. Undifferentiated small round cell carcinoma with t(4;19)(q35;q13.1) CIC-DUX4 fusion. Am. J. Surg. Pathol. 2013, 37, 1379–1386. [Google Scholar] [CrossRef]
- Yoshimoto, T.; Tanaka, M.; Homme, M.; Yamazaki, Y.; Takazawa, Y.; Antonescu, C.R.; Nakamura, T. CIC-DUX4 induces small round cell sarcomas distinct from Ewing sarcoma. Cancer Res. 2017, 77, 2927–2937. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le Guellec, S.; Velasco, V.; Perot, G.; Watson, S.; Tirode, F.; Coindre, J.M. ETV4 is a useful marker for the diagnosis of CIC-rearranged undifferentiated round-cell sarcomas: A study of 127 cases including mimicking lesions. Mod. Pathol. 2016, 29, 1523–1531. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.C.; Palanisamy, N.; Martin, E.; Almenara, J.; McHugh, J.B.; Choi, E.K.; Lucas, D.R.; Betz, B.L.; Thomas, D.; Patel, R.M. The utility of ETV1, ETV4 and ETV5 RNA in-situ hybridization in the diagnosis of CIC-DUX sarcomas. Histopathology. 2017, 70, 657–663. [Google Scholar] [CrossRef]
- Hung, Y.P.; Fletcher, C.D.M.; Hornick, J.L. Evaluation of ETV4 and WT1 expression in CIC-rearranged sarcomas and histological mimics. Mod. Pathol. 2016, 29, 1324–1334. [Google Scholar] [CrossRef]
- Kondo, T. Current status and future outlook for patient-derived cancer models from a rare cancer research perspective. Cancer Sci. 2021, 112, 953–961. [Google Scholar] [CrossRef]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Komatsu, A.; Matsumoto, K.; Yoshimatsu, Y.; Sin, Y.; Kubota, A.; Saito, T.; Mizumoto, A.; Ohashi, S.; Muto, M.; Noguchi, R.; et al. The CAM Model for CIC-DUX4 Sarcoma and Its Potential Use for Precision Medicine. Cells 2021, 10, 2613. https://doi.org/10.3390/cells10102613
Komatsu A, Matsumoto K, Yoshimatsu Y, Sin Y, Kubota A, Saito T, Mizumoto A, Ohashi S, Muto M, Noguchi R, et al. The CAM Model for CIC-DUX4 Sarcoma and Its Potential Use for Precision Medicine. Cells. 2021; 10(10):2613. https://doi.org/10.3390/cells10102613
Chicago/Turabian StyleKomatsu, Aoi, Kotaro Matsumoto, Yuki Yoshimatsu, Yooksil Sin, Arisa Kubota, Tomoki Saito, Ayaka Mizumoto, Shinya Ohashi, Manabu Muto, Rei Noguchi, and et al. 2021. "The CAM Model for CIC-DUX4 Sarcoma and Its Potential Use for Precision Medicine" Cells 10, no. 10: 2613. https://doi.org/10.3390/cells10102613
APA StyleKomatsu, A., Matsumoto, K., Yoshimatsu, Y., Sin, Y., Kubota, A., Saito, T., Mizumoto, A., Ohashi, S., Muto, M., Noguchi, R., Kondo, T., & Tamanoi, F. (2021). The CAM Model for CIC-DUX4 Sarcoma and Its Potential Use for Precision Medicine. Cells, 10(10), 2613. https://doi.org/10.3390/cells10102613






