Cell Transdifferentiation and Reprogramming in Disease Modeling: Insights into the Neuronal and Cardiac Disease Models and Current Translational Strategies
Abstract
1. Introduction
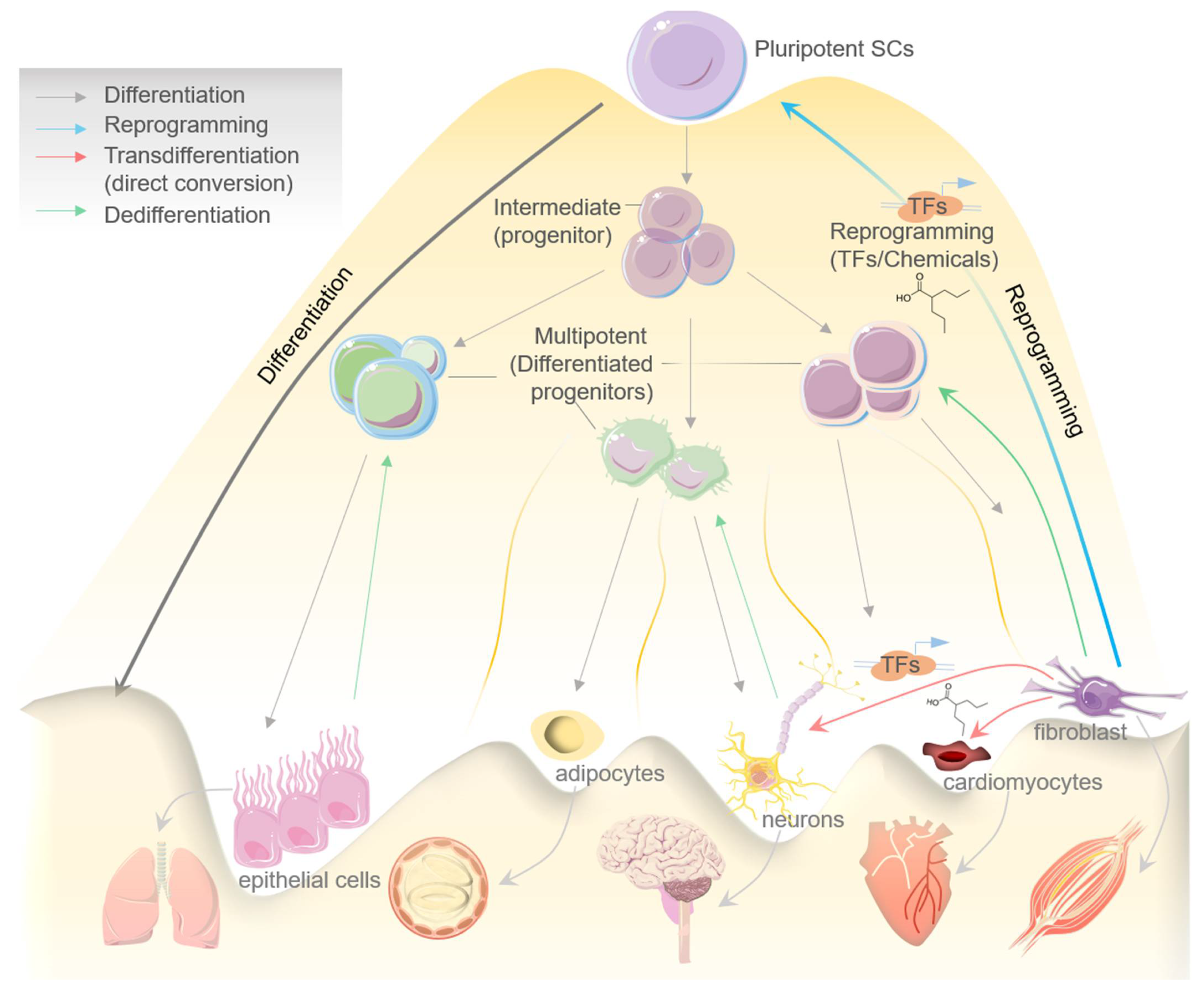
2. Cell Transdifferentiation: An Overview
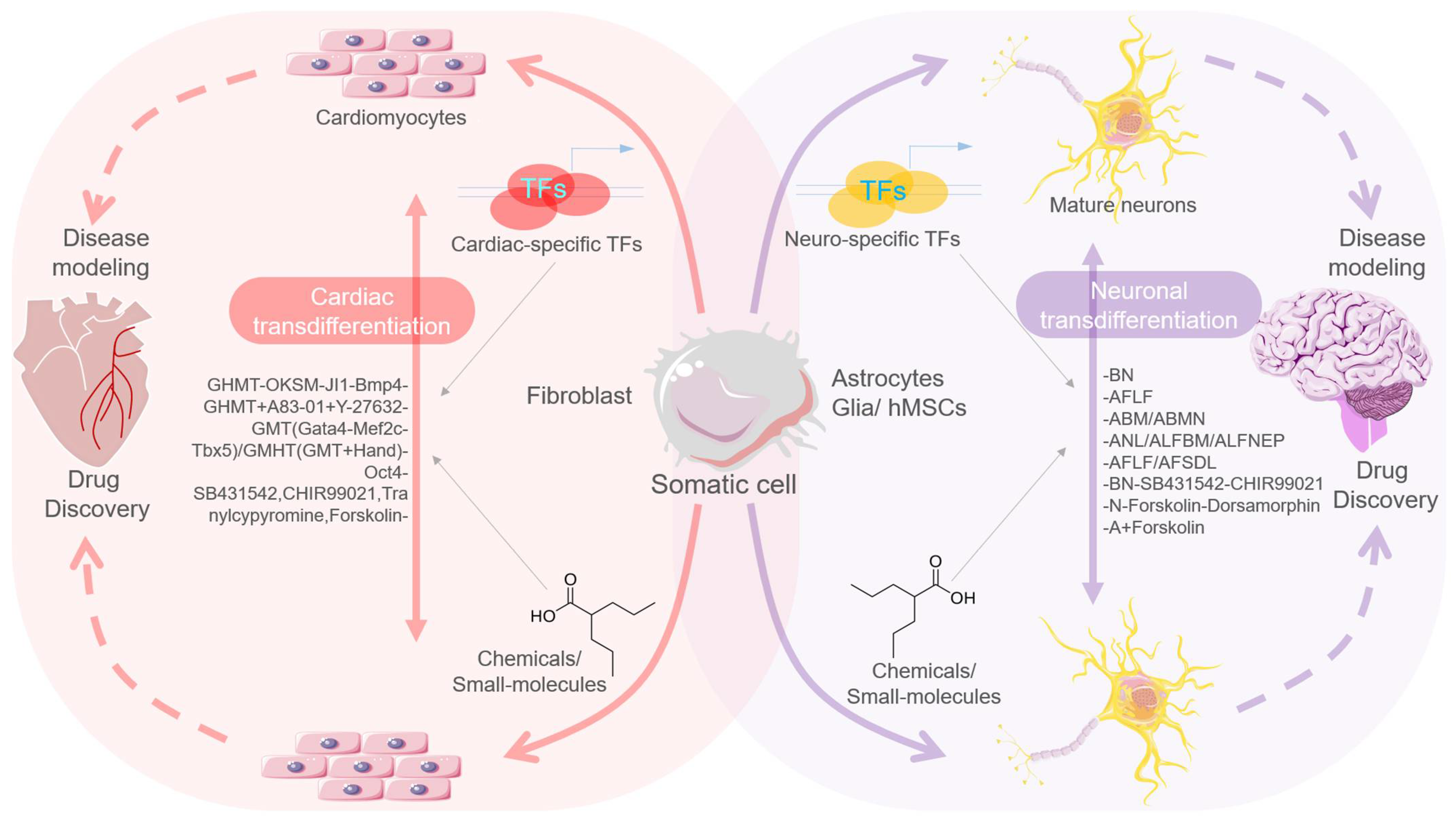
2.1. TFs-Mediated Transdifferentiation and Scope in Disease Modeling
2.1.1. TFs-Mediated Neural Stem/Progenitor Cell Transdifferentiation
2.1.2. TFs-Mediated Cardiomyocytes’ Transdifferentiation
2.2. Chemicals/Small Molecule-Mediated Transdifferentiation
2.2.1. Chemical/Small Molecule-Mediated Neural Stem/Progenitor Cell Transdifferentiation
2.2.2. Chemical/Small Molecule-Mediated Neuronal Transdifferentiation
2.2.3. Other Chemical/Small Molecule-Mediated Transdifferentiation
3. Cellular Reprogramming in Disease Modeling
3.1. Chemical/Small Molecule-Induced Cellular Reprogramming in Cardiomyocytes
3.2. Embryonic and Induced Pluripotent Stem Cells for Disease Modeling
3.3. Cellular Reprogramming in Neuronal and Cardiac Disease Modeling
3.4. Present Status, Developments, and Emerging Reprogramming Trends
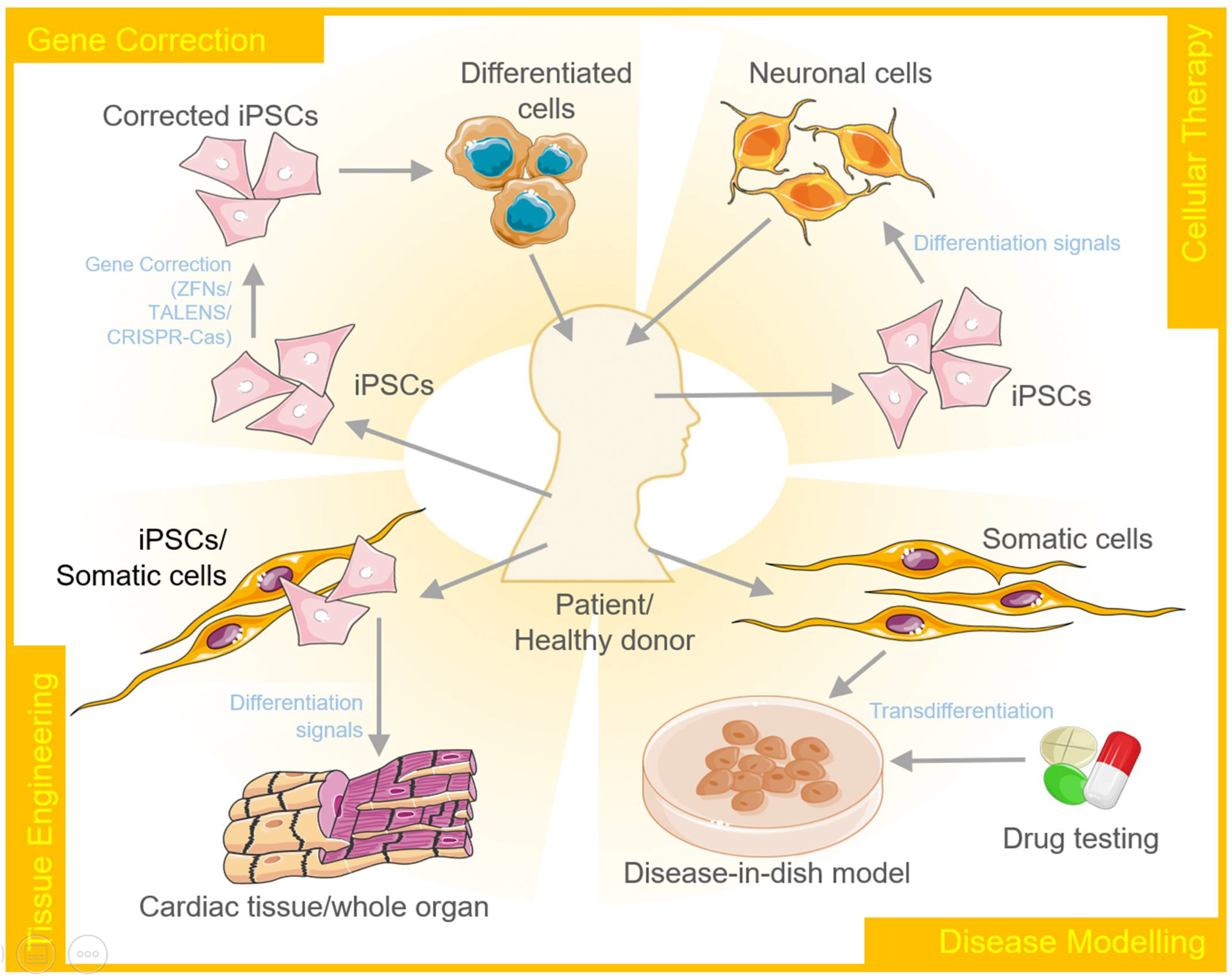
4. Therapeutic Applications of Transdifferentiation and Cellular Reprogramming
4.1. Disease Modeling and Testing Therapeutics
4.2. Regenerative Medicine
4.3. Tissue Engineering
4.4. Gene Editing
4.5. Personalized Medicine
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Graf, T. Historical Origins of Transdifferentiation and Reprogramming. Cell Stem Cell 2011, 9, 504–516. [Google Scholar] [CrossRef] [PubMed]
- Slack, J.M.W. Metaplasia and transdifferentiation: From pure biology to the clinic. Nat. Rev. Mol. Cell Biol. 2007, 8, 369–378. [Google Scholar] [CrossRef] [PubMed]
- Jopling, C.; Boue, S.; Belmonte, J.C.I. Dedifferentiation, transdifferentiation and reprogramming: Three routes to regeneration. Nat. Rev. Mol. Cell Biol. 2011, 12, 79–89. [Google Scholar] [CrossRef] [PubMed]
- Roccio, M. Directed differentiation and direct reprogramming: Applying stem cell technologies to hearing research. Stem Cells 2021, 39, 375–388. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Wang, H.; Hu, X. Reprogramming and Transdifferentiation Shift the Landscape of Regenerative Medicine. DNA Cell Biol. 2013, 32, 565–572. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Xu, M.; Wu, M.; Tian, X.; Zhang, C.; Fu, X. Transdifferentiation of Fibroblasts by Defined Factors. Cell. Reprogramm. 2015, 17, 151–159. [Google Scholar] [CrossRef]
- Davis, R.L.; Weintraub, H.; Lassar, A.B. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 1987, 51, 987–1000. [Google Scholar] [CrossRef]
- Murry, C.E.; Kay, M.A.; Bartosek, T.; Hauschka, S.D.; Schwartz, S.M. Muscle differentiation during repair of myocardial necrosis in rats via gene transfer with MyoD. J. Clin. Investig. 1996, 98, 2209–2217. [Google Scholar] [CrossRef] [PubMed]
- Mollinari, C.; Zhao, J.; Lupacchini, L.; Garaci, E.; Merlo, D.; Pei, G. Transdifferentiation: A new promise for neurodegenerative diseases. Cell Death Dis. 2018, 9, 830. [Google Scholar] [CrossRef]
- Barrett, N.R. The lower esophagus lined by columnar epithelium. Surgery 1957, 41, 881–894. [Google Scholar]
- Tsukamoto, H.; She, H.; Hazra, S.; Cheng, J.; Miyahara, T. Anti-adipogenic regulation underlies hepatic stellate cell transdifferentiation. J. Gastroenterol. Hepatol. 2006, 21 (Suppl. 3), S102–S105. [Google Scholar] [CrossRef]
- Hay, E.D.; Zuk, A. Transformations between epithelium and mesenchyme: Normal, pathological, and experimentally induced. Am. J. Kidney Dis. 1995, 26, 678–690. [Google Scholar] [CrossRef]
- Li, Y.; Huard, J. Differentiation of Muscle-Derived Cells into Myofibroblasts in Injured Skeletal Muscle. Am. J. Pathol. 2002, 161, 895–907. [Google Scholar] [CrossRef]
- Zhang, R.; Han, P.; Yang, H.; Ouyang, K.; Lee, D.; Lin, Y.-F.; Ocorr, K.; Kang, G.; Chen, J.; Stainier, D.Y.R.; et al. In vivo cardiac reprogramming contributes to zebrafish heart regeneration. Nature 2013, 498, 497–501. [Google Scholar] [CrossRef]
- He, J.; Lu, H.; Zou, Q.; Luo, L. Regeneration of Liver After Extreme Hepatocyte Loss Occurs Mainly via Biliary Transdifferentiation in Zebrafish. Gastroenterology 2014, 146, 789–800. [Google Scholar] [CrossRef] [PubMed]
- Suetsugu-Maki, R.; Maki, N.; Nakamura, K.; Sumanas, S.; Zhu, J.; Del Rio-Tsonis, K.; Tsonis, P.A. Lens regeneration in axolotl: New evidence of developmental plasticity. BMC Biol. 2012, 10, 103. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Chai, R.; Kim, G.S.; Pham, N.; Jansson, L.; Nguyen, D.-H.; Kuo, B.; May, L.A.; Zuo, J.; Cunningham, L.; et al. Lgr5+ cells regenerate hair cells via proliferation and direct transdifferentiation in damaged neonatal mouse utricle. Nat. Commun. 2015, 6, 6613. [Google Scholar] [CrossRef] [PubMed]
- Chambers, S.M.; Studer, L. Cell Fate Plug and Play: Direct Reprogramming and Induced Pluripotency. Cell 2011, 145, 827–830. [Google Scholar] [CrossRef]
- Cherry, A.B.; Daley, G.Q. Reprogramming Cellular Identity for Regenerative Medicine. Cell 2012, 148, 1110–1122. [Google Scholar] [CrossRef]
- Baranek, M.; Belter, A.; Naskręt-Barciszewska, M.Z.; Stobiecki, M.; Markiewicz, W.T.; Barciszewski, J. Effect of small molecules on cell reprogramming. Mol. BioSyst. 2016, 13, 277–313. [Google Scholar] [CrossRef]
- Julian, L.M.; McDonald, A.C.; Stanford, W.L. Direct reprogramming with SOX factors: Masters of cell fate. Curr. Opin. Genet. Dev. 2017, 46, 24–36. [Google Scholar] [CrossRef] [PubMed]
- Cheung, C.T.; Singh, R.; Kalra, R.S.; Kaul, S.C.; Wadhwa, R. Collaborator of ARF (CARF) Regulates Proliferative Fate of Human Cells by Dose-dependent Regulation of DNA Damage Signaling. J. Biol. Chem. 2014, 289, 18258–18269. [Google Scholar] [CrossRef]
- De Lázaro, I.; Kostarelos, K. Engineering Cell Fate for Tissue Regeneration by In Vivo Transdifferentiation. Stem Cell Rev. Rep. 2015, 12, 129–139. [Google Scholar] [CrossRef] [PubMed]
- Suman, S.; Domingues, A.; Ratajczak, J.; Ratajczak, M.Z. Potential Clinical Applications of Stem Cells in Regenerative Medicine. Adv. Exp. Med. Biol. 2019, 1201, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Kalra, R.S.; Chaudhary, A.; Omar, A.; Cheung, C.T.; Garg, S.; Kaul, S.C.; Wadhwa, R. Stress-induced changes in CARF expression determine cell fate to death, survival, or malignant transformation. Cell Stress Chaperones 2020, 25, 481–494. [Google Scholar] [CrossRef]
- Wadhwa, R.; Kalra, R.S.; Kaul, S.C. CARF is a multi-module regulator of cell proliferation and a molecular bridge between cellular senescence and carcinogenesis. Mech. Ageing Dev. 2017, 166, 64–68. [Google Scholar] [CrossRef] [PubMed]
- Torper, O.; Pfisterer, U.; Wolf, D.A.; Pereira, M.; Lau, S.; Jakobsson, J.; Björklund, A.; Grealish, S.; Parmar, M. Generation of induced neurons via direct conversion in vivo. Proc. Natl. Acad. Sci. USA 2013, 110, 7038–7043. [Google Scholar] [CrossRef]
- Niu, W.; Zang, T.; Zou, Y.; Fang, S.; Smith, D.K.; Bachoo, R.; Zhang, C.-L. In vivo reprogramming of astrocytes to neuroblasts in the adult brain. Nat. Cell Biol. 2013, 15, 1164–1175. [Google Scholar] [CrossRef]
- Niu, W.; Zang, T.; Smith, D.K.; Vue, T.Y.; Zou, Y.; Bachoo, R.; Johnson, J.E.; Zhang, C.-L. SOX2 Reprograms Resident Astrocytes into Neural Progenitors in the Adult Brain. Stem Cell Rep. 2015, 4, 780–794. [Google Scholar] [CrossRef]
- Su, Z.; Niu, W.; Liu, M.-L.; Zou, Y.; Zhang, C.-L. In vivo conversion of astrocytes to neurons in the injured adult spinal cord. Nat. Commun. 2014, 5, 3338. [Google Scholar] [CrossRef]
- Yiu, G.; He, Z. Glial inhibition of CNS axon regeneration. Nat. Rev. Neurosci. 2006, 7, 617–627. [Google Scholar] [CrossRef]
- Guo, Z.; Zhang, L.; Wu, Z.; Chen, Y.; Wang, F.; Chen, G. In Vivo Direct Reprogramming of Reactive Glial Cells into Functional Neurons after Brain Injury and in an Alzheimer’s Disease Model. Cell Stem Cell 2014, 14, 188–202. [Google Scholar] [CrossRef]
- Rouaux, C.; Arlotta, P. Direct lineage reprogramming of post-mitotic callosal neurons into corticofugal neurons in vivo. Nat. Cell Biol. 2013, 15, 214–221. [Google Scholar] [CrossRef] [PubMed]
- De La Rossa, A.; Bellone, C.; Golding, B.J.; Vitali, I.; Moss, J.; Toni, N.; Lüscher, C.; Jabaudon, D. In vivo reprogramming of circuit connectivity in postmitotic neocortical neurons. Nat. Neurosci. 2013, 16, 193–200. [Google Scholar] [CrossRef] [PubMed]
- Qin, H.; Zhao, A.-D.; Sun, M.-L.; Ma, K.; Fu, X.-B. Direct conversion of human fibroblasts into dopaminergic neuron-like cells using small molecules and protein factors. Mil. Med. Res. 2020, 7, 52. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Ashok, A.; Williams, D.; Kaufman, M.; Duff, K.; Sproul, A. Efficient Derivation of Excitatory and Inhibitory Neurons from Human Pluripotent Stem Cells Stably Expressing Direct Reprogramming Factors. Curr. Protoc. 2021, 1, e141. [Google Scholar] [CrossRef]
- Qian, L.; Huang, Y.; Spencer, C.I.; Foley, A.; Vedantham, V.; Liu, L.; Conway, S.J.; Fu, J.; Srivastava, D. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nat. Cell Biol. 2012, 485, 593–598. [Google Scholar] [CrossRef]
- Inagawa, K.; Miyamoto, K.; Yamakawa, H.; Muraoka, N.; Sadahiro, T.; Umei, T.; Wada, R.; Katsumata, Y.; Kaneda, R.; Nakade, K.; et al. Induction of Cardiomyocyte-Like Cells in Infarct Hearts by Gene Transfer of Gata4, Mef2c, and Tbx5. Circ. Res. 2012, 111, 1147–1156. [Google Scholar] [CrossRef]
- Song, K.; Nam, Y.-J.; Luo, X.; Qi, X.; Tan, W.; Huang, G.; Acharya, A.; Smith, C.L.; Tallquist, M.; Neilson, E.G.; et al. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nat. Cell Biol. 2012, 485, 599–604. [Google Scholar] [CrossRef]
- Jayawardena, T.M.; Finch, E.A.; Zhang, L.; Zhang, H.; Hodgkinson, C.P.; Pratt, R.E.; Rosenberg, P.; Mirotsou, M.; Dzau, V.J. MicroRNA Induced Cardiac Reprogramming In Vivo: Evidence for mature cardiac myocytes and improved cardiac function. Circ. Res. 2015, 116, 418–424. [Google Scholar] [CrossRef]
- Jayawardena, T.M.; Egemnazarov, B.; Finch, E.A.; Zhang, L.; Payne, J.A.; Pandya, K.; Zhang, Z.; Rosenberg, P.; Mirotsou, M.; Dzau, V.J. MicroRNA-Mediated In Vitro and In Vivo Direct Reprogramming of Cardiac Fibroblasts to Cardiomyocytes. Circ. Res. 2012, 110, 1465–1473. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.-F.; Dawkins, J.F.; Cho, H.C.; Marbán, E.; Cingolani, E. Biological pacemaker created by minimally invasive somatic reprogramming in pigs with complete heart block. Sci. Transl. Med. 2014, 6, 245ra94. [Google Scholar] [CrossRef]
- Kapoor, N.; Liang, W.; Marbán, E.; Cho, H.C. Direct conversion of quiescent cardiomyocytes to pacemaker cells by expression of Tbx18. Nat. Biotechnol. 2013, 31, 54–62. [Google Scholar] [CrossRef]
- Mozaffarian, D.; Benjamin, E.J.; Go, A.S.; Arnett, D.K.; Blaha, M.J.; Cushman, M.; de Ferranti, S.; Després, J.-P.; Fullerton, H.J.; Howard, V.J.; et al. Heart Disease and Stroke Statistics—2015 Update: A report from the American Heart Association. Circulation 2015, 131, e29–e322. [Google Scholar] [CrossRef]
- Christoforou, N.; Chellappan, M.; Adler, A.F.; Kirkton, R.D.; Wu, T.; Addis, R.C.; Bursac, N.; Leong, K.W. Transcription Factors MYOCD, SRF, Mesp1 and SMARCD3 Enhance the Cardio-Inducing Effect of GATA4, TBX5, and MEF2C during Direct Cellular Reprogramming. PLoS ONE 2013, 8, e63577. [Google Scholar] [CrossRef]
- Christoforou, N.; Chakraborty, S.; Kirkton, R.D.; Adler, A.F.; Addis, R.C.; Leong, K.W. Core Transcription Factors, MicroRNAs, and Small Molecules Drive Transdifferentiation of Human Fibroblasts Towards the Cardiac Cell Lineage. Sci. Rep. 2017, 7, 40285. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Brown, J.; Kanarek, A.; Rajagopal, J.; Melton, D.A. In vivo reprogramming of adult pancreatic exocrine cells to β-cells. Nat. Cell Biol. 2008, 455, 627–632. [Google Scholar] [CrossRef]
- Mathison, M.; Singh, V.P.; Chiuchiolo, M.J.; Sanagasetti, D.; Mao, Y.; Patel, V.B.; Yang, J.; Kaminsky, S.; Crystal, R.G.; Rosengart, T.K. In situ reprogramming to transdifferentiate fibroblasts into cardiomyocytes using adenoviral vectors: Implications for clinical myocardial regeneration. J. Thorac. Cardiovasc. Surg. 2017, 153, 329–339.e3. [Google Scholar] [CrossRef]
- Cheng, L.; Hu, W.; Qiu, B.; Zhao, J.; Yu, Y.; Guan, W.; Wang, M.; Yang, W.; Pei, G. Generation of neural progenitor cells by chemical cocktails and hypoxia. Cell Res. 2014, 24, 665–679. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Lin, Y.-H.; Sun, Y.J.; Zhu, S.; Zheng, J.; Liu, K.; Cao, N.; Li, K.; Huang, Y.; Ding, S. Pharmacological Reprogramming of Fibroblasts into Neural Stem Cells by Signaling-Directed Transcriptional Activation. Cell Stem Cell 2016, 18, 653–667. [Google Scholar] [CrossRef] [PubMed]
- Medina, L.V.C.; Pasantes-Morales, H.; Aguilera-Castrejon, A.; Picones, A.; Lara-Figueroa, C.O.; Luis, E.; Montesinos, J.J.; Cortés-Morales, V.A.; Ruiz, M.P.D.L.R.; Hernández-Estévez, E.; et al. Neuronal Transdifferentiation Potential of Human Mesenchymal Stem Cells from Neonatal and Adult Sources by a Small Molecule Cocktail. Stem Cells Int. 2019, 2019, 7627148. [Google Scholar] [CrossRef]
- Li, X.; Zuo, X.; Jing, J.; Ma, Y.; Wang, J.; Liu, D.; Zhu, J.; Du, X.; Xiong, L.; Du, Y.; et al. Small-Molecule-Driven Direct Reprogramming of Mouse Fibroblasts into Functional Neurons. Cell Stem Cell 2015, 17, 195–203. [Google Scholar] [CrossRef]
- Hu, W.; Qiu, B.; Guan, W.; Wang, Q.; Wang, M.; Li, W.; Gao, L.; Shen, L.; Huang, Y.; Xie, G.; et al. Direct Conversion of Normal and Alzheimer’s Disease Human Fibroblasts into Neuronal Cells by Small Molecules. Cell Stem Cell 2015, 17, 204–212. [Google Scholar] [CrossRef]
- Zhang, L.; Yin, J.; Yeh, H.; Ma, N.-X.; Lee, G.; Chen, X.A.; Wang, Y.; Lin, L.; Chen, L.; Jin, P.; et al. Small Molecules Efficiently Reprogram Human Astroglial Cells into Functional Neurons. Cell Stem Cell 2015, 17, 735–747. [Google Scholar] [CrossRef]
- Gautam, S.; Biswas, S.; Singh, B.; Guo, Y.; Deng, P.; Deng, W. Urine Cells-derived iPSCs: An Upcoming Frontier in Regenerative Medicine. Curr. Med. Chem. 2021, 28, 1. [Google Scholar] [CrossRef]
- Xu, G.; Wu, F.; Gu, X.; Zhang, J.; You, K.; Chen, Y.; Getachew, A.; Zhuang, Y.; Zhong, X.; Lin, Z.; et al. Direct Conversion of Human Urine Cells to Neurons by Small Molecules. Sci. Rep. 2019, 9, 16707. [Google Scholar] [CrossRef] [PubMed]
- Fomina-Yadlin, D.; Kubicek, S.; Walpita, D.; Dančik, V.; Hecksher-Sørensen, J.; Bittker, J.A.; Sharifnia, T.; Shamji, A.; Clemons, P.A.; Wagner, B.K.; et al. Small-molecule inducers of insulin expression in pancreatic -cells. Proc. Natl. Acad. Sci. USA 2010, 107, 15099–15104. [Google Scholar] [CrossRef] [PubMed]
- Koestenbauer, S.; Zech, N.H.; Juch, H.; Vanderzwalmen, P.; Schoonjans, L.; Dohr, G. Embryonic Stem Cells: Similarities and Differences Between Human and Murine Embryonic Stem Cells. Am. J. Reprod. Immunol. 2006, 55, 169–180. [Google Scholar] [CrossRef]
- Katsuda, T.; Kawamata, M.; Hagiwara, K.; Takahashi, R.-U.; Yamamoto, Y.; Camargo, F.D.; Ochiya, T. Conversion of Terminally Committed Hepatocytes to Culturable Bipotent Progenitor Cells with Regenerative Capacity. Cell Stem Cell 2017, 20, 41–55. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Qin, J.; Wang, S.; Zhang, W.; Duan, J.; Zhang, J.; Wang, X.; Yan, F.; Chang, M.; Liu, X.; et al. Conversion of Human Gastric Epithelial Cells to Multipotent Endodermal Progenitors using Defined Small Molecules. Cell Stem Cell 2016, 19, 449–461. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Yamanaka, S. Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of Pluripotent Stem Cells from Adult Human Fibroblasts by Defined Factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.S.; Macadangdang, J.; Leung, W.; Laflamme, M.; Kim, D.-H. Human iPSC-derived cardiomyocytes and tissue engineering strategies for disease modeling and drug screening. Biotechnol. Adv. 2017, 35, 77–94. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Zhang, L.-H.; Xie, X. iPSCs and small molecules: A reciprocal effort towards better approaches for drug discovery. Acta Pharmacol. Sin. 2013, 34, 765–776. [Google Scholar] [CrossRef] [PubMed]
- Doss, M.X.; Sachinidis, A. Current Challenges of iPSC-Based Disease Modeling and Therapeutic Implications. Cells 2019, 8, 403. [Google Scholar] [CrossRef]
- Bai, X. Stem Cell-Based Disease Modeling and Cell Therapy. Cells 2020, 9, 2193. [Google Scholar] [CrossRef]
- Saito-Diaz, K.; Zeltner, N. Induced pluripotent stem cells for disease modeling, cell therapy and drug discovery in genetic autonomic disorders: A review. Clin. Auton. Res. 2019, 29, 367–384. [Google Scholar] [CrossRef]
- Baillie-Benson, P.; Moris, N.; Arias, A.M. Pluripotent stem cell models of early mammalian development. Curr. Opin. Cell Biol. 2020, 66, 89–96. [Google Scholar] [CrossRef]
- Ford, E.; Pearlman, J.; Ruan, T.; Manion, J.; Waller, M.; Neely, G.G.; Caron, L. Human Pluripotent Stem Cells-Based Therapies for Neurodegenerative Diseases: Current Status and Challenges. Cells 2020, 9, 2517. [Google Scholar] [CrossRef]
- Kim, S.-Y.; Kim, Y.-R.; Park, W.-J.; Kim, H.S.; Jung, S.-C.; Woo, S.-Y.; Jo, I.; Ryu, K.-H.; Park, J.-W. Characterisation of insulin-producing cells differentiated from tonsil derived mesenchymal stem cells. Differentiation 2015, 90, 27–39. [Google Scholar] [CrossRef]
- González, J.F.; Alcántara, A.R.; Doadrio, A.L.; Sánchez-Montero, J.M. Developments with multi-target drugs for Alzheimer’s disease: An overview of the current discovery approaches. Expert Opin. Drug Discov. 2019, 14, 879–891. [Google Scholar] [CrossRef]
- Pinheiro, E.A.; Fetterman, K.A.; Burridge, P.W. hiPSCs in cardio-oncology: Deciphering the genomics. Cardiovasc. Res. 2019, 115, 935–948. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Huang, C.; Xu, X.; Gu, H.; Ye, Y.; Jiang, C.; Qiu, Z.; Xie, X. Direct reprogramming of mouse fibroblasts into cardiomyocytes with chemical cocktails. Cell Res. 2015, 25, 1013–1024. [Google Scholar] [CrossRef] [PubMed]
- Cao, N.; Huang, Y.; Zheng, J.; Spencer, C.I.; Zhang, Y.; Fu, J.-D.; Nie, B.; Xie, M.; Zhang, M.; Wang, H.; et al. Conversion of human fibroblasts into functional cardiomyocytes by small molecules. Science 2016, 352, 1216–1220. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.P.; Pinnamaneni, J.P.; Pugazenthi, A.; Sanagasetti, D.; Mathison, M.; Wang, K.; Yang, J.; Rosengart, T.K. Enhanced Generation of Induced Cardiomyocytes Using a Small-Molecule Cocktail to Overcome Barriers to Cardiac Cellular Reprogramming. J. Am. Heart Assoc. 2020, 9, e015686. [Google Scholar] [CrossRef] [PubMed]
- Vierbuchen, T.; Ostermeier, A.; Pang, Z.; Kokubu, Y.; Südhof, T.C.; Wernig, M. Direct conversion of fibroblasts to functional neurons by defined factors. Nat. Cell Biol. 2010, 463, 1035–1041. [Google Scholar] [CrossRef] [PubMed]
- Rossant, J.; Tam, P.P. Opportunities and challenges with stem cell-based embryo models. Stem Cell Rep. 2021, 16, 1031–1038. [Google Scholar] [CrossRef] [PubMed]
- Smith, L.A.; Liu, X.; Hu, J.; Wang, P.; Ma, P.X. Enhancing Osteogenic Differentiation of Mouse Embryonic Stem Cells by Nanofibers. Tissue Eng. Part A 2009, 15, 1855–1864. [Google Scholar] [CrossRef] [PubMed]
- Xu, N.; Papagiannakopoulos, T.; Pan, G.; Thomson, J.A.; Kosik, K.S. MicroRNA-145 Regulates OCT4, SOX2, and KLF4 and Represses Pluripotency in Human Embryonic Stem Cells. Cell 2009, 137, 647–658. [Google Scholar] [CrossRef]
- Kingham, E.; Oreffo, R.O. Embryonic and Induced Pluripotent Stem Cells: Understanding, Creating, and Exploiting the Nano-Niche for Regenerative Medicine. ACS Nano 2013, 7, 1867–1881. [Google Scholar] [CrossRef]
- Pang, Z.; Yang, N.; Vierbuchen, T.; Ostermeier, A.; Fuentes, D.; Yang, T.Q.; Citri, A.; Sebastiano, V.; Marro, S.; Südhof, T.C.; et al. Induction of human neuronal cells by defined transcription factors. Nat. Cell Biol. 2011, 476, 220–223. [Google Scholar] [CrossRef]
- Marro, S.; Pang, Z.; Yang, N.; Tsai, M.-C.; Qu, K.; Chang, H.Y.; Südhof, T.C.; Wernig, M. Direct Lineage Conversion of Terminally Differentiated Hepatocytes to Functional Neurons. Cell Stem Cell 2011, 9, 374–382. [Google Scholar] [CrossRef] [PubMed]
- Caiazzo, M.; Dell’Anno, M.T.; Dvoretskova, E.; Lazarevic, D.; Taverna, S.; Leo, D.; Sotnikova, T.D.; Menegon, A.; Roncaglia, P.; Colciago, G.; et al. Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nat. Cell Biol. 2011, 476, 224–227. [Google Scholar] [CrossRef] [PubMed]
- Karow, M.; Sánchez, R.; Schichor, C.; Masserdotti, G.; Ortega, F.; Heinrich, C.; Gascón, S.; Khan, M.A.; Lie, D.C.; Dellavalle, A.; et al. Reprogramming of Pericyte-Derived Cells of the Adult Human Brain into Induced Neuronal Cells. Cell Stem Cell 2012, 11, 471–476. [Google Scholar] [CrossRef]
- Liu, X.; Li, F.; Stubblefield, E.A.; Blanchard, B.; Richards, T.L.; Larson, G.A.; He, Y.; Huang, Q.; Tan, A.C.; Zhang, D.; et al. Direct reprogramming of human fibroblasts into dopaminergic neuron-like cells. Cell Res. 2011, 22, 321–332. [Google Scholar] [CrossRef]
- Xu, Z.; Jiang, H.; Zhong, P.; Yan, Z.; Chen, S.; Feng, J. Direct conversion of human fibroblasts to induced serotonergic neurons. Mol. Psychiatry 2015, 21, 62–70. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhang, W.; Nam, Y.-J. Stoichiometric optimization of Gata4, Hand2, Mef2c, and Tbx5 expression for contractile cardiomyocyte reprogramming. Sci. Rep. 2019, 9, 14970. [Google Scholar] [CrossRef] [PubMed]
- Hirai, H.; Katoku-Kikyo, N.; Keirstead, S.A.; Kikyo, N. Accelerated direct reprogramming of fibroblasts into cardiomyocyte-like cells with the MyoD transactivation domain. Cardiovasc. Res. 2013, 100, 105–113. [Google Scholar] [CrossRef]
- Ifkovits, J.L.; Addis, R.C.; Epstein, J.A.; Gearhart, J.D. Inhibition of TGFβ Signaling Increases Direct Conversion of Fibroblasts to Induced Cardiomyocytes. PLoS ONE 2014, 9, e89678. [Google Scholar] [CrossRef]
- Zhao, Y.; Londono, P.; Cao, Y.; Sharpe, E.J.; Proenza, C.; O’Rourke, R.; Jones, K.L.; Jeong, M.Y.; Walker, L.; Buttrick, P.M.; et al. High-efficiency reprogramming of fibroblasts into cardiomyocytes requires suppression of pro-fibrotic signalling. Nat. Commun. 2015, 6, 8243. [Google Scholar] [CrossRef]
- Zhou, Y.; Wang, L.; Vaseghi, H.R.; Liu, Z.; Lu, R.; Alimohamadi, S.; Yin, C.; Fu, J.; Wang, G.G.; Liu, J.; et al. Bmi1 Is a Key Epigenetic Barrier to Direct Cardiac Reprogramming. Cell Stem Cell 2016, 18, 382–395. [Google Scholar] [CrossRef]
- Mohamed, T.M.A.; Stone, N.R.; Berry, E.C.; Radzinsky, E.; Huang, Y.; Pratt, K.; Ang, Y.-S.; Yu, P.; Wang, H.; Tang, S.; et al. Chemical Enhancement of In Vitro and In Vivo Direct Cardiac Reprogramming. Circulation 2017, 135, 978–995. [Google Scholar] [CrossRef]
- Haratizadeh, S.; Bojnordi, M.N.; Darabi, S.; Karimi, N.; Naghikhani, M.; Hamidabadi, H.G.; Seifi, M. Condition medium of cerebrospinal fluid and retinoic acid induces the transdifferentiation of human dental pulp stem cells into neuroglia and neural like cells. Anat. Cell Biol. 2017, 50, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Wada, R.; Muraoka, N.; Inagawa, K.; Yamakawa, H.; Miyamoto, K.; Sadahiro, T.; Umei, T.; Kaneda, R.; Suzuki, T.; Kamiya, K.; et al. Induction of human cardiomyocyte-like cells from fibroblasts by defined factors. Proc. Natl. Acad. Sci. USA 2013, 110, 12667–12672. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Stone, N.R.; Liu, L.; Spencer, C.I.; Qian, L.; Hayashi, Y.; Olguín, P.D.; Ding, S.; Bruneau, B.G.; Srivastava, D. Direct Reprogramming of Human Fibroblasts toward a Cardiomyocyte-like State. Stem Cell Rep. 2013, 1, 235–247. [Google Scholar] [CrossRef] [PubMed]
- Muraoka, N.; Yamakawa, H.; Miyamoto, K.; Sadahiro, T.; Umei, T.; Isomi, M.; Nakashima, H.; Akiyama, M.; Wada, R.; Inagawa, K.; et al. MiR-133 promotes cardiac reprogramming by directly repressing Snai1 and silencing fibroblast signatures. EMBO J. 2014, 33, 1565–1581. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.P.; Mathison, M.; Patel, V.; Sanagasetti, D.; Gibson, B.W.; Yang, J.; Rosengart, T.K. MiR-590 Promotes Transdifferentiation of Porcine and Human Fibroblasts Toward a Cardiomyocyte-Like Fate by Directly Repressing Specificity Protein 1. J. Am. Heart Assoc. 2016, 5, e003922. [Google Scholar] [CrossRef] [PubMed]
- Sivandzade, F.; Cucullo, L. Regenerative Stem Cell Therapy for Neurodegenerative Diseases: An Overview. Int. J. Mol. Sci. 2021, 22, 2153. [Google Scholar] [CrossRef]
- Takahashi, K.; Yamanaka, S. A decade of transcription factor-mediated reprogramming to pluripotency. Nat. Rev. Mol. Cell Biol. 2016, 17, 183–193. [Google Scholar] [CrossRef]
- Kim, J.; Ambasudhan, R.; Ding, S. Direct lineage reprogramming to neural cells. Curr. Opin. Neurobiol. 2012, 22, 778–784. [Google Scholar] [CrossRef][Green Version]
- Musunuru, K.; Sheikh, F.; Gupta, R.M.; Houser, S.R.; Maher, K.O.; Milan, D.J.; Terzic, A.; Wu, J.C. Induced Pluripotent Stem Cells for Cardiovascular Disease Modeling and Precision Medicine: A Scientific Statement From the American Heart Association. Circ. Genom. Precis. Med. 2018, 11, e000043. [Google Scholar] [CrossRef] [PubMed]
- Preininger, M.; Jha, R.; Maxwell, J.T.; Wu, Q.; Singh, M.; Wang, B.; Dalal, A.; Mceachin, Z.T.; Rossoll, W.; Hales, C.M.; et al. A human pluripotent stem cell model of catecholaminergic polymorphic ventricular tachycardia recapitulates patient-specific drug responses. Dis. Model. Mech. 2016, 9, 927–939. [Google Scholar] [CrossRef] [PubMed]
- Ben Jehuda, R.; Shemer, Y.; Binah, O. Genome Editing in Induced Pluripotent Stem Cells using CRISPR/Cas9. Stem Cell Rev. Rep. 2018, 14, 323–336. [Google Scholar] [CrossRef] [PubMed]
- Wattanapanitch, M. Recent Updates on Induced Pluripotent Stem Cells in Hematological Disorders. Stem Cells Int. 2019, 2019, 5171032. [Google Scholar] [CrossRef]
- Nishizawa, M.; Chonabayashi, K.; Nomura, M.; Tanaka, A.; Nakamura, M.; Inagaki, A.; Nishikawa, M.; Takei, I.; Oishi, A.; Tanabe, K.; et al. Epigenetic Variation between Human Induced Pluripotent Stem Cell Lines Is an Indicator of Differentiation Capacity. Cell Stem Cell 2016, 19, 341–354. [Google Scholar] [CrossRef]
- Guerrero-Aspizua, S.; Carretero, M.; Conti, C.J.; Del Río, M. The importance of immunity in the development of reliable animal models for psoriasis and atopic dermatitis. Immunol. Cell Biol. 2020, 98, 626–638. [Google Scholar] [CrossRef]
- Brandão, K.O.; Tabel, V.A.; Atsma, D.E.; Mummery, C.; Davis, R.P. Human pluripotent stem cell models of cardiac disease: From mechanisms to therapies. Dis. Models Mech. 2017, 10, 1039–1059. [Google Scholar] [CrossRef]
- Lindvall, O.; Kokaia, Z.; Martinez-Serrano, A. Stem cell therapy for human neurodegenerative disorders—How to make it work. Nat. Med. 2004, 10, S42–S50. [Google Scholar] [CrossRef]
- Hunsberger, J.G.; Rao, M.; Kurtzberg, J.; Bulte, J.W.M.; Atala, A.; LaFerla, F.M.; Greely, H.T.; Sawa, A.; Gandy, S.; Schneider, L.S.; et al. Accelerating stem cell trials for Alzheimer’s disease. Lancet Neurol. 2016, 15, 219–230. [Google Scholar] [CrossRef]
- Sharma, P.; Ando, D.; Daub, A.; Kaye, J.A.; Finkbeiner, S. High-Throughput Screening in Primary Neurons. Methods Enzymol. 2012, 506, 331–360. [Google Scholar] [CrossRef]
- Nguyen, H.V.; Tsang, S.H.; Schwartz, S.D.; Nagiel, A.; Lanza, R.; Blumenkranz, M.S. (Eds.) “Disease in a Dish” Modeling of Retinal Diseases. In Cellular Therapies for Retinal Disease; Springer: Cham, Switzerland, 2017; pp. 107–115. [Google Scholar]
- Carpenter, M.K.; Rao, M.S. Concise Review: Making and Using Clinically Compliant Pluripotent Stem Cell Lines. Stem Cells Transl. Med. 2015, 4, 381–388. [Google Scholar] [CrossRef]
- Danter, W.R. DeepNEU: Cellular reprogramming comes of age—A machine learning platform with application to rare diseases research. Orphanet J. Rare Dis. 2019, 14, 13. [Google Scholar] [CrossRef]
- Sterneckert, J.L.; Reinhardt, P.; Scholer, H.R. Investigating human disease using stem cell models. Nat. Rev. Genet. 2014, 15, 625–639. [Google Scholar] [CrossRef]
- Eiges, R.; Urbach, A.; Malcov, M.; Frumkin, T.; Schwartz, T.; Amit, A.; Yaron, Y.; Eden, A.; Yanuka, O.; Benvenisty, N.; et al. Developmental Study of Fragile X Syndrome Using Human Embryonic Stem Cells Derived from Preimplantation Genetically Diagnosed Embryos. Cell Stem Cell 2007, 1, 568–577. [Google Scholar] [CrossRef] [PubMed]
- Ebert, A.D.; Yu, J.; Rose, F.F.; Mattis, V.B.; Lorson, C.L.; Thomson, J.A.; Svendsen, C.N. Induced pluripotent stem cells from a spinal muscular atrophy patient. Nat. Cell Biol. 2008, 457, 277–280. [Google Scholar] [CrossRef] [PubMed]
- Chiang, C.-H.; Su, Y.; Wen, Z.; Yoritomo, N.; Ross, C.A.; Margolis, R.L.; Song, H.; Ming, G.-L. Integration-free induced pluripotent stem cells derived from schizophrenia patients with a DISC1 mutation. Mol. Psychiatry 2011, 16, 358–360. [Google Scholar] [CrossRef] [PubMed]
- Marchetto, M.C.; Carromeu, C.; Acab, A.; Yu, D.; Yeo, G.W.; Mu, Y.; Chen, G.; Gage, F.H.; Muotri, A.R. A Model for Neural Development and Treatment of Rett Syndrome Using Human Induced Pluripotent Stem Cells. Cell 2010, 143, 527–539. [Google Scholar] [CrossRef]
- Huo, H.-Q.; Qu, Z.-Y.; Yuan, F.; Ma, L.; Yao, L.; Xu, M.; Hu, Y.; Ji, J.; Bhattacharyya, A.; Zhang, S.-C.; et al. Modeling Down Syndrome with Patient iPSCs Reveals Cellular and Migration Deficits of GABAergic Neurons. Stem Cell Rep. 2018, 10, 1251–1266. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Danes, A.; Richaud-Patin, Y.; Carballo-Carbajal, I.; Delgado, S.J.; Caig, C.; Mora, S.; Di Guglielmo, C.; Ezquerra, M.; Patel, B.; Giralt, A.; et al. Disease-specific phenotypes in dopamine neurons from human iPS-based models of genetic and sporadic Parkinson’s disease. EMBO Mol. Med. 2012, 4, 380–395. [Google Scholar] [CrossRef]
- Yagi, T.; Ito, D.; Okada, Y.; Akamatsu, W.; Nihei, Y.; Yoshizaki, T.; Yamanaka, S.; Okano, H.; Suzuki, N. Modeling familial Alzheimer’s disease with induced pluripotent stem cells. Hum. Mol. Genet. 2011, 20, 4530–4539. [Google Scholar] [CrossRef]
- Parrotta, E.I.; Lucchino, V.; Scaramuzzino, L.; Scalise, S.; Cuda, G. Modeling Cardiac Disease Mechanisms Using Induced Pluripotent Stem Cell-Derived Cardiomyocytes: Progress, Promises and Challenges. Int. J. Mol. Sci. 2020, 21, 4354. [Google Scholar] [CrossRef]
- Jagadeesan, S.; Workman, M.J.; Herland, A.; Svendsen, C.N.; Vatine, G.D. Generation of a Human iPSC-Based Blood-Brain Barrier Chip. J. Vis. Exp. 2020, 157, e60925. [Google Scholar] [CrossRef]
- Ramme, A.P.; Koenig, L.; Hasenberg, T.; Schwenk, C.; Magauer, C.; Faust, D.; Lorenz, A.K.; Krebs, A.-C.; Drewell, C.; Schirrmann, K.; et al. Autologous induced pluripotent stem cell-derived four-organ-chip. Future Sci. OA 2019, 5, FSO413. [Google Scholar] [CrossRef]
- Lacerda, A.E.; Kuryshev, Y.A.; Chen, Y.; Renganathan, M.; Eng, H.; Danthi, S.J.; Kramer, J.W.; Yang, T.; Brown, A.M. Alfuzosin Delays Cardiac Repolarization by a Novel Mechanism. J. Pharmacol. Exp. Ther. 2007, 324, 427–433. [Google Scholar] [CrossRef]
- Navarrete, E.G.; Liang, P.; Lan, F.; Sanchez-Freire, V.; Simmons, C.; Gong, T.; Sharma, A.; Burridge, P.W.; Patlolla, B.; Lee, A.S.; et al. Screening Drug-Induced Arrhythmia Using Human Induced Pluripotent Stem Cell–Derived Cardiomyocytes and Low-Impedance Microelectrode Arrays. Circulation 2013, 128, S3–S13. [Google Scholar] [CrossRef] [PubMed]
- Moretti, A.; Bellin, M.; Welling, A.; Jung, C.B.; Lam, J.T.; Bott-Flügel, L.; Dorn, T.; Goedel, A.; Höhnke, C.; Hofmann, F.; et al. Patient-Specific Induced Pluripotent Stem-Cell Models for Long-QT Syndrome. N. Engl. J. Med. 2010, 363, 1397–1409. [Google Scholar] [CrossRef] [PubMed]
- Itzhaki, I.; Maizels, L.; Huber, I.; Zwi-Dantsis, L.; Caspi, O.; Winterstern, A.; Feldman, O.; Gepstein, A.; Arbel, G.; Hammerman, H.; et al. Modelling the long QT syndrome with induced pluripotent stem cells. Nat. Cell Biol. 2011, 471, 225–229. [Google Scholar] [CrossRef] [PubMed]
- Kumari, D.; Swaroop, M.; Southall, N.; Huang, W.; Zheng, W.; Usdin, K. High-Throughput Screening to Identify Compounds That Increase Fragile X Mental Retardation Protein Expression in Neural Stem Cells Differentiated from Fragile X Syndrome Patient-Derived Induced Pluripotent Stem Cells. Stem Cells Transl. Med. 2015, 4, 800–808. [Google Scholar] [CrossRef] [PubMed]
- Sirenko, O.; Cromwell, E.F.; Crittenden, C.; Wignall, J.A.; Wright, F.A.; Rusyn, I. Assessment of beating parameters in human induced pluripotent stem cells enables quantitative in vitro screening for cardiotoxicity. Toxicol. Appl. Pharmacol. 2013, 273, 500–507. [Google Scholar] [CrossRef] [PubMed]
- Pointon, A.; Harmer, A.R.; Dale, I.L.; Abi-Gerges, N.; Bowes, J.; Pollard, C.; Garside, H. Assessment of Cardiomyocyte Contraction in Human-Induced Pluripotent Stem Cell-Derived Cardiomyocytes. Toxicol. Sci. 2015, 144, 227–237. [Google Scholar] [CrossRef] [PubMed]
- Blinova, K.; Stohlman, J.; Vicente, J.; Chan, D.; Johannesen, L.; Hortigon-Vinagre, M.P.; Zamora, V.; Smith, G.; Crumb, W.J.; Pang, L.; et al. Comprehensive Translational Assessment of Human-Induced Pluripotent Stem Cell Derived Cardiomyocytes for Evaluating Drug-Induced Arrhythmias. Toxicol. Sci. 2017, 155, 234–247. [Google Scholar] [CrossRef]
- Reynolds, J.G.; Geretti, E.; Hendriks, B.S.; Lee, H.; Leonard, S.C.; Klinz, S.G.; Noble, C.O.; Lücker, P.B.; Zandstra, P.W.; Drummond, D.C.; et al. HER2-targeted liposomal doxorubicin displays enhanced anti-tumorigenic effects without associated cardiotoxicity. Toxicol. Appl. Pharmacol. 2012, 262, 1–10. [Google Scholar] [CrossRef]
- Colwell, A.S.; Longaker, M.T.; Lorenz, H.P. Mammalian Fetal Organ Regeneration. Blue Biotechnol. 2005, 93, 83–100. [Google Scholar] [CrossRef]
- Frantz, S. Embryonic stem cell pioneer Geron exits field, cuts losses. Nat. Biotechnol. 2012, 30, 12–13. [Google Scholar] [CrossRef]
- Oh, J.; Lee, K.-I.; Kim, H.-T.; You, Y.; Yoon, D.H.; Song, K.Y.; Cheong, E.; Ha, Y.; Hwang, D.-Y. Human-induced pluripotent stem cells generated from intervertebral disc cells improve neurologic functions in spinal cord injury. Stem Cell Res. Ther. 2015, 6, 125. [Google Scholar] [CrossRef]
- Sundberg, M.; Bogetofte, H.; Lawson, T.; Jansson, J.; Smith, G.; Astradsson, A.; Moore, M.; Osborn, T.; Cooper, O.; Spealman, R.; et al. Improved Cell Therapy Protocols for Parkinson’s Disease Based on Differentiation Efficiency and Safety of hESC-, hiPSC-, and Non-Human Primate iPSC-Derived Dopaminergic Neurons. Stem Cells 2013, 31, 1548–1562. [Google Scholar] [CrossRef] [PubMed]
- Jeon, K.; Lim, H.; Kim, J.-H.; Van Thuan, N.; Park, S.H.; Lim, Y.-M.; Choi, H.-Y.; Lee, E.-R.; Kim, J.-H.; Lee, M.-S.; et al. Differentiation and Transplantation of Functional Pancreatic Beta Cells Generated from Induced Pluripotent Stem Cells Derived from a Type 1 Diabetes Mouse Model. Stem Cells Dev. 2012, 21, 2642–2655. [Google Scholar] [CrossRef]
- Shiba, Y.; Fernandes, S.; Zhu, W.-Z.; Filice, D.; Muskheli, V.; Kim, J.; Palpant, N.; Gantz, J.; Moyes, K.W.; Reinecke, H.; et al. Human ES-cell-derived cardiomyocytes electrically couple and suppress arrhythmias in injured hearts. Nat. Cell Biol. 2012, 489, 322–325. [Google Scholar] [CrossRef] [PubMed]
- Ieda, M.; Fu, J.; Olguín, P.D.; Vedantham, V.; Hayashi, Y.; Bruneau, B.; Srivastava, D. Direct Reprogramming of Fibroblasts into Functional Cardiomyocytes by Defined Factors. Cell 2010, 142, 375–386. [Google Scholar] [CrossRef] [PubMed]
- Mathison, M.; Gersch, R.P.; Nasser, A.; Lilo, S.; Korman, M.; Fourman, M.; Hackett, N.; Shroyer, K.; Yang, J.; Ma, Y.; et al. In Vivo Cardiac Cellular Reprogramming Efficacy Is Enhanced by Angiogenic Preconditioning of the Infarcted Myocardium with Vascular Endothelial Growth Factor. J. Am. Heart Assoc. 2012, 1, e005652. [Google Scholar] [CrossRef]
- Berthiaume, F.; Maguire, T.J.; Yarmush, M.L. Tissue Engineering and Regenerative Medicine: History, Progress, and Challenges. Annu. Rev. Chem. Biomol. Eng. 2011, 2, 403–430. [Google Scholar] [CrossRef]
- Wang, A.; Tang, Z.; Park, I.-H.; Zhu, Y.; Patel, S.; Daley, G.Q.; Li, S. Induced pluripotent stem cells for neural tissue engineering. Biomaterials 2011, 32, 5023–5032. [Google Scholar] [CrossRef]
- Margariti, A.; Winkler, B.; Karamariti, E.; Zampetaki, A.; Tsai, T.-N.; Baban, D.; Ragoussis, J.; Huang, Y.; Han, J.-D.J.; Zeng, L.; et al. Direct reprogramming of fibroblasts into endothelial cells capable of angiogenesis and reendothelialization in tissue-engineered vessels. Proc. Natl. Acad. Sci. USA 2012, 109, 13793–13798. [Google Scholar] [CrossRef] [PubMed]
- Hong, X.; Margariti, A.; Le Bras, A.; Jacquet, L.; Kong, W.; Hu, Y.; Xu, Q. Transdifferentiated Human Vascular Smooth Muscle Cells are a New Potential Cell Source for Endothelial Regeneration. Sci. Rep. 2017, 7, 5590. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.; He, Z.; Ji, S.; Sun, H.; Xiang, D.; Liu, C.; Hu, Y.; Wang, X.; Hui, L. Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nat. Cell Biol. 2011, 475, 386–389. [Google Scholar] [CrossRef]
- Limaye, P.B.; Bowen, W.C.; Orr, A.; Apte, U.M.; Michalopoulos, G.K. Expression of hepatocytic- and biliary-specific transcription factors in regenerating bile ducts during hepatocyte-to-biliary epithelial cell transdifferentiation. Comp. Hepatol. 2010, 9, 9. [Google Scholar] [CrossRef] [PubMed]
- Berg, C.W.V.D.; Ritsma, L.; Avramut, M.C.; Wiersma, L.E.; Berg, B.M.V.D.; Leuning, D.G.; Lievers, E.; Koning, M.; Vanslambrouck, J.M.; Koster, A.J.; et al. Renal Subcapsular Transplantation of PSC-Derived Kidney Organoids Induces Neo-vasculogenesis and Significant Glomerular and Tubular Maturation In Vivo. Stem Cell Rep. 2018, 10, 751–765. [Google Scholar] [CrossRef]
- An, M.C.; Zhang, N.; Scott, G.; Montoro, D.; Wittkop, T.; Mooney, S.; Melov, S.; Ellerby, L.M. Genetic Correction of Huntington’s Disease Phenotypes in Induced Pluripotent Stem Cells. Cell Stem Cell 2012, 11, 253–263. [Google Scholar] [CrossRef]
- Yusa, K.; Rashid, S.T.; Strick-Marchand, H.; Varela, I.; Liu, P.-Q.; Paschon, D.E.; Miranda, E.; Ordóñez, A.; Hannan, N.; Rouhani, F.J.; et al. Targeted gene correction of α1-antitrypsin deficiency in induced pluripotent stem cells. Nat. Cell Biol. 2011, 478, 391–394. [Google Scholar] [CrossRef]
- Zou, J.; Mali, P.; Huang, X.; Dowey, S.N.; Cheng, L. Site-specific gene correction of a point mutation in human iPS cells derived from an adult patient with sickle cell disease. Blood 2011, 118, 4599–4608. [Google Scholar] [CrossRef]
- Fong, H.; Wang, C.; Knoferle, J.; Walker, D.; Balestra, M.E.; Tong, L.M.; Leung, L.; Ring, K.L.; Seeley, W.W.; Karydas, A.; et al. Genetic Correction of Tauopathy Phenotypes in Neurons Derived from Human Induced Pluripotent Stem Cells. Stem Cell Rep. 2013, 1, 226–234. [Google Scholar] [CrossRef]
- Corti, S.; Nizzardo, M.; Simone, C.; Falcone, M.; Nardini, M.; Ronchi, D.; Donadoni, C.; Salani, S.; Riboldi, G.; Magri, F.; et al. Genetic Correction of Human Induced Pluripotent Stem Cells from Patients with Spinal Muscular Atrophy. Sci. Transl. Med. 2012, 4, 165ra162. [Google Scholar] [CrossRef]
- Wang, Y.; Zheng, C.-G.; Jiang, Y.; Zhang, J.; Chen, J.; Yao, C.; Zhao, Q.; Liu, S.; Chen, K.; Du, J.; et al. Genetic correction of β-thalassemia patient-specific iPS cells and its use in improving hemoglobin production in irradiated SCID mice. Cell Res. 2012, 22, 637–648. [Google Scholar] [CrossRef]
- Ding, Q.; Lee, Y.-K.; Schaefer, E.; Peters, D.T.; Veres, A.; Kim, K.; Kuperwasser, N.; Motola, D.L.; Meissner, T.B.; Hendriks, W.T.; et al. A TALEN Genome-Editing System for Generating Human Stem Cell-Based Disease Models. Cell Stem Cell 2013, 12, 238–251. [Google Scholar] [CrossRef] [PubMed]
- Liang, P.; Lan, F.; Lee, A.S.; Gong, T.; Sanchez-Freire, V.; Wang, Y.; Diecke, S.; Sallam, K.; Knowles, J.W.; Wang, P.J.; et al. Drug Screening Using a Library of Human Induced Pluripotent Stem Cell–Derived Cardiomyocytes Reveals Disease-Specific Patterns of Cardiotoxicity. Circulation 2013, 127, 1677–1691. [Google Scholar] [CrossRef]
- Braam, S.; Tertoolen, L.; Casini, S.; Matsa, E.; Lu, H.; Teisman, A.; Passier, R.; Denning, C.; Gallacher, D.; Towart, R.; et al. Repolarization reserve determines drug responses in human pluripotent stem cell derived cardiomyocytes. Stem Cell Res. 2013, 10, 48–56. [Google Scholar] [CrossRef] [PubMed]
- Terrenoire, C.; Wang, K.; Tung, K.W.C.; Chung, W.K.; Pass, R.H.; Lu, J.T.; Jean, J.-C.; Omari, A.; Sampson, K.J.; Kotton, D.; et al. Induced pluripotent stem cells used to reveal drug actions in a long QT syndrome family with complex genetics. J. Gen. Physiol. 2012, 141, 61–72. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Burridge, P.W.; McKeithan, W.L.; Serrano, R.; Shukla, P.; Sayed, N.; Churko, J.M.; Kitani, T.; Wu, H.; Holmström, A.; et al. High-throughput screening of tyrosine kinase inhibitor cardiotoxicity with human induced pluripotent stem cells. Sci. Transl. Med. 2017, 9, eaaf2584. [Google Scholar] [CrossRef]
- Sharma, A.; Marceau, C.; Hamaguchi, R.; Burridge, P.W.; Rajarajan, K.; Churko, J.M.; Wu, H.; Sallam, K.I.; Matsa, E.; Sturzu, A.C.; et al. Human Induced Pluripotent Stem Cell–Derived Cardiomyocytes as an In Vitro Model for Coxsackievirus B3–Induced Myocarditis and Antiviral Drug Screening Platform. Circ. Res. 2014, 115, 556–566. [Google Scholar] [CrossRef]
| Disease Model | Species/Tissue | Source (Cell Type) | Transdifferentiated Cell (Converted) | Transdifferentiation Factors/TFs | Delivery/Vehicle | Efficiency | Results/Physiological Outcome | References | |
|---|---|---|---|---|---|---|---|---|---|
| CNS/Brain | - | Mouse brain | Astrocyte, Fibroblast | Induced neuron (iN) | Myt1l, Ascl1, Brn2a | Transduced in vitro/Doxycycline in drinking water/Lentiviral | 0.4% to 5.9% | iNs in tissue | [27] |
| CPN (Colossal projection neuron) | Corticofugal projection neuron (CFuPN) | Fezf2 | Electroporation (in utero)/Plasmid DNA | 75.6% of Fezf2-expressing CPN (+ve for CFuPN marker) | Morphological change, gene and protein expression (until P3). Axon connectivity change (until E17.5) | [33] | |||
| L4 neuron | L5B neuron | Fezf2 | Electroporation (in utero)/Plasmid DNA | 50% for Fezf2+ L4 and L5B neurons | Morphological change, gene and protein expression (until P1) | [34] | |||
| Astrocyte | Induced adult neuroblast (iANB) | Sox2 | Stereotactic (brain)/Lentiviral | 41% of YFP+ cells expressed NG2; 23.2% of GFP+ cells were +ve for DCX | Transdifferentiated iANBs in tissue. Transdifferentiated mature neurons (+BNTP/Noggin or VPA) | [28,29] | |||
| Stab injury in AD | Mouse brain, Human cortical astrocytes | Astrocyte NG2 cell | Induced neuron (iN) | NeuroD1 | Stereotactic (brain)/Retroviral | 90% | Transdifferentiated iN in tissue. Functional or behavioural data not acquired | [32] | |
| Spinal cord injury | Mouse Spinal cord | Astrocyte | Induced adult neuroblast (iANB) | Sox2 | Stereotactic (spinal cord)/Lentiviral | 3–6% were reprogrammed by SOX2 | Transdifferentiated iANBs in tissue. Mature neurons (+VPA) synapses with resident neurons. Functional data not acquired | [30] | |
| Cardiac | Freeze–thaw injury | Rat | Cardiac fibroblasts | Skeletal myofibers | MyoD | Intramyocardial/Adenovirus | 2–14% | Produced myofibers (immature) in tissue | [8] |
| Myocardial infarction | Mouse | Cardiac fibroblast | Cardiomyocytes | GMT (Gata4, Mef2c, Tbx5) | Intramyocardial/Retrovirus | 10–15% | Reduced infarct size. Significant decrease in cardiac dysfunction | [37] | |
| Cardiac fibroblast | Cardiomyocytes | GMT (Gata4, Mef2c, Tbx5) | Intramyocardial/Retrovirus | 3–7% | Cardiomyocytelike cells in fibrotic area. No phsiological functional results | [38] | |||
| Cardiac fibroblast | Cardiomyocytes | GHMT (Gata4, Hand2, Mef2c, Tbx5) | Intramyocardial/Retrovirus | ~7% | Reduced infarct size. Significant decrease in cardiac dysfunction | [39] | |||
| Cardiac fibroblast | Cardiomyocytes | microRNAs 1, 133, 208 & 499 | Intramyocardial/Lentivirus | 12–25% | Fibroblast transdifferentiation into cardiomyocyte in the infarct spot/area. Moderate decrease in cardiac dysfunction | [40,41] | |||
| Complete heart blockage | Pig | Ventricular cardiomyocytes | Pacemaker cell-induced sinoatrial node cells | Tbx18 | Percutaneous, to heart ventricule/Adenovirus | 24.5% | Constituting a biological pacemaker. Improvement of bradycardia | [42] | |
| Ventricular cardiomyocytes | Pacemaker cell-induced sinoatrial node cell | Tbx18 | Intramyocardial/Adenovirus | 9.2% | Constituting a biological pacemaker. Improvement of bradycardia | [43] |
| Disease Model for Cardiac Dysfunctions | Impacted/Mutated Genes |
|---|---|
| Left ventricular noncompaction | TBX20, GATA4 |
| Familial hypercholesterolemia | LDLR, PCSK9 |
| Timothy syndrome | CACNA1C |
| Dilated cardiomyopathy | TTN, TNNT2, LMNA, DES |
| Duchenne muscular dystrophy | DMD |
| Arrhythmogenic right ventricular dysplasia | PKP2 |
| Long-QT syndrome type 1 | KCNQ1 |
| Jervell and Lange-Nielsen syndrome | KCNQ1 |
| Catecholaminergic polymorphic ventricular tachycardia type 1 | RYR2 |
| Catecholaminergic polymorphic ventricular tachycardia type 2 | CASQ2 |
| Brugada syndrome | SCN5A |
| Calcific aortic valve | NOTCH1 |
| Williams–Beuren syndrome | ELN |
| Familial pulmonary hypertension | BMPR2 |
| Barth syndrome | TAZ |
| Hypertrophic cardiomyopathy | MYH7 |
| Maturity-onset diabetes of the young type 2 | GCK |
| Insulin resistance | AKT2 |
| Familial hypobetalipoproteinemia | PCSK9 |
| Long-QT syndrome type 2 | KCNH2 |
| Long-QT syndrome type 3 | SCN5A |
| Tangier disease | ABCA1 |
| Dyslipidemia | SORT1 |
| Hypoinsulinemic hypoglycemia and hemihypertrophy | AKT2 |
| Chemicals/Small Molecules | Molecular Activity/Induced Mechanism(s) | Cellular Reprogramming Function(s) | References |
|---|---|---|---|
| RepSox (E-616452) | TGF-βRI (ALK5) inhibitor | CiNPC, CiN, CiCM | [49,53,73] |
| TTNPB | RAR ligand | CiCM, CiN | [54,73] |
| Forskolin | Adenylyl cyclase activator | CiN, CiCM | [52,53,73] |
| CHIR99021 | GSK3 inhibitor | CiNPC, CiNSLCe, CiNf, CiCM | [52,53,54,73,74,76] |
| VPA | HDAC inhibitor | CiPSCa, CiNPCb, CiNc, CiCMd | [49,53,54,73] |
| LiCl and Li2CO3 | GSK3 inhibitor | CiNPC | [49] |
| SB431542 | TGF-βRI inhibitor | CiPSC, CiNPC, CiN, hiEndoPC | [49,54] |
| NaB | HDAC inhibitor | CiNPC | [49] |
| Tranilast | Inhibit TGF-β1 secretion | CiNPC | [49] |
| TSA (Trichostatin A) | HDAC inhibitor | CiNPC | [49] |
| RG108 | DNA methyltransferase inhibitor | CiNSLC | [50] |
| A-83-01 | TGF-βRI (ALK4/5/7) inhibitor | CiNSLC, CiCM | [50,74] |
| Hh-Ag 1.5 | Smoothened agonist | CiNSLC | [50] |
| SMER28 | Autophagy modulator | CiNSLC | [50] |
| Retinoic acid | RAR ligand | CiNSLC | [50] |
| LDN193189 | BMP type I receptor (ALK2/3) inhibitor | CiNSLC | [50] |
| GO6983 | PKC inhibitor | CiN | [53] |
| ISX9 | neurogenesis inducer | CiN | [52] |
| Dorsomorphin | AMPK and BMP I receptor inhibitor | CiN | [53] |
| I-BET151 | BET inhibitor | CiN | [52] |
| SP600125 | JNK inhibitor | CiN | [53] |
| SAG | Smoothened agonist | CiN | [54] |
| Y-27632 | ROCK inhibitor | CiN, CiCM | [53,74] |
| Purmorphamine | Smoothened agonist | CiN | [54] |
| DAPT | Gamma-secretase inhibitor | CiN | [54] |
| SC1 | ERK1 and RasGAP inhibitor | CiCM | [74] |
| Thiazovivin | ROCK inhibitor | CiN | [54] |
| OAC2 | Epigenetic modulation | CiCM | [74] |
| AS8351 | Epigenetic modulator | CiCM | [74] |
| SU16F | PDGFR-β inhibitor | CiCM | [74] |
| JNJ10198409 | PDGFR-α and PDGFR-β inhibitor | CiCM | [74] |
| Bix01294 | Histone methyl transferase inhibitor | CiCM | [74] |
| Reprogramming Factors (TFs) | Species/Model/Cell Type | Obtained Cell Types | Efficiency | Results/Functional Outcome | References | |
|---|---|---|---|---|---|---|
| Neuronal | Brn2, Myt1l, Zic1, Olig2, and Ascl1 | Mouse embryonic and postnatal fibroblast cells | iN (mostly GABAergic and glutamatergic neurons) | ∼50% | Synaptic maturation, functional electrophysiology | [76] |
| Ascl1, Brn2 and Myt1l | iN (mostly excitatory neurons) | 19.50% | Synaptic maturation, functional electrophysiology | [76,81] | ||
| Forskolin, ISX9, CHIR99021 and SB431542 | Mouse fibroblast cells | iN | >90% | Functional electrophysiology | [52] | |
| Ascl1, Brn2, Myt1l | Mouse hepatocytes | iN | >90% | Functional electrophysiology | [82] | |
| Mash1, Nurr1 and Lmx1a | Mouse and human cells/fibroblast cells | iN (mostly dopaminergic neurons) | High | - | [83] | |
| Ascl1, Brn2 and Myt1l | neurons | 20% | Functional | [27] | ||
| Sox2 and Mash1 | Pericyte-derived cells of the adult human cerebral cortex | GABAergic neurons | ∼50% | Obtained iN acquire the ability of action potential firing, synaptic targets for neurons | [84] | |
| LDN193189, SB431542, TTNPB, Tzv, CHIR99021, VPA, DAPT, SAG, Purmo | Human astrocytes | Functional neurons (mainly glutamatergic neurons) | >90% | Functional | [54] | |
| ASCL1, NGN2, SOX2, NURR1 and PITX3 | Human fibroblast cells | iN (mostly dopaminergic neurons) | ∼80% | Functional electrophysiology | [85] | |
| NeuroD1, Ascl1, Brn2, and Mytl1 | iN | ∼60% | Functional neurons | [81] | ||
| Ascl1, Lmx1a, FoxA2, and FEV | serotonergic (i5HT) neurons | ∼25% | Showed spontaneous electrophysiological activity, Active synaptic transmission observed | [86] | ||
| Cardiac | GATA4, MEF2C, TBX5, HAND2 | Mouse | iCMs from MEFs | ~70–80% | Spontaneous beating, Ca2+ transients | [87] |
| GATA4, MYOD-MEF2C, TBX5, HAND2 | iCMs from embryonic head fibroblasts | 10-20% | Spontaneous beating, Ca2+ transients | [88] | ||
| GATA4, MEF2C, TBX5, HAND2, NKX2.5, SB431542 | iCMs from MEFs | 17% | Spontaneous beating, Ca2+ transients | [89] | ||
| MEF2C, GATA4, TBX5 | iCMs from CFs | ~10% | Action potentials, spontaneous beating, Ca2+ transients | [38] | ||
| GATA4, MEF2C, TBX5, HAND2, miR-1, miR-133, A83-01, Y-27632 | iCMs from MEFs | 60% | Action potentials, spontaneous beating, Ca2+ transients | [90] | ||
| GATA4, MEF2C, TBX5, (HAND2), Bmi1 shRNA | iCMs from CFs | 22% | Spontaneous beating, Ca2+ transients | [91] | ||
| GATA4, MEF2C, TBX5, SB431542, XAV939 | iCMs from CFs | ~30% | Spontaneous beating, Ca2+ transients | [92] | ||
| GATA4, MEF2C, TBX5, HAND2, DAPT | iCMs from MEFs | ~38% | Ca2+ transients, spontaneous beating | [93] | ||
| GATA4, MEF2C, TBX5, MESP1, MYOCD | Human | iCMs from HCFs | 5.90% | Ca2+ transients, action potentials | [94] | |
| GATA4, MEF2C, TBX5, ESRGG, MESP1, MYOCD, ZFPM2 | iCMs from hESC-derived fibroblasts | 13% | Ca2+ transients, action potentials | [95] | ||
| GATA4, MEF2C, TBX5 (+ MESP1, MYOCD) with miR-133 | iCMs from HCFs | 27.80% | Ca2+ transients | [96] | ||
| GATA4, MEF2C, TBX5, (HAND2, MYOCD or miR-590) | Human, rat, porcine | iCMs from adult HCFs | ~40% | No spontaneous beating in human iCMs | [97] |
| S. No. | NCT Number | Title | Disease Condition | Phase | Location of Study | |
|---|---|---|---|---|---|---|
| Disease modelling | 1 | NCT02564484 | iPSC Neurons From Adult Survivors of Childhood Cancer Who Have Persistent Vincristine-Induced Neuropathy | Leukemia|Lymphoma | Unknown | St. Jude Children’s Research Hospital, Memphis, Tennessee, United States |
| 2 | NCT01860898 | A Phase I Study of iPS Cell Generation From Patients With COPD | Thoracic Diseases|Respiratory Tract Diseases|Cancer of Lung|Cancer of the Lung|Lung Cancer|Lung Diseases, Obstructive|COPD|Pulmonary Emphysema|Neoplasms, Lung|Neoplasms, Pulmonary|Pulmonary Cancer|Pulmonary Neoplasms|Carcinoma, Non-Small-Cell Lung|Carcinoma, Small Cell | Not Applicable | Mayo Clinic in Rochester, Rochester, Minnesota, United States | |
| 3 | NCT02980302 | Development of the Tool “ iPSC “ for the Functional Study of Mutations Responsible for Mental Retardation | Intellectual Deficiency|Asymptomatic Carrier of the Mutation of the Gene MYT1L|Healthy Volunteers | Not Applicable | UniversityHospitalGrenoble, La Tronche, France | |
| 4 | NCT02193724 | Feasibility of Generating Pluripotent Stem Cells From Patients With Familial Retinoblastoma | Retinoblastoma | Unknown | St. Jude Children’s Research Hospital, Memphis, Tennessee, United States | |
| 5 | NCT02162953 | Stem Cell Models of Best Disease and Other Retinal Degenerative Diseases | Retinal Disease|Bestrophinopathy|Best Vitelliform Macular Dystrophy|Adult Onset Vitelliform Macular Dystrophy|Autosomal Dominant Vitreoretinalchoroidopathy | Unknown | Mayo Clinic, Rochester, Minnesota, United States | |
| 6 | NCT03883750 | Induced Pluripotent Stem Cells for Niemann Pick Disease | Niemann–Pick Diseases | Unknown | Childrens Hospital and Institute of Child Health, Ferozepur Road, Lahore, Pakistan | |
| 7 | NCT03867526 | Establishment of Human Cellular Disease Models for Wilson Disease | Wilson Disease | Unknown | Childrens Hospital and Institute of Child Health, Ferozepur Road, Lahore, Pakistan | |
| 8 | NCT03754088 | In vitro Model of the Cystic Fibrosis Bronchial Epithelium Via iPS Technology | Cystic Fibrosis | Unknown | HÃ’pital Arnaud de Villeneuve—CHU de Montpellier, Montpellier, France | |
| 9 | NCT01534624 | Stem Cell Study of Genetics and Drug Addiction | Induced Pluripotent Stem Cells | Unknown | National Institute on Drug Abuse, Baltimore, Maryland, United States | |
| 10 | NCT01865981 | Investigating Hereditary Cardiac Disease by Reprogramming Skin Cells to Heart Muscle | Eletrophysiology of iPS-derived Cardiomyocytes | Unknown | University of Dundee, Dundee, Angus, United Kingdom | |
| 11 | NCT03872713 | Establishment of Human Cellular Disease Models for Morquio Disease | Morquio Disease | Unknown | Childrens Hospital and Institute of Child Health, Ferozepur Road, Lahore, Pakistan | |
| 12 | NCT01639391 | Creation of a Bank of Fibroblast From Patients With Amyotrophic Lateral Sclerosis: Pilot Study | Amyotrophic Lateral Sclerosis | Not Applicable | Centre référent maladies rares SLA, Paris, France | |
| 13 | NCT03898817 | Pathology of Helicases and Premature Aging: Study by Derivation of hiPS | Age Problem | Unknown | University Hospital Montpellier, Montpellier, France | |
| 14 | NCT01517425 | Evaluating Cardiovascular Phenotypes Using Induced Pluripotent Stem Cells | Coronary Artery Disease | Unknown | Scripps Translational Science Institute, La Jolla, California, United States | |
| 15 | NCT02413450 | Derivation of Human Induced Pluripotent Stem (iPS) Cells to Heritable Cardiac Arrhythmias | Inherited Cardiac Arrythmias|Long QT Syndrome (LQTS)|Brugada Syndrome (BrS)|Catecholaminergic Polymorphic Ventricular Tachycardia (CPVT)|Early Repolarization Syndrome (ERS)|Arrhythmogenic Cardiomyopathy (AC, ARVD/C)|Hypertrophic Cardiomyopathy (HCM)|Dilated Cardiomyopathy (DCM)|Muscular Dystrophies (Duchenne, Becker, Myotonic Dystrophy)|Normal Control Subjects | Unknown | Johns Hopkins Medical Institute, Baltimore, Maryland, United States | |
| 16 | NCT03682458 | Study of Neurodegenerative Diseases Induced Stem Cells in Patients and Healthy Family Controls. | Neurodegenerative Diseases | Unknown | Stefano Gambardella, Pozzilli, Isernia, Italy | |
| 17 | NCT03635294 | Multi-Omics and IPSCs to Improve Diagnosis of Rare Intellectual Disabilities | Rare Intellectual Disabilities | Not Applicable | CHU Angers, Angers, France|HCL Lyon, Bron, France|CHU de Bourgogne, Dijon, France|CHU Nantes, Nantes, France|CHU Poitiers, Poitiers, France|CHU Rennes, Rennes, France | |
| 18 | NCT03181204 | Modeling Bronchial Epithelium Modifications Associated With COPD Using iPS | Pulmonary Disease, Chronic Obstructive|Smoking | Unknown | Centre Hospitalier Universitaire de Montpellier, Montpellier, France | |
| 19 | NCT02772367 | Generation of Heart Muscle Cells From Blood or Skin Cells of Breast Cancer Patients | Breast Cancer | Unknown | Memorial Sloan-Kettering Cancer Center, New York, New York, United States | |
| 20 | NCT00895271 | Establishing Fibroblast-Derived Cell Lines From Skin Biopsies of Patients With Immunodeficiency or Immunodysregulation Disorders | Primary Immunodeficiency|DOCK8 | Unknown | National Institutes of Health Clinical Center, 9000 Rockville Pike, Bethesda, Maryland, United States | |
| Drug screening | 21 | NCT01943383 | Pharmacogenomic Evaluation of Antihypertensive Responses in Induced Pluripotent Stem (iPS) Cells Study | Hypertension | Unknown | University of Florida, Gainesville, Florida, United States |
| 22 | NCT04744532 | iPSC-based Drug Repurposing for ALS Medicine (iDReAM) Study | Amyotrophic Lateral Sclerosis | Phase 1 | Kyoto University, Kyoto, Japan|Kitasato University, Sagamihara, Japan|Tokushima University, Tokushima, Japan|Tottori University, Yonago, Japan | |
| 23 | NCT04097275 | Induced Pluripotent Stem Cells for the Development of Novel Drug Therapies for Inborn Errors of Metabolism (iPSC-IEM) | Inborn Errors of Metabolism | Unknown | Children Hospital and Institute of Child Health, Lahore, Pakistan | |
| 24 | NCT03407040 | Generation of Cancer Antigen-Specific T-cells From Human Induced Pluripotent Stem Cells (iPSC) for Research and Potential FutureTherapy | Gastrointestinal Cancers|Breast Cancer|Pancreatic Cancer|Melanoma|Lung Cancer | Unknown | National Institutes of Health Clinical Center, Bethesda, Maryland, United States | |
| Therapy-based studies | 25 | NCT04945018 | A Study of iPS Cell-derived Cardiomyocyte Spheroids (HS-001) in Patients With Heart Failure (LAPiS Study) | Heart Failure|Ischemic Heart Disease | Phase 1|Phase 2 | St. Marianna University Hospital, Kawasaki, Japan|Nihon University Itabashi Hospital, Tokyo, Japan|The University of Tokyo Hospital, Tokyo, Japan|Tokyo Medical and Dental University Medical Hospital, Tokyo, Japan|Tokyo Metropolitan Geriatric Medical Center, Tokyo, Japan |
| 26 | NCT03403699 | Human iPSC for Repair of Vasodegenerative Vessels in Diabetic Retinopathy | Diabetes Complications|Diabetic Retinopathy | Unknown | University of Alabama at Birmingham, Birmingham, Alabama, United States | |
| 27 | NCT04696328 | Clinical Trial of Human (Allogeneic) iPS Cell-derived Cardiomyocytes Sheet for Ischemic Cardiomyopathy | Myocardial Ischemia | Phase 1 | Osaka University Hospital, Suita, Osaka, Japan | |
| 28 | NCT04339764 | Autologous Transplantation of Induced Pluripotent Stem Cell-Derived Retinal Pigment Epithelium for Geographic Atrophy Associated With Age-Related Macular Degeneration | Age-Related Macular Degeneration | Phase 1|Phase 2 | National Institutes of Health Clinical Center, Bethesda, Maryland, United States | |
| 29 | NCT04982081 | Treating Congestive HF With hiPSC-CMs Through Endocardial Injection | Cardiovascular Diseases|Congestive Heart Failure|Dilated Cardiomyopathy | Phase 1 | Help Therapeutics, Nanjing, Jiangsu, China | |
| 30 | NCT03971812 | Organoids Derived From Induced-Pluripotent Stem Cells (iPS) From Patients With High Grade Astrocytoma | Glioma | Unknown | Assistance Publique Hôpitaux de Marseille, Marseille, France | |
| 31 | NCT03763136 | Treating Heart Failure With hPSC-CMs | Heart Failure | Phase 1|Phase 2 | HelpThera, Nanjing, Jiangsu, China |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kalra, R.S.; Dhanjal, J.K.; Das, M.; Singh, B.; Naithani, R. Cell Transdifferentiation and Reprogramming in Disease Modeling: Insights into the Neuronal and Cardiac Disease Models and Current Translational Strategies. Cells 2021, 10, 2558. https://doi.org/10.3390/cells10102558
Kalra RS, Dhanjal JK, Das M, Singh B, Naithani R. Cell Transdifferentiation and Reprogramming in Disease Modeling: Insights into the Neuronal and Cardiac Disease Models and Current Translational Strategies. Cells. 2021; 10(10):2558. https://doi.org/10.3390/cells10102558
Chicago/Turabian StyleKalra, Rajkumar Singh, Jaspreet Kaur Dhanjal, Mriganko Das, Birbal Singh, and Rajesh Naithani. 2021. "Cell Transdifferentiation and Reprogramming in Disease Modeling: Insights into the Neuronal and Cardiac Disease Models and Current Translational Strategies" Cells 10, no. 10: 2558. https://doi.org/10.3390/cells10102558
APA StyleKalra, R. S., Dhanjal, J. K., Das, M., Singh, B., & Naithani, R. (2021). Cell Transdifferentiation and Reprogramming in Disease Modeling: Insights into the Neuronal and Cardiac Disease Models and Current Translational Strategies. Cells, 10(10), 2558. https://doi.org/10.3390/cells10102558







