Role of Notch Receptors in Hematologic Malignancies
Abstract
1. Introduction
2. Notch 1
2.1. Physiology of Notch 1 Signaling in the Immune System Cells
2.2. Notch 1 in T-Cell Acute and Chronic Lymphoblastic Leukemia
2.3. Notch 1 and B-Cell Malignancies
3. Notch 2
3.1. Physiology of Notch 2 Signaling in the Immune System Cells
3.2. Notch 2 Mutations in B-Cell Lymphomas
3.3. Notch 2 in B-Cell Acute and Chronic Lymphocytic Leukemia
4. Notch 3
Notch 3 from Physiological to Oncogenic Pathways
5. Notch 4
Notch 4 from Physiological to Oncogenic Pathways
6. Conclusions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| T-ALL | acute lymphoblastic leukemia |
| B-CLL | B-cell chronic lymphocytic leukemia |
| CLL | B-cell chronic lymphocytic leukemia |
| DLL4 | Delta-like ligand 4 |
| MCL | mantle cells lymphoma |
| RS | Richter Syndrome |
| ALCL | anaplastic large cell lymphoma |
| MM | multiple myeloma |
| SMZL | splenic marginal zone lymphoma |
| DLBCL | diffuse large B-cell lymphoma |
| MALT | mucosa-associated lymphoid tissue |
| FL | follicular lymphoma |
| MZB | marginal zone B |
| AML | acute myeloid leukemia |
| NHL | Non-Hodgkin Lymphoma |
References
- Artavanis-Tsakonas, S.; Matsuno, K.; Fortini, M.E. Notch signaling. Science 1995, 268, 225–232. [Google Scholar] [CrossRef] [PubMed]
- Fortini, M.E.; Artavanis-Tsakonas, S. Notch: Neurogenesis is only part of the picture. Cell 1993, 75, 1245–1247. [Google Scholar] [CrossRef]
- Kovall, R.A.; Blacklow, S.C. Mechanistic Insights into Notch Receptor Signaling from Structural and Biochemical Studies. Curr. Top. Dev. Biol. 2010, 92, 31–71. [Google Scholar] [CrossRef] [PubMed]
- Van Tetering, G.; Van Diest, P.; Verlaan, I.; Van Der Wall, E.; Kopan, R.; Vooijs, M. Metalloprotease ADAM10 Is Required for Notch 1 Site 2 Cleavage. J. Biol. Chem. 2009, 284, 31018–31027. [Google Scholar] [CrossRef] [PubMed]
- Christian, L.M. The ADAM family. Fly 2012, 6, 30–34. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mumm, J.S.; Kopan, R. Notch Signaling: From the Outside In. Dev. Biol. 2000, 228, 151–165. [Google Scholar] [CrossRef] [PubMed]
- Petcherski, A.G.; Kimble, J. Mastermind is a putative activator for Notch. Curr. Biol. 2000, 10, R471–R473. [Google Scholar] [CrossRef]
- Kopan, R. Notch Signaling. Cold Spring Harb. Perspect. Biol. 2012, 4, a011213. [Google Scholar] [CrossRef]
- Phng, L.-K.; Gerhardt, H. Angiogenesis: A Team Effort Coordinated by Notch. Dev. Cell 2009, 16, 196–208. [Google Scholar] [CrossRef]
- Kojika, S.; Griffin, J.D. Notch receptors and hematopoiesis. Exp. Hematol. 2001, 29, 1041–1052. [Google Scholar] [CrossRef]
- Aster, J.C.; Pear, W.S.; Blacklow, S.C. The Varied Roles of Notch in Cancer. Annu. Rev. Pathol. Mech. Dis. 2017, 12, 245–275. [Google Scholar] [CrossRef] [PubMed]
- Chiang, M.Y.; Radojcic, V.; Maillard, I. Oncogenic Notch signaling in T-cell and B-cell lymphoproliferative disorders. Curr. Opin. Hematol. 2016, 23, 362–370. [Google Scholar] [CrossRef] [PubMed]
- Arruga, F.; Vaisitti, T.; Deaglio, S. The NOTCH Pathway and Its Mutations in Mature B Cell Malignancies. Front. Oncol. 2018, 8, 550. [Google Scholar] [CrossRef] [PubMed]
- Pancewicz-Wojtkiewicz, J.; Nicot, C. Current views on the role of Notch signaling and the pathogenesis of human leukemia. BMC Cancer 2011, 11, 502. [Google Scholar] [CrossRef]
- Onaindia, A.; Medeiros, L.J.; Patel, K.P. Clinical utility of recently identified diagnostic, prognostic, and predictive molecular biomarkers in mature B-cell neoplasms. Mod. Pathol. 2017, 30, 1338–1366. [Google Scholar] [CrossRef]
- Bray, S.J. Notch signalling in context. Nat. Rev. Mol. Cell Biol. 2016, 17, 722–735. [Google Scholar] [CrossRef]
- Koch, U.; Fiorini, E.; Benedito, R.; Besseyrias, V.; Schuster-Gossler, K.; Pierres, M.; Manley, N.R.; Duarte, A.; Macdonald, H.R.; Radtke, F. Delta-like 4 is the essential, nonredundant ligand for Notch 1 during thymic T cell lineage commitment. J. Exp. Med. 2008, 205, 2515–2523. [Google Scholar] [CrossRef]
- Hozumi, K.; Mailhos, C.; Negishi, N.; Hirano, K.-I.; Yahata, T.; Ando, K.; Zuklys, S.; Holländer, G.A.; Shima, D.T.; Habu, S. Delta-like 4 is indispensable in thymic environment specific for T cell development. J. Exp. Med. 2008, 205, 2507–2513. [Google Scholar] [CrossRef]
- Hellström, M.; Phng, L.-K.; Hofmann, J.J.; Wallgard, E.; Coultas, L.; Lindblom, P.; Alva, J.A.; Nilsson, A.-K.; Karlsson, L.; Gaiano, N.; et al. Dll4 signalling through Notch 1 regulates formation of tip cells during angiogenesis. Nat. Cell Biol. 2007, 445, 776–780. [Google Scholar] [CrossRef]
- Yuan, J.S.; Kousis, P.C.; Suliman, S.; Visan, I.; Guidos, C.J. Functions of Notch Signaling in the Immune System: Consensus and Controversies. Annu. Rev. Immunol. 2010, 28, 343–365. [Google Scholar] [CrossRef]
- Radtke, F.; Wilson, A.; Stark, G.; Bauer, M.; Van Meerwijk, J.; Macdonald, H.; Aguet, M. Deficient T Cell Fate Specification in Mice with an Induced Inactivation of Notch. Immunity 1999, 10, 547–558. [Google Scholar] [CrossRef]
- García-León, M.J.; Fuentes, P.; De La Pompa, J.L.; Toribio, M.L. Dynamic regulation of NOTCH 1 activation and Notch ligand expression in human thymus development. Development 2018, 145, dev165597. [Google Scholar] [CrossRef] [PubMed]
- Morath, A.; Schamel, W.W. αβ and γδ T cell receptors: Similar but different. J. Leukoc. Biol. 2020, 107, 1045–1055. [Google Scholar] [CrossRef] [PubMed]
- Silva-Santos, B.; Pennington, D.J.; Hayday, A.C. Lymphotoxin-Mediated Regulation of Cell Differentiation by T Cell Progenitors. Science 2005, 307, 925–928. [Google Scholar] [CrossRef]
- Chien, Y.-H.; Meyer, C.; Bonneville, M. γδT Cells: First Line of Defense and Beyond. Annu. Rev. Immunol. 2014, 32, 121–155. [Google Scholar] [CrossRef]
- García-Peydró, M.; De Yébenes, V.G.; Toribio, M.L. Sustained Notch 1 signaling instructs the earliest human intrathymic precursors to adopt a γδ T-cell fate in fetal thymus organ culture. Blood 2003, 102, 2444–2451. [Google Scholar] [CrossRef]
- Van De Walle, I.; De Smet, G.; De Smedt, M.; Vandekerckhove, B.; Leclercq, G.; Plum, J.; Taghon, T. An early decrease in Notch activation is required for human TCR-αβ lineage differentiation at the expense of TCR-γδ T cells. Blood 2009, 113, 2988–2998. [Google Scholar] [CrossRef]
- Ikawa, T.; Hirose, S.; Masuda, K.; Kakugawa, K.; Satoh, R.; Shibano-Satoh, A.; Kominami, R.; Katsura, Y.; Kawamoto, H.; Pocivavsek, L.; et al. An Essential Developmental Checkpoint for Production of the T Cell Lineage. Science 2010, 329, 93–96. [Google Scholar] [CrossRef]
- Li, L.; Leid, M.; Rothenberg, E.V. An Early T Cell Lineage Commitment Checkpoint Dependent on the Transcription Factor Bcl11b. Science 2010, 329, 89–93. [Google Scholar] [CrossRef]
- Dolens, A.; Durinck, K.; Lavaert, M.; Van Der Meulen, J.; Velghe, I.; De Medts, J.; Weening, K.; Roels, J.; De Mulder, K.; Volders, P.; et al. Distinct Notch 1 and BCL11B requirements mediate human γδ/αβ T cell development. EMBO Rep. 2020, 21, e49006. [Google Scholar] [CrossRef]
- Ciofani, M.; Zúñiga-Pflücker, J.C. Notch promotes survival of pre-T cells at the β-selection checkpoint by regulating cellular metabolism. Nat. Immunol. 2005, 6, 881–888. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Yan, R.; Pinnell, N.; McCarter, A.C.; Oh, Y.; Liu, Y.; Sha, C.; Garber, N.F.; Chen, Y.; Wu, Q.; et al. Stage-specific roles for Zmiz1 in Notch -dependent steps of early T-cell development. Blood 2018, 132, 1279–1292. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.-A.; Kim, W.-S.; Park, S.-G. Notch 1 is an important mediator for enhancing of B-cell activation and antibody secretion by Notch ligand. Immunology 2014, 143, 550–559. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.; Wang, X.; Xiao, H.; Liu, X.; Fang, Y.; Zhai, B.; Xu, R.; Han, G.; Chen, G.; Hou, C.; et al. Both Notch 1 and its ligands in B cells promote antibody production. Mol. Immunol. 2017, 91, 17–23. [Google Scholar] [CrossRef]
- Palomero, T.; Lim, W.K.; Odom, D.T.; Sulis, M.L.; Real, P.J.; Margolin, A.; Barnes, K.C.; O’Neil, J.; Neuberg, D.; Weng, A.P.; et al. NOTCH 1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc. Natl. Acad. Sci. USA 2006, 103, 18261–18266. [Google Scholar] [CrossRef]
- Jarriault, S.; Le Bail, O.; Hirsinger, E.; Pourquié, O.; Logeat, F.; Strong, C.F.; Brou, C.; Seidah, N.G.; Israël, A. Delta-1 Activation of Notch -1 Signaling Results inHES-1 Transactivation. Mol. Cell. Biol. 1998, 18, 7423–7431. [Google Scholar] [CrossRef]
- Yang, Z.; Qi, Y.; Lai, N.; Zhang, J.; Chen, Z.; Liu, M.; Zhang, W.; Luo, R.; Kang, S. Notch 1 signaling in melanoma cells promoted tumor-induced immunosuppression via upregulation of TGF-β. J. Exp. Clin. Cancer Res. 2018, 37, 1–13. [Google Scholar] [CrossRef]
- Guo, J.; Fu, W.; Xiang, M.; Zhang, Y.; Zhou, K.; Xu, C.-R.; Li, L.; Kuang, D.; Ye, F. Notch 1 Drives the Formation and Proliferation of Intrahepatic Cholangiocarcinoma. Curr. Med. Sci. 2019, 39, 929–937. [Google Scholar] [CrossRef]
- Rice, M.A.; Hsu, E.-C.; Aslan, M.; Ghoochani, A.; Su, A.; Stoyanova, T. Loss of Notch 1 Activity Inhibits Prostate Cancer Growth and Metastasis and Sensitizes Prostate Cancer Cells to Antiandrogen Therapies. Mol. Cancer Ther. 2019, 18, 1230–1242. [Google Scholar] [CrossRef]
- Yu, L.; Xia, K.; Gao, T.; Chen, J.; Zhang, Z.; Sun, X.; Simões, B.M.; Eyre, R.; Fan, Z.; Guo, W.; et al. The Notch Pathway Promotes Osteosarcoma Progression through Activation of Ephrin Reverse Signaling. Mol. Cancer Res. 2019, 17, 2383–2394. [Google Scholar] [CrossRef]
- Ellisen, L.W.; Bird, J.; West, D.C.; Soreng, A.; Reynolds, T.C.; Smith, S.D.; Sklar, J. TAN-1, the human homolog of the Drosophila Notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell 1991, 66, 649–661. [Google Scholar] [CrossRef]
- Weng, A.P.; Nam, Y.; Wolfe, M.S.; Pear, W.S.; Griffin, J.D.; Blacklow, S.C.; Aster, J.C. Growth Suppression of Pre-T Acute Lymphoblastic Leukemia Cells by Inhibition of Notch Signaling. Mol. Cell. Biol. 2003, 23, 655–664. [Google Scholar] [CrossRef] [PubMed]
- Weng, A.P.; Ferrando, A.A.; Lee, W.; Morris IV, J.P.; Silverman, L.B.; Sanchez-Irizarry, C.; Blacklow, S.C.; Look, A.T.; Aster, J.C. Activating Mutations of NOTCH 1 in Human T Cell Acute Lymphoblastic Leukemia. Science 2004, 306, 269–271. [Google Scholar] [CrossRef] [PubMed]
- Sulis, M.L.; Williams, O.; Palomero, T.; Tosello, V.; Pallikuppam, S.; Real, P.; Barnes, K.; Zuurbier, L.; Meijerink, J.; Ferrando, A.A. NOTCH 1 extracellular juxtamembrane expansion mutations in T-ALL. Blood 2008, 112, 733–740. [Google Scholar] [CrossRef] [PubMed]
- Kumar, D.; Ganeshpurkar, A.; Kumar, D.; Modi, G.; Gupta, S.K.; Singh, S.K. Secretase inhibitors for the treatment of Alzheimer’s disease: Long road ahead. Eur. J. Med. Chem. 2018, 148, 436–452. [Google Scholar] [CrossRef] [PubMed]
- Ranganathan, P.; Weaver, K.L.; Capobianco, A.J. Notch signalling in solid tumours: A little bit of everything but not all the time. Nat. Rev. Cancer 2011, 11, 338–351. [Google Scholar] [CrossRef] [PubMed]
- Milano, J.; McKay, J.; Dagenais, C.; Foster-Brown, L.; Pognan, F.; Gadient, R.; Jacobs, R.T.; Zacco, A.; Greenberg, B.D.; Ciaccio, P.J. Modulation of Notch Processing by γ-Secretase Inhibitors Causes Intestinal Goblet Cell Metaplasia and Induction of Genes Known to Specify Gut Secretory Lineage Differentiation. Toxicol. Sci. 2004, 82, 341–358. [Google Scholar] [CrossRef] [PubMed]
- Wong, G.T.; Manfra, D.J.; Poulet, F.M.; Zhang, Q.; Josien, H.B.; Bara, T.; Engstrom, L.; Pinzon-Ortiz, M.; Fine, J.S.; Lee, H.-J.J.; et al. Chronic Treatment with the γ-Secretase Inhibitor LY-411,575 Inhibits β-Amyloid Peptide Production and Alters Lymphopoiesis and Intestinal Cell Differentiation. J. Biol. Chem. 2004, 279, 12876–12882. [Google Scholar] [CrossRef]
- Habets, R.A.; De Bock, C.E.; Serneels, L.; Lodewijckx, I.; Verbeke, D.; Nittner, D.; Narlawar, R.; Demeyer, S.; Dooley, J.; Liston, A.; et al. Safe targeting of T cell acute lymphoblastic leukemia by pathology-specific NOTCH inhibition. Sci. Transl. Med. 2019, 11, eaau6246. [Google Scholar] [CrossRef]
- Takebe, N.; Nguyen, D.; Yang, S.X. Targeting Notch signaling pathway in cancer: Clinical development advances and challenges. Pharmacol. Ther. 2014, 141, 140–149. [Google Scholar] [CrossRef]
- Borthakur, G.; Martinelli, G.; Raffoux, E.; Chevallier, P.; Chromik, J.; Lithio, A.; Ms, C.L.S.; Yuen, E.; Iii, G.J.O.; Benhadji, K.A.; et al. Phase 1 study to evaluate Crenigacestat (LY3039478) in combination with dexamethasone in patients with T-cell acute lymphoblastic leukemia and lymphoma. Cancer 2020. [Google Scholar] [CrossRef] [PubMed]
- Even, C.; Lassen, U.; Merchan, J.; Le Tourneau, C.; Soria, J.-C.; Ferte, C.; Ricci, F.; Diener, J.T.; Yuen, E.; Smith, C.; et al. Safety and clinical activity of the Notch inhibitor, crenigacestat (LY3039478), in an open-label phase I trial expansion cohort of advanced or metastatic adenoid cystic carcinoma. Investig. New Drugs 2019, 38, 402–409. [Google Scholar] [CrossRef] [PubMed]
- Doi, T.; Tajimi, M.; Mori, J.; Asou, H.; Inoue, K.; Benhadji, K.A.; Naito, Y. A phase 1 study of crenigacestat (LY3039478), the Notch inhibitor, in Japanese patients with advanced solid tumors. Investig. New Drugs 2020, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Palomero, T.; Sulis, M.L.; E Cortina, M.; Real, P.J.; Barnes, K.; Ciofani, M.; Caparros, E.; Buteau, J.; Brown, K.J.; Perkins, S.L.; et al. Mutational loss of PTEN induces resistance to NOTCH 1 inhibition in T-cell leukemia. Nat. Med. 2007, 13, 1203–1210. [Google Scholar] [CrossRef] [PubMed]
- Sulis, M.L. PTEN: From pathology to biology. Trends Cell Biol. 2003, 13, 478–483. [Google Scholar] [CrossRef]
- Palomero, T.; Dominguez, M.; Ferrando, A.A. The role of the PTEN/AKT Pathway in NOTCH 1-induced leukemia. Cell Cycle 2008, 7, 965–970. [Google Scholar] [CrossRef]
- O’Neil, J.; Grim, J.; Strack, P.; Rao, S.; Tibbitts, D.; Winter, C.; Hardwick, J.; Welcker, M.; Meijerink, J.P.; Pieters, R.; et al. FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to γ-secretase inhibitors. J. Exp. Med. 2007, 204, 1813–1824. [Google Scholar] [CrossRef]
- Öberg, C.; Li, J.; Pauley, A.; Wolf, E.; Gurney, M.; Lendahl, U. The Notch Intracellular Domain Is Ubiquitinated and Negatively Regulated by the Mammalian Sel-10 Homolog. J. Biol. Chem. 2001, 276, 35847–35853. [Google Scholar] [CrossRef]
- Erbilgin, Y.; Sayitoglu, M.; Ng, O.H.; Dogru, O.; Akcay, A.; Tuysuz, G.; Celkan, T.; Aydogan, G.; Salcioglu, Z.; Timur, C.; et al. Prognostic Significance of NOTCH 1 and FBXW7 Mutations in Pediatric T-ALL. Dis. Markers 2010, 28, 353–360. [Google Scholar] [CrossRef]
- Kimura, S.; Seki, M.; Yoshida, K.; Shiraishi, Y.; Akiyama, M.; Koh, K.; Imamura, T.; Manabe, A.; Hayashi, Y.; Kobayashi, M.; et al. NOTCH 1 pathway activating mutations and clonal evolution in pediatric T-cell acute lymphoblastic leukemia. Cancer Sci. 2019, 110, 784–794. [Google Scholar] [CrossRef]
- Valliyammai, N.; Nancy, N.K.; Sagar, T.G.; Rajkumar, T. Study of NOTCH 1 and FBXW7 Mutations and Its Prognostic Significance in South Indian T-Cell Acute Lymphoblastic Leukemia. J. Pediatr. Hematol. 2018, 40, e1–e8. [Google Scholar] [CrossRef]
- Yonekura, S.; Itoh, M.; Shiratori, E.; Ohtaka, M.; Tohda, S. FOXP3 knockdown inhibits the proliferation and reduces NOTCH 1 expression of T cell acute lymphoblastic leukemia cells. BMC Res. Notes 2018, 11, 582. [Google Scholar] [CrossRef] [PubMed]
- Jin, Q.-H.; Martinez, C.A.; Arcipowski, K.M.; Zhu, Y.; Gutierrez-Diaz, B.T.; Wang, K.K.; Johnson, M.R.; Volk, A.; Wang, F.; Wu, J.; et al. USP7 Cooperates with NOTCH 1 to Drive the Oncogenic Transcriptional Program in T-Cell Leukemia. Clin. Cancer Res. 2018, 25, 222–239. [Google Scholar] [CrossRef] [PubMed]
- Dastur, A.; Choi, A.; Costa, C.; Yin, X.; Williams, A.; McClanaghan, J.D.; Greenberg, M.; Roderick, J.; Patel, N.U.; Boisvert, J.L.; et al. NOTCH 1 Represses MCL-1 Levels in GSI-resistant T-ALL, Making them Susceptible to ABT-263. Clin. Cancer Res. 2018, 25, 312–324. [Google Scholar] [CrossRef]
- Passaro, D.; Irigoyen, M.; Catherinet, C.; Gachet, S.; Jesus, C.D.C.D.; Lasgi, C.; Quang, C.T.; Ghysdael, J. CXCR4 Is Required for Leukemia-Initiating Cell Activity in T Cell Acute Lymphoblastic Leukemia. Cancer Cell 2015, 27, 769–779. [Google Scholar] [CrossRef] [PubMed]
- Pitt, L.A.; Tikhonova, A.N.; Hu, H.; Trimarchi, T.; King, B.; Gong, Y.; Sanchez-Martin, M.; Tsirigos, A.; Littman, D.R.; Ferrando, A.A.; et al. CXCL12-Producing Vascular Endothelial Niches Control Acute T Cell Leukemia Maintenance. Cancer Cell 2015, 27, 755–768. [Google Scholar] [CrossRef]
- De Bie, J.; Demeyer, S.; Alberti-Servera, L.; Geerdens, E.; Segers, H.; Broux, M.; De Keersmaecker, K.; Michaux, L.; Vandenberghe, P.; Voet, T.; et al. Single-cell sequencing reveals the origin and the order of mutation acquisition in T-cell acute lymphoblastic leukemia. Leukemia 2018, 32, 1358–1369. [Google Scholar] [CrossRef]
- Mori, N.; Fujii, M.; Ikeda, S.; Yamada, Y.; Tomonaga, M.; Ballard, D.W.; Yamamoto, N. Constitutive activation of NF-kappaB in primary adult T-cell leukemia cells. Blood 1999, 93, 2360–2368. [Google Scholar]
- Jost, P.J.; Ruland, J. Aberrant NF-κB signaling in lymphoma: Mechanisms, consequences, and therapeutic implications. Blood 2006, 109, 2700–2707. [Google Scholar] [CrossRef]
- Vilimas, T.; Mascarenhas, J.; Palomero, T.; Mandal, M.; Buonamici, S.; Meng, F.; Thompson, B.; Spaulding, C.; Macaroun, S.; Alegre, M.-L.; et al. Targeting the NF-κB signaling pathway in Notch 1-induced T-cell leukemia. Nat. Med. 2007, 13, 70–77. [Google Scholar] [CrossRef]
- Shin, H.M.; Minter, L.M.; Cho, O.H.; Gottipati, S.; Fauq, A.H.; E Golde, T.; E Sonenshein, G.; Osborne, B.A. Notch 1 augments NF-κB activity by facilitating its nuclear retention. EMBO J. 2005, 25, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Zong, W.-X.; Edelstein, L.C.; Chen, C.; Bash, J.; Gélinas, C. The prosurvival Bcl-2 homolog Bfl-1/A1 is a direct transcriptional target of NF-kappa B that blocks TNFalpha -induced apoptosis. Genes Dev. 1999, 13, 382–387. [Google Scholar] [CrossRef] [PubMed]
- Mandal, M.; Borowski, C.; Palomero, T.; Ferrando, A.A.; Oberdoerffer, P.; Meng, F.; Ruiz-Vela, A.; Ciofani, M.; Zuniga-Pflucker, J.-C.; Screpanti, I.; et al. The BCL2A1 gene as a pre–T cell receptor–induced regulator of thymocyte survival. J. Exp. Med. 2005, 201, 603–614. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Nie, L.; Zhang, L.; Li, Y. The Notch pathway promotes NF-κB activation through Asb2 in T cell acute lymphoblastic leukemia cells. Cell. Mol. Biol. Lett. 2018, 23, 37. [Google Scholar] [CrossRef] [PubMed]
- Rosati, E.; Sabatini, R.; Rampino, G.; Tabilio, A.; Di Ianni, M.; Fettucciari, K.; Bartoli, A.; Coaccioli, S.; Screpanti, I.; Marconi, P. Constitutively activated Notch signaling is involved in survival and apoptosis resistance of B-CLL cells. Blood 2009, 113, 856–865. [Google Scholar] [CrossRef] [PubMed]
- Puente, X.S.; Pinyol, M.; Quesada, V.; Conde, L.; Ordóñez, G.R.; Villamor, N.; Escaramis, G.; Jares, P.; Beà, S.; González-Díaz, M.; et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature 2011, 475, 101–105. [Google Scholar] [CrossRef] [PubMed]
- Zou, Y.; Fan, L.; Xia, Y.; Miao, Y.; Wu, W.; Cao, L.; Wu, J.; Zhu, H.; Qiao, C.; Wang, L.; et al. NOTCH 1 mutation and its prognostic significance in Chinese chronic lymphocytic leukemia: A retrospective study of 317 cases. Cancer Med. 2018, 7, 1689–1696. [Google Scholar] [CrossRef]
- Willander, K.; Dutta, R.K.; Ungerbäck, J.; Gunnarsson, R.; Juliusson, G.; Fredrikson, M.; Linderholm, M.; Söderkvist, P. NOTCH 1 mutations influence survival in chronic lymphocytic leukemia patients. BMC Cancer 2013, 13, 274. [Google Scholar] [CrossRef]
- Del Giudice, I.; Rossi, D.; Chiaretti, S.; Marinelli, M.; Tavolaro, S.; Gabrielli, S.; Laurenti, L.; Marasca, R.; Rasi, S.; Fangazio, M.; et al. NOTCH 1 mutations in +12 chronic lymphocytic leukemia (CLL) confer an unfavorable prognosis, induce a distinctive transcriptional profiling and refine the intermediate prognosis of +12 CLL. Haematologica 2011, 97, 437–441. [Google Scholar] [CrossRef]
- Rossi, D.; Rasi, S.; Fabbri, G.; Spina, V.; Fangazio, M.; Forconi, F.; Marasca, R.; Laurenti, L.; Bruscaggin, A.; Cerri, M.; et al. Mutations of NOTCH 1 are an independent predictor of survival in chronic lymphocytic leukemia. Blood 2012, 119, 521–529. [Google Scholar] [CrossRef]
- Close, V.; Close, W.; Kugler, S.J.; Reichenzeller, M.; Yosifov, D.Y.; Bloehdorn, J.; Pan, L.; Tausch, E.; Westhoff, M.-A.; Döhner, H.; et al. FBXW7 mutations reduce binding of NOTCH 1, leading to cleaved NOTCH 1 accumulation and target gene activation in CLL. Blood 2019, 133, 830–839. [Google Scholar] [CrossRef] [PubMed]
- Nefedova, Y.; Cheng, P.; Alsina, M.; Dalton, W.S.; Gabrilovich, D.I. Involvement of Notch -1 signaling in bone marrow stroma-mediated de novo drug resistance of myeloma and other malignant lymphoid cell lines. Blood 2004, 103, 3503–3510. [Google Scholar] [CrossRef] [PubMed]
- López-Guerra, M.; Xargay-Torrent, S.; Fuentes, P.; Roldán, J.; González-Farré, B.; Rosich, L.; Silkenstedt, E.; García-León, M.J.; Lee-Vergés, E.; Giménez, N.; et al. Specific NOTCH 1 antibody targets DLL4-induced proliferation, migration, and angiogenesis in NOTCH 1-mutated CLL cells. Oncogene 2020, 39, 1185–1197. [Google Scholar] [CrossRef] [PubMed]
- Villamor, N.; Conde, L.; Martínez-Trillos, A.; Cazorla, M.; Navarro, A.; Bea, S.; López, C.; Colomer, D.; Pinyol, M.; Aymerich, M.; et al. NOTCH 1 mutations identify a genetic subgroup of chronic lymphocytic leukemia patients with high risk of transformation and poor outcome. Leukemia 2012, 27, 1100–1106. [Google Scholar] [CrossRef] [PubMed]
- Arima, H.; Nishikori, M.; Otsuka, Y.; Kishimoto, W.; Izumi, K.; Yasuda, K.; Yoshimoto, T.; Takaori-Kondo, A. B cells with aberrant activation of Notch 1 signaling promote Treg and Th2 cell–dominant T-cell responses via IL-33. Blood Adv. 2018, 2, 2282–2295. [Google Scholar] [CrossRef]
- Kridel, R.; Meissner, B.; Rogic, S.; Boyle, M.; Telenius, A.; Woolcock, B.; Gunawardana, J.; Jenkins, C.; Cochrane, C.; Ben-Neriah, S.; et al. Whole transcriptome sequencing reveals recurrent NOTCH 1 mutations in mantle cell lymphoma. Blood 2012, 119, 1963–1971. [Google Scholar] [CrossRef]
- Bea, S.; Mas, R.M.V.; Navarro, A.; Salaverria, I.; Martín-Garcia, D.; Jares, P.; Giné, E.; Pinyol, M.; Royo, C.; Nadeu, F.; et al. Landscape of somatic mutations and clonal evolution in mantle cell lymphoma. Proc. Natl. Acad. Sci. USA 2013, 110, 18250–18255. [Google Scholar] [CrossRef]
- Silkenstedt, E.; Arenas, F.; Colom-Sanmartí, B.; Xargay-Torrent, S.; Higashi, M.; Giró, A.; Rodriguez, V.; Fuentes, P.; Aulitzky, W.E.; Van Der Kuip, H.; et al. Notch 1 signaling in NOTCH 1-mutated mantle cell lymphoma depends on Delta-Like ligand 4 and is a potential target for specific antibody therapy. J. Exp. Clin. Cancer Res. 2019, 38, 1–15. [Google Scholar] [CrossRef]
- Jain, P.; Zhang, S.; Kanagal-Shamanna, R.; Ok, C.Y.; Nomie, K.; Gonzalez, G.N.; Gonzalez-Pagan, O.; Hill, H.A.; Lee, H.J.; Fayad, L.; et al. Genomic profiles and clinical outcomes of de novo blastoid/pleomorphic MCL are distinct from those of transformed MCL. Blood Adv. 2020, 4, 1038–1050. [Google Scholar] [CrossRef]
- Hansen, M.C.; Cédile, O.; Blum, M.K.; Hansen, S.V.; Ebbesen, L.H.; Bentzen, H.H.N.; Thomassen, M.; Kruse, T.A.; Kavan, S.; Kjeldsen, E.; et al. Molecular characterization of sorted malignant B cells from patients clinically identified with mantle cell lymphoma. Exp. Hematol. 2020, 84, 7–18.e12. [Google Scholar] [CrossRef]
- LaRose, H.; Prokoph, N.; Matthews, J.D.; Schlederer, M.; Högler, S.; Alsulami, A.F.; Ducray, S.P.; Nuglozeh, E.; Fazaludeen, F.M.; Elmouna, A.; et al. Whole Exome Sequencing reveals NOTCH 1 mutations in anaplastic large cell lymphoma and points to Notch both as a key pathway and a potential therapeutic target. Haematologica 2020. [Google Scholar] [CrossRef] [PubMed]
- Rossi, D.; Trifonov, V.; Fangazio, M.; Bruscaggin, A.; Rasi, S.; Spina, V.; Monti, S.; Vaisitti, T.; Arruga, F.; Famà, R.; et al. The coding genome of splenic marginal zone lymphoma: Activation of NOTCH 2 and other pathways regulating marginal zone development. J. Exp. Med. 2012, 209, 1537–1551. [Google Scholar] [CrossRef] [PubMed]
- Arcaini, L.; Rossi, D.; Lucioni, M.; Nicola, M.; Bruscaggin, A.; Fiaccadori, V.; Riboni, R.; Ramponi, A.; Ferretti, V.V.; Cresta, S.; et al. The NOTCH pathway is recurrently mutated in diffuse large B-cell lymphoma associated with hepatitis C virus infection. Haematologica 2014, 100, 246–252. [Google Scholar] [CrossRef] [PubMed]
- Mensah, A.A.; Rinaldi, A.; Ponzoni, M.; Canzonieri, V.; Uccella, S.; Rossi, D.; Bhagat, G.; Gaidano, G.; Zucca, E.; Bertoni, F. Absence of NOTCH 1 gene mutations in MALT lymphomas. Br. J. Haematol. 2011, 157, 382–384. [Google Scholar] [CrossRef]
- Johansson, P.; Klein-Hitpass, L.; Grabellus, F.; Arnold, G.; Klapper, W.; Pförtner, R.; Dührsen, U.; Eckstein, A.; Dürig, J.; Küppers, R. Recurrent mutations in NF-κB pathway components, KMT2D, and NOTCH 1/2 in ocular adnexal MALT-type marginal zone lymphomas. Oncotarget 2016, 7, 62627–62639. [Google Scholar] [CrossRef]
- Huh, S.J.; Oh, S.Y.; Lee, S.; Lee, J.H.; Kim, S.H.; Pak, M.K.; Kim, H.-J. Mutational analysis of extranodal marginal zone lymphoma using next generation sequencing. Oncol. Lett. 2020, 20, 1. [Google Scholar] [CrossRef]
- Karube, K.; Martínez, D.; Royo, C.; Navarro, A.; Pinyol, M.; Cazorla, M.; Castillo, P.; Valera, A.; Carrió, A.; Costa, D.; et al. Recurrent mutations ofNOTCH genes in follicular lymphoma identify a distinctive subset of tumours. J. Pathol. 2014, 234, 423–430. [Google Scholar] [CrossRef]
- Lee, S.-Y.; Kumano, K.; Nakazaki, K.; Sanada, M.; Matsumoto, A.; Yamamoto, G.; Nannya, Y.; Suzuki, R.; Ota, S.; Ota, Y.; et al. Gain-of-function mutations and copy number increases of Notch 2 in diffuse large B-cell lymphoma. Cancer Sci. 2009, 100, 920–926. [Google Scholar] [CrossRef]
- Trøen, G.; Wlodarska, I.; Warsame, A.; Llodrà, S.H.; De Wolf-Peeters, C.; Delabie, J. NOTCH 2 mutations in marginal zone lymphoma. Haematologica 2008, 93, 1107–1109. [Google Scholar] [CrossRef]
- Kiel, M.J.; Velusamy, T.; Betz, B.L.; Zhao, L.; Weigelin, H.G.; Chiang, M.Y.; Huebner-Chan, D.R.; Bailey, N.G.; Yang, D.T.; Bhagat, G.; et al. Whole-genome sequencing identifies recurrent somatic NOTCH 2 mutations in splenic marginal zone lymphoma. J. Exp. Med. 2012, 209, 1553–1565. [Google Scholar] [CrossRef]
- Kamga, P.T.; Bassi, G.; Cassaro, A.; Midolo, M.; Di Trapani, M.; Gatti, A.; Carusone, R.; Resci, F.; Perbellini, O.; Gottardi, M.; et al. Notch signalling drives bone marrow stromal cell-mediated chemoresistance in acute myeloid leukemia. Oncotarget 2016, 7, 21713–21727. [Google Scholar] [CrossRef] [PubMed]
- Mangolini, M.; Götte, F.; Moore, A.; Ammon, T.; Oelsner, M.; Lutzny-Geier, G.; Klein-Hitpass, L.; Williamson, J.C.; Lehner, P.J.; Dürig, J.; et al. Notch 2 controls non-autonomous Wnt-signalling in chronic lymphocytic leukaemia. Nat. Commun. 2018, 9, 1–17. [Google Scholar] [CrossRef] [PubMed]
- DiRaimondo, F.; Albitar, M.; Huh, Y.; O’Brien, S.; Montillo, M.; Tedeschi, A.; Kantarjian, H.; Lerner, S.; Giustolisi, R.; Keating, M. The clinical and diagnostic relevance of CD23 expression in the chronic lymphoproliferative disease. Cancer 2002, 94, 1721–1730. [Google Scholar] [CrossRef] [PubMed]
- Hubmann, R.; Schwarzmeier, J.D.; Shehata, M.; Hilgarth, M.; Duechler, M.; Dettke, M.; Berger, R. Notch 2 is involved in the overexpression of CD23 in B-cell chronic lymphocytic leukemia. Blood 2002, 99, 3742–3747. [Google Scholar] [CrossRef]
- Hubmann, R.; Düchler, M.; Schnabl, S.; Hilgarth, M.; Demirtas, D.; Mitteregger, D.; Hölbl, A.; Vanura, K.; Le, T.; Look, T.; et al. NOTCH 2 links protein kinase C delta to the expression of CD23 in chronic lymphocytic leukaemia (CLL) cells. Br. J. Haematol. 2010, 148, 868–878. [Google Scholar] [CrossRef]
- Kamdje, A.H.N.; Mosna, F.; Bifari, F.; Lisi, V.; Bassi, G.; Malpeli, G.; Ricciardi, M.; Perbellini, O.; Scupoli, M.T.; Pizzolo, G.; et al. Notch -3 and Notch -4 signaling rescue from apoptosis human B-ALL cells in contact with human bone marrow–derived mesenchymal stromal cells. Blood 2011, 118, 380–389. [Google Scholar] [CrossRef]
- Bernasconi-Elias, P.; Hu, T.; Jenkins, D.; Firestone, B.; Gans, S.; Kurth, E.; Capodieci, P.; Deplazes-Lauber, J.; Petropoulos, K.; Thiel, P.; et al. Characterization of activating mutations of NOTCH 3 in T cell acute lymphoblastic leukemia and anti-leukemic activity of NOTCH 3 inhibitory antibodies. Oncogene 2016, 35, 6077–6086. [Google Scholar] [CrossRef]
- Ferrandino, F.; Bernardini, G.; Tsaouli, G.; Grazioli, P.; Campese, A.F.; Noce, C.; Ciuffetta, A.; Vacca, A.; Besharat, Z.M.; Bellavia, D.; et al. Intrathymic Notch 3 and CXCR4 combinatorial interplay facilitates T-cell leukemia propagation. Oncogene 2018, 37, 6285–6298. [Google Scholar] [CrossRef]
- Jespersen, D.S.; Schönherz, A.A.; Due, H.; Bøgsted, M.; Sondergaard, T.E.; Dybkaer, K. Expression of NOTCH 3 exon 16 differentiates Diffuse Large B-cell Lymphoma into molecular subtypes and is associated with prognosis. Sci. Rep. 2019, 9, 1–11. [Google Scholar] [CrossRef]
- Kamdje, A.H.N.; Bassi, G.S.; Pacelli, L.; Malpeli, G.; Amati, E.; Nichele, I.; Pizzolo, G.; Krampera, M. Role of stromal cell-mediated Notch signaling in CLL resistance to chemotherapy. Blood Cancer J. 2012, 2, e73. [Google Scholar] [CrossRef]
- Gragnani, L.; Fognani, E.; De Re, V.; Libra, M.; Garozzo, A.; Caini, P.; Cerretelli, G.; Giovannelli, A.; Lorini, S.; Monti, M.; et al. Notch 4 and mhc class II polymorphisms are associated with hcv-related benign and malignant lymphoproliferative diseases. Oncotarget 2017, 8, 71528–71535. [Google Scholar] [CrossRef] [PubMed]
- Ye, Q.; Shieh, J.-H.; Morrone, G.; Moore, M.A. Expression of constitutively active Notch 4 (Int-3) modulates myeloid proliferation and differentiation and promotes expansion of hematopoietic progenitors. Leukemia 2004, 18, 777–787. [Google Scholar] [CrossRef] [PubMed]
- Artavanis-Tsakonas, S.; Simpson, P. Choosing a cell fate: A view from the Notch locus. Trends Genet. 1991, 7, 403–408. [Google Scholar] [CrossRef]
- Wang, J.; Dong, M.; Xu, Z.; Song, X.; Zhang, S.; Qiao, Y.; Che, L.; Gordan, J.; Hu, K.; Liu, Y.; et al. Notch 2 controls hepatocyte-derived cholangiocarcinoma formation in mice. Oncogene 2018, 37, 3229–3242. [Google Scholar] [CrossRef]
- Saito, T.; Chiba, S.; Ichikawa, M.; Kunisato, A.; Asai, T.; Shimizu, K.; Yamaguchi, T.; Yamamoto, G.; Seo, S.; Kumano, K.; et al. Notch 2 Is Preferentially Expressed in Mature B Cells and Indispensable for Marginal Zone B Lineage Development. Immunity 2003, 18, 675–685. [Google Scholar] [CrossRef]
- Lewis, K.L.; Caton, M.L.; Bogunovic, M.; Greter, M.; Grajkowska, L.T.; Ng, D.; Klinakis, A.; Charo, I.F.; Jung, S.; Gommerman, J.L.; et al. Notch 2 Receptor Signaling Controls Functional Differentiation of Dendritic Cells in the Spleen and Intestine. Immunity 2011, 35, 780–791. [Google Scholar] [CrossRef]
- Martin, F.; Kearney, J.F. Marginal-zone B cells. Nat. Rev. Immunol. 2002, 2, 323–335. [Google Scholar] [CrossRef]
- Loder, B.F.; Mutschler, B.; Ray, R.J.; Paige, C.J.; Sideras, P.; Torres, R.; Lamers, M.C.; Carsetti, R. B Cell Development in the Spleen Takes Place in Discrete Steps and Is Determined by the Quality of B Cell Receptor–Derived Signals. J. Exp. Med. 1999, 190, 75–90. [Google Scholar] [CrossRef]
- Briseño, C.G.; Satpathy, A.T.; Iv, J.T.D.; Ferris, S.T.; Durai, V.; Bagadia, P.; O’Connor, K.W.; Theisen, D.J.; Murphy, T.L.; Murphy, K.M. Notch 2-dependent DC2s mediate splenic germinal center responses. Proc. Natl. Acad. Sci. USA 2018, 115, 10726–10731. [Google Scholar] [CrossRef]
- Amsen, D.; Blander, J.; Lee, G.R.; Tanigaki, K.; Honjo, T.; Flavell, R.A. Instruction of Distinct CD4 T Helper Cell Fates by Different Notch Ligands on Antigen-Presenting Cells. Cell 2004, 117, 515–526. [Google Scholar] [CrossRef]
- Sun, J.; Krawczyk, C.J.; Pearce, E.J. Suppression of Th2 cell development by Notch ligands Delta 1 and Delta 4. J. Immunol. 2008, 180, 1655–1661. [Google Scholar] [CrossRef]
- Maekawa, Y.; Minato, Y.; Ishifune, C.; Kurihara, T.; Kitamura, A.; Kojima, H.; Yagita, H.; Sakata-Yanagimoto, M.; Saito, T.; Taniuchi, I.; et al. Notch 2 integrates signaling by the transcription factors RBP-J and CREB1 to promote T cell cytotoxicity. Nat. Immunol. 2008, 9, 1140–1147. [Google Scholar] [CrossRef]
- Taylor, A.K.; Klisak, I.; Mohandas, T.; Sparkes, R.S.; Li, C.; Gaynor, R.; Lusis, A.J. Assignment of the human gene for CREB1 to chromosome 2q32.3–q34. Genomics 1990, 7, 416–421. [Google Scholar] [CrossRef]
- Sugimoto, K.; Maekawa, Y.; Kitamura, A.; Nishida, J.; Koyanagi, A.; Yagita, H.; Kojima, H.; Chiba, S.; Shimada, M.; Yasutomo, K. Notch 2 Signaling Is Required for Potent Antitumor Immunity In Vivo. J. Immunol. 2010, 184, 4673–4678. [Google Scholar] [CrossRef] [PubMed]
- Varnum-Finney, B.; Halasz, L.M.; Sun, M.; Gridley, T.; Radtke, F.; Bernstein, I.D. Notch 2 governs the rate of generation of mouse long- and short-term repopulating stem cells. J. Clin. Investig. 2011, 121, 1207–1216. [Google Scholar] [CrossRef] [PubMed]
- Witt, C.M.; Hurez, V.; Swindle, C.S.; Hamada, Y.; Klug, C.A. Activated Notch 2 Potentiates CD8 Lineage Maturation and Promotes the Selective Development of B1 B Cells. Mol. Cell. Biol. 2003, 23, 8637–8650. [Google Scholar] [CrossRef] [PubMed]
- Rechsteiner, M.; Rogers, S.W. PEST sequences and regulation by proteolysis. Trends Biochem. Sci. 1996, 21, 267–271. [Google Scholar] [CrossRef]
- Kralovics, R.; Passamonti, F.; Buser, A.S.; Teo, S.-S.; Tiedt, R.; Passweg, J.R.; Tichelli, A.; Cazzola, M.; Skoda, R.C. A Gain-of-Function Mutation ofJAK2in Myeloproliferative Disorders. New Engl. J. Med. 2005, 352, 1779–1790. [Google Scholar] [CrossRef]
- Arcaini, L.; Rossi, D.; Paulli, M. Splenic marginal zone lymphoma: From genetics to management. Blood 2016, 127, 2072–2081. [Google Scholar] [CrossRef]
- Kumar, C.C. Genetic Abnormalities and Challenges in the Treatment of Acute Myeloid Leukemia. Genes Cancer 2011, 2, 95–107. [Google Scholar] [CrossRef]
- Hubmann, R.; Hilgarth, M.; Schnabl, S.; Ponath, E.; Reiter, M.; Demirtas, D.; Sieghart, W.; Valent, P.; Zielinski, C.; Jäger, U.; et al. Gliotoxin is a potent NOTCH 2 transactivation inhibitor and efficiently induces apoptosis in chronic lymphocytic leukaemia (CLL) cells. Br. J. Haematol. 2013, 160, 618–629. [Google Scholar] [CrossRef] [PubMed]
- Hubmann, R.; Schnabl, S.; Araghi, M.; Schmidl, C.; Rendeiro, A.F.; Hilgarth, M.; Demirtas, D.; Ali, F.; Staber, P.B.; Valent, P.; et al. Targeting Nuclear NOTCH 2 by Gliotoxin Recovers a Tumor-Suppressor NOTCH 3 Activity in CLL. Cells 2020, 9, 1484. [Google Scholar] [CrossRef] [PubMed]
- Inder, S.; O’Rourke, S.; McDermott, N.; Manecksha, R.P.; Finn, S.; Lynch, T.; Marignol, L. The Notch -3 receptor: A molecular switch to tumorigenesis? Cancer Treat. Rev. 2017, 60, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Leontovich, A.A.; Jalalirad, M.; Salisbury, J.L.; Mills, L.; Haddox, C.; Schroeder, M.A.; Tuma, A.; Guicciardi, M.; Zammataro, L.; Gambino, M.W.; et al. NOTCH 3 expression is linked to breast cancer seeding and distant metastasis. Breast Cancer Res. 2018, 20, 105. [Google Scholar] [CrossRef] [PubMed]
- O’Rourke, C.J.; Matter, M.S.; Nepal, C.; Caetano-Oliveira, R.; Ton, P.T.; Factor, V.M.; Andersen, J.B. Identification of a Pan-Gamma-Secretase Inhibitor Response Signature for Notch -Driven Cholangiocarcinoma. Hepatology 2020, 71, 196–213. [Google Scholar] [CrossRef]
- Bs, Y.R.S.; Yu, X.-M.; Lloyd, R.V.; Leverson, G.; Aburjania, Z.; Jang, S.; Jaskula-Sztul, R.; Chen, H. Notch 3 expression correlates with thyroid cancer differentiation, induces apoptosis, and predicts disease prognosis. Cancer 2017, 123, 769–782. [Google Scholar] [CrossRef]
- Mizugaki, H.; Sakakibarakonishi, J.; Ikezawa, Y.; Kikuchi, J.; Kikuchi, E.; Oizumi, S.; Dang, T.P.; Nishimura, M. γ-Secretase inhibitor enhances antitumour effect of radiation in Notch -expressing lung cancer. Br. J. Cancer 2012, 106, 1953–1959. [Google Scholar] [CrossRef]
- Kamga, P.T.; Collo, G.D.; Midolo, M.; Adamo, A.; Delfino, P.; Mercuri, A.; Cesaro, S.; Mimiola, E.; Bonifacio, M.; Andreini, A.; et al. Inhibition of Notch Signaling Enhances Chemosensitivity in B-cell Precursor Acute Lymphoblastic Leukemia. Cancer Res. 2019, 79, 639–649. [Google Scholar] [CrossRef]
- Aburjania, Z.; Jang, S.; Whitt, J.; Jaskula-Stzul, R.; Chen, H.; Rose, J.B. The Role ofNotch 3in Cancer. Oncologist 2018, 23, 900–911. [Google Scholar] [CrossRef]
- Felli, M.P.; Maroder, M.; Mitsiadis, T.A.; Campese, A.F.; Bellavia, D.; Vacca, A.; Mann, R.S.; Frati, L.; Lendahl, U.; Gulino, A.; et al. Expression pattern of Notch 1, 2 and 3 and Jagged1 and 2 in lymphoid and stromal thymus components: Distinct ligand–receptor interactions in intrathymic T cell development. Int. Immunol. 1999, 11, 1017–1025. [Google Scholar] [CrossRef]
- Bellavia, D.; Campese, A.F.; Checquolo, S.; Balestri, A.; Biondi, A.; Cazzaniga, G.; Lendahl, U.; Fehling, H.J.; Hayday, A.C.; Frati, L.; et al. Combined expression of pTα and Notch 3 in T cell leukemia identifies the requirement of preTCR for leukemogenesis. Proc. Natl. Acad. Sci. USA 2002, 99, 3788–3793. [Google Scholar] [CrossRef] [PubMed]
- Koyanagi, A.; Sekine, C.; Yagita, H. Expression of Notch receptors and ligands on immature and mature T cells. Biochem. Biophys. Res. Commun. 2012, 418, 799–805. [Google Scholar] [CrossRef] [PubMed]
- Suliman, S.; Tan, J.; Xu, K.; Kousis, P.C.; Kowalski, P.E.; Chang, G.; Egan, S.E.; Guidos, C. Notch 3 Is Dispensable for Thymocyte β-Selection and Notch 1-Induced T Cell Leukemogenesis. PLoS ONE 2011, 6, e24937. [Google Scholar] [CrossRef] [PubMed]
- Giuli, M.V.; Diluvio, G.; Giuliani, E.; Franciosa, G.; Di Magno, L.; Pignataro, M.G.; Tottone, L.; Nicoletti, C.; Besharat, Z.M.; Peruzzi, G.; et al. Notch 3 contributes to T-cell leukemia growth via regulation of the unfolded protein response. Oncogenesis 2020, 9, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Pelullo, M.; Quaranta, R.; Talora, C.; Checquolo, S.; Cialfi, S.; Felli, M.P.; Kronnie, G.T.; Borga, C.; Besharat, Z.M.; Palermo, R.; et al. Notch 3/Jagged1 Circuitry Reinforces Notch Signaling and Sustains T-ALL. Neoplasia 2014, 16, 1007–1017. [Google Scholar] [CrossRef]
- Choi, S.H.; Severson, E.; Pear, W.S.; Liu, X.S.; Aster, J.C.; Blacklow, S.C. The common oncogenomic program of NOTCH 1 and NOTCH 3 signaling in T-cell acute lymphoblastic leukemia. PLoS ONE 2017, 12, e0185762. [Google Scholar] [CrossRef]
- Tottone, L.; Zhdanovskaya, N.; Carmona Pestaña, Á.; Zampieri, M.; Simeoni, F.; Lazzari, S.; Ruocco, V.; Pelullo, M.; Caiafa, P.; Felli, M.P.; et al. Histone Modifications Drive Aberrant Notch 3 Expression/Activity and Growth in T-ALL. Front. Oncol. 2019, 9, 198. [Google Scholar] [CrossRef]
- Pinazza, M.; Ghisi, M.; Minuzzo, S.; Agnusdei, V.; Fossati, G.; Ciminale, V.; Pezzè, L.; Ciribilli, Y.; Pilotto, G.; Venturoli, C.; et al. Histone deacetylase 6 controls Notch 3 trafficking and degradation in T-cell acute lymphoblastic leukemia cells. Oncogene 2018, 37, 3839–3851. [Google Scholar] [CrossRef]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef]
- Vacca, A.; Ribatti, D.; Roncali, L.; Ranieri, G.; Di Serio, F.; Silvestris, F.; Dammacco, F. Bone marrow angiogenesis and progression in multiple myeloma. Br. J. Haematol. 1994, 87, 503–508. [Google Scholar] [CrossRef]
- Ribatti, D.; Nico, B.; Ranieri, G.; Specchia, G.; Vacca, A. The Role of Angiogenesis in Human Non-Hodgkin Lymphomas. Neoplasia 2013, 15, 231–238. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Hendzel, M.; Allalunis-Turner, J. Notch signaling as a therapeutic target for breast cancer treatment? Breast Cancer Res. 2011, 13, 210. [Google Scholar] [CrossRef] [PubMed]
- James, A.; Szot, J.; Iyer, K.; Major, J.; Pursglove, S.; Chapman, G.; Dunwoodie, S.L. Notch 4 reveals a novel mechanism regulating Notch signal transduction. Biochim. et Biophys. Acta (BBA) Bioenerg. 2014, 1843, 1272–1284. [Google Scholar] [CrossRef] [PubMed]
- Karanu, F.N.; Yuefei, L.; Gallacher, L.; Sakano, S.; Bhatia, M. Differential response of primitive human CD34− and CD34+ hematopoietic cells to the Notch ligand Jagged-1. Leukemia 2003, 17, 1366–1374. [Google Scholar] [CrossRef] [PubMed]
- Uyttendaele, H.; Marazzi, G.; Wu, G.; Yan, Q.; Sassoon, D.; Kitajewski, J. Notch 4/int-3, a mammary proto-oncogene, is an endothelial cell-specific mammalian Notch gene. Development 1996, 122, 2251–2259. [Google Scholar] [PubMed]
- Vercauteren, S.; Sutherland, H.J. Constitutively active Notch 4 promotes early human hematopoietic progenitor cell maintenance while inhibiting differentiation and causes lymphoid abnormalities in vivo. Blood 2004, 104, 2315–2322. [Google Scholar] [CrossRef]
- Zignego, A.L.; Wojcik, G.L.; Cacoub, P.; Visentini, M.; Casato, M.; Mangia, A.; Latanich, R.; Charles, E.D.; Gragnani, L.; Terrier, B.; et al. Genome-wide association study of hepatitis C virus- and cryoglobulin-related vasculitis. Genes Immun. 2014, 15, 500–505. [Google Scholar] [CrossRef]
- Chen, D.-P.; Chang, S.-W.; Wang, P.-N.; Hus, F.-P.; Tseng, C.-P. Association between single nucleotide polymorphisms within HLA region and disease relapse for patients with hematopoietic stem cell transplantation. Sci. Rep. 2019, 9, 1–8. [Google Scholar] [CrossRef]
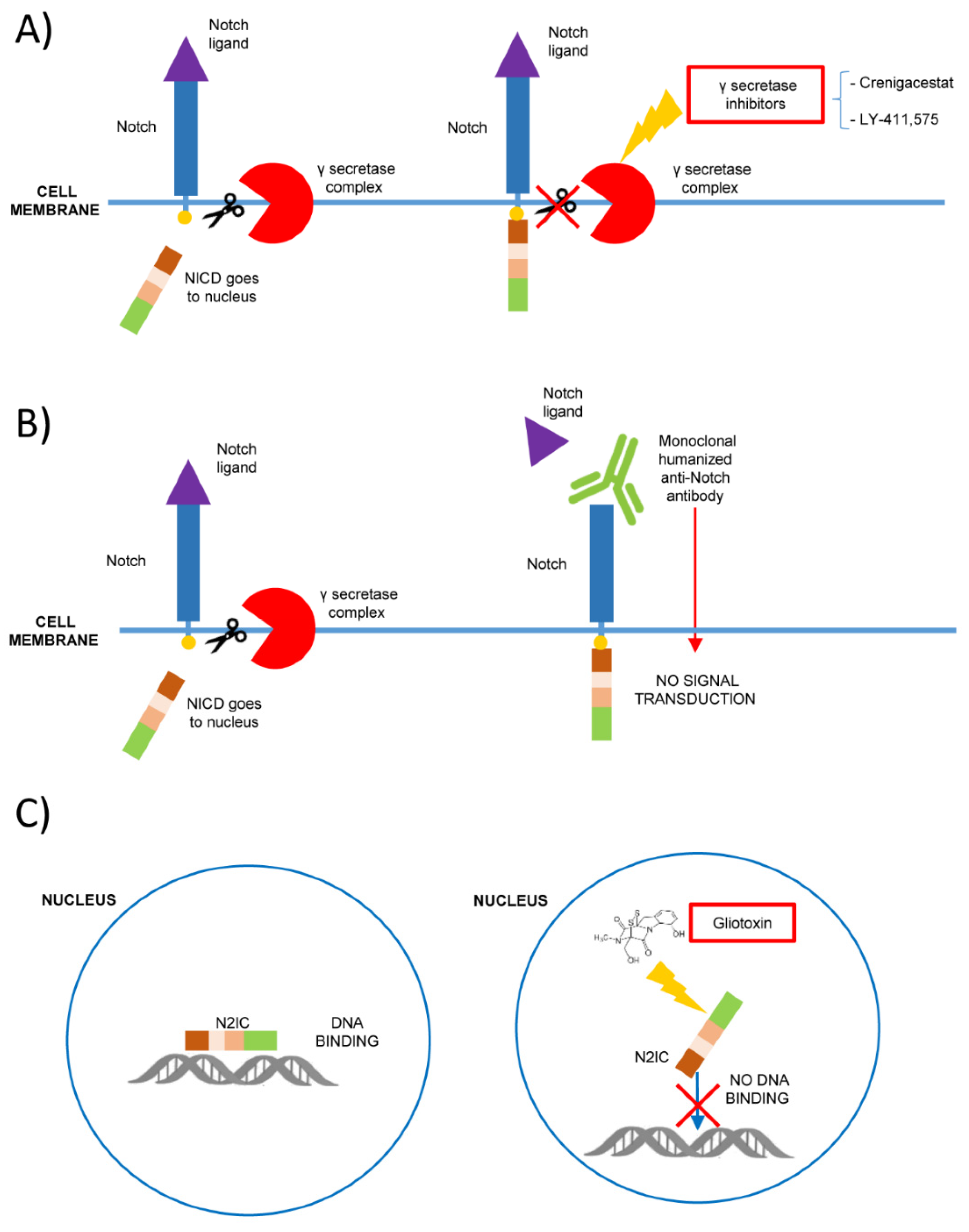
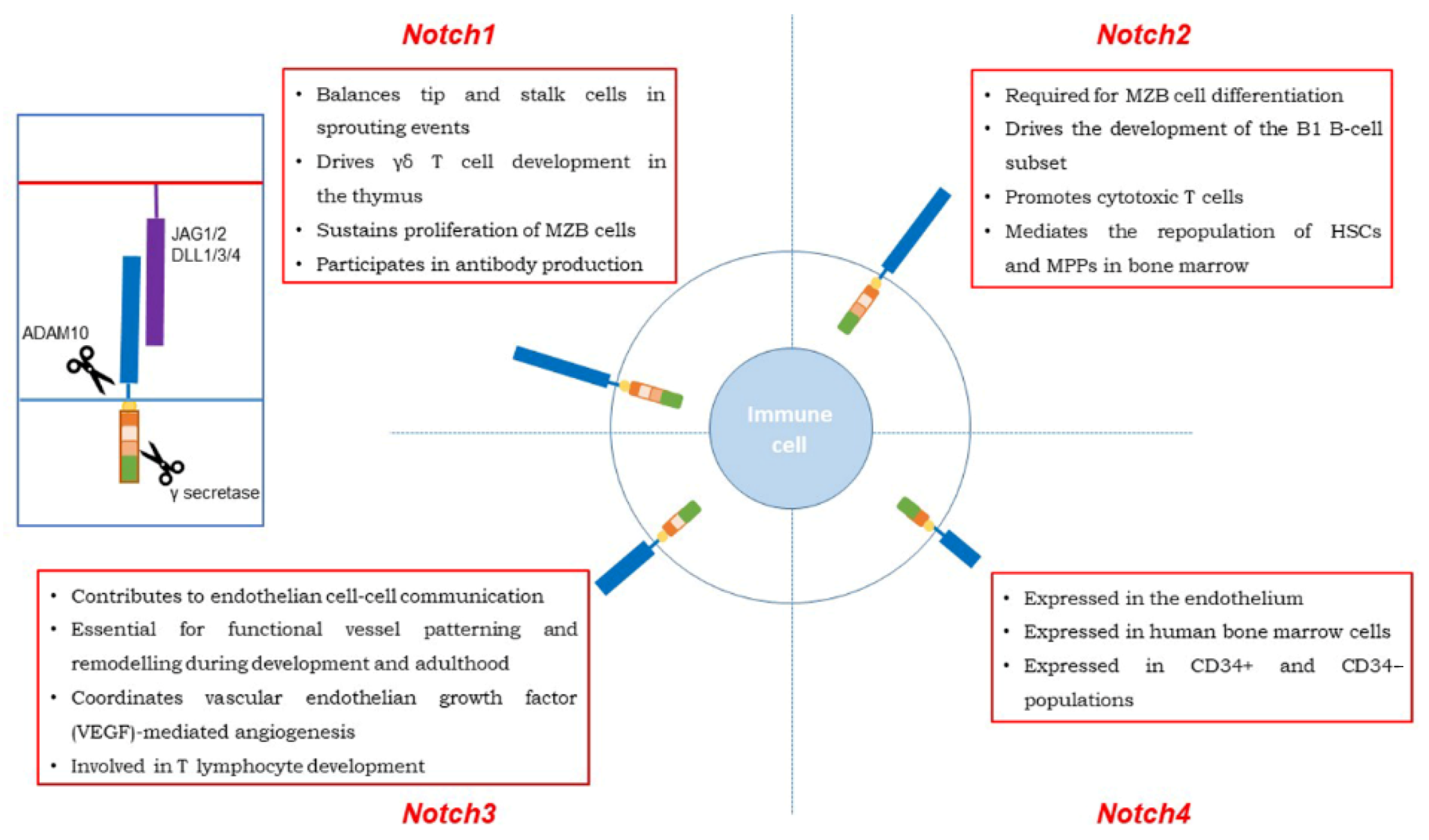
| Notch Receptor | Tumor Type | Type of Alterations | Ref |
|---|---|---|---|
| NOTCH 1 | RS | 30% of cases carry Notch 1 genetic alteration with overexpression and MYC activation | [13] |
| T-ALL | T(7;9) is responsible for abnormal Notch 1 signaling | [42] | |
| Activating mutations in HD and PEST domain | [43] | ||
| Presenilin-1, a γ-secretase, is overexpressed in T-ALL cells | [49] | ||
| FBXW7 mutations avoid NICD degradation sustaining Notch 1 signaling | [57] | ||
| NICD binds to NF-kB promoters increasing protein expression | [71] | ||
| B-CLL | Constitutively expressed, confers resistance to apoptosis | [75] | |
| CLL | Somatic mutations generate stable and activated forms of the protein and are associated with advanced tumor stage | [76] | |
| Mutation in FBXW7 reduce degradation of Notch intracellular domain | [81] | ||
| MM | Notch 1-JAG1 interaction resulted in apoptosis protection | [82] | |
| DLL4 interaction in CLL with activating Notch 1 mutations promotes proliferation, migration and angiogenesis | [83] | ||
| MCL | Notch 1 mutations associated with severe prognosis | [86,87,89,90] | |
| Notch 1 pathway overexpression by DLL4 interaction in patients with Notch 1 mutation | [88] | ||
| ALCL | Mutation in extracellular domain enhance cell proliferation | [91] | |
| SMZL | Notch 1 mutations were present in 5% of cases | [92] | |
| DLBCL | Genetic Notch 1 alteration in small subset of HCV positive patients are associated with a worse prognosis | [93] | |
| MALT | Notch 1 mutations were found in some cases but further analyses are needed to explain biological meaning | [95,96] | |
| FL | Mutations are associated with splenic involvement | [97] | |
| NOTCH 2 | DLBCL | Mutated PEST domain is associated with severe prognosis | [93,98] |
| MALT | Notch 2 mutations were found in some cases but further analysis is needed to explain biological meaning | [95] | |
| FL | Mutations are associated with splenic involvement | [97] | |
| MZB | Activating mutations are associated with poor outcomes | [99,100] | |
| AML | Overexpression promotes cell survival and chemo-resistance | [101] | |
| CLL | Notch 2 boosts Wnt signaling in stromal cells | [102] | |
| B-CLL | High concentration of soluble CD23 by Notch 2 signaling promotes disease progression | [103,104,105] | |
| NOTCH 3 | MCL | Mutations are associated with severe prognosis in variants with high levels of Ki-67, a protein associated with proliferation | [89] |
| T-ALL | Increased crosstalk with stromal cells mediating an antiapoptotic effect on leukemia cells | [106] | |
| Activating mutation in non-regulatory region and PEST domain enhanced Notch 3 signaling | [107] | ||
| Notch 3 boosts CXCR4 expression and promotes cell migration | [108] | ||
| DLBCL | An alternatively spliced variant, with differential exon 16 depletion (-exon 16) was described among different subtypes | [109] | |
| NOTCH 4 | CLL | Stromal cell stimulation of Notch 4 determines survival and resistance to chemotherapy | [110] |
| B-ALL | Stromal cell induces an anti-apoptotic effect | [106] | |
| NHL | Notch 4 germline single nucleotide polymorphism was reported as associated to an HCV-related NHL | [111] | |
| DLBCL | A truncated variant of Notch 4 named Int-3 is constitutively active in human myeloid leukemia HL-60 cell line inhibiting differentiation and promoting the expansion of hematopoietic stem/progenitor cells | [112] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gragnani, L.; Lorini, S.; Marri, S.; Zignego, A.L. Role of Notch Receptors in Hematologic Malignancies. Cells 2021, 10, 16. https://doi.org/10.3390/cells10010016
Gragnani L, Lorini S, Marri S, Zignego AL. Role of Notch Receptors in Hematologic Malignancies. Cells. 2021; 10(1):16. https://doi.org/10.3390/cells10010016
Chicago/Turabian StyleGragnani, Laura, Serena Lorini, Silvia Marri, and Anna Linda Zignego. 2021. "Role of Notch Receptors in Hematologic Malignancies" Cells 10, no. 1: 16. https://doi.org/10.3390/cells10010016
APA StyleGragnani, L., Lorini, S., Marri, S., & Zignego, A. L. (2021). Role of Notch Receptors in Hematologic Malignancies. Cells, 10(1), 16. https://doi.org/10.3390/cells10010016







