Abstract
Roots are crucial for adaptation to drought stress. However, phenotyping root systems is a difficult and time-consuming task due to the special feature of the traits in the process of being analyzed. Correlations between root system architecture (RSA) at the early stages of development and in adult plants have been reported. In this study, the seminal RSA was analysed on a collection of 160 durum wheat landraces from 21 Mediterranean countries and 18 modern cultivars. The landraces showed large variability in RSA, and differences in root traits were found between previously identified genetic subpopulations. Landraces from the eastern Mediterranean region, which is the driest and warmest within the Mediterranean Basin, showed the largest seminal root size in terms of root length, surface, and volume and the widest root angle, whereas landraces from eastern Balkan countries showed the lowest values. Correlations were found between RSA and yield-related traits in a very dry environment. The identification of molecular markers linked to the traits of interest detected 233 marker-trait associations for 10 RSA traits and grouped them in 82 genome regions named marker-train association quantitative trait loci (MTA-QTLs). Our results support the use of ancient local germplasm to widen the genetic background for root traits in breeding programs.
1. Introduction
Wheat is estimated to have been first cultivated around 10,000 years before present (BP) in the Fertile Crescent region. It spread to the west of the Mediterranean Basin and reached the Iberian Peninsula around 7000 years BP [1]. During this migration, both natural and human selection resulted in the development of local landraces considered to be very well adapted to the regions where they were grown and containing the largest genetic diversity within the species [2]. From the middle of the 20th century, as a consequence of the Green Revolution, the cultivation of local landraces was progressively abandoned and replaced by the improved, more productive, and genetically uniform semi-dwarf cultivars. However, scientists are convinced that local landraces may provide new alleles to improve commercially valuable traits [3]. Introgression of these alleles into modern cultivars can be very useful, especially in breeding for suboptimal environments.
Drought is the most important environmental factor limiting wheat productivity in many parts of the world. Therefore, improving yield under water-limited conditions is one of the major challenges for wheat production worldwide. Breeding for adaptation to drought is extremely challenging due to the complexity of the target environments and the stress-adaptive mechanisms adopted by plants to withstand and mitigate the negative effects of a water deficit [4]. These mechanisms allow the plant to escape (e.g., early flowering date), avoid (e.g., root system), and/or tolerate (e.g., osmolyte accumulation) the negative effects of drought, which plays a role in determining final crop performance [5]. The crop traits to be considered as selection targets under drought conditions must be genetically correlated with yield and should have a greater heritability than yield itself [6,7]. Among these traits, early vigour, leaf area duration, crop water status, radiation use efficiency, and root architecture have been identified to be associated with yield under rainfed conditions (reviewed by Reference [8]).
Root system architecture (RSA) is crucial for wheat adaptation to drought stress. Roots exhibit a high level of morphological plasticity in response to soil conditions, which allows plants to adapt better, which is particularly under drought conditions. However, evaluating root architecture in the field is very difficult, expensive, and time-consuming, especially when a large number of plants need to be phenotyped. Several studies have reported a correlation of RSA in the early stages of development with RSA in adult plants [9], Manschadi et al. [10] reported that adult root geometry is strongly related to seminal root angle (SRA). Wasson et al. [11] described a relationship of root vigor between plants grown in the field and controlled conditions. Several systems have been adopted to enable early screening of the RSA in wheat [12].
Identifying quantitative trait loci (QTLs) and using marker-assisted selection is an efficient way to increase selection efficiency and boost genetic gains in breeding programs. However, while numerous studies have reported QTLs for RSA in bi-parental crosses [13], very few of them were based on association mapping [12,14,15,16,17,18]. Association mapping is a complementary approach to bi-parental linkage analysis and provides broader allelic coverage with higher mapping resolution. Association mapping is based on linkage disequilibrium, defined as the non-random association of alleles at different loci, and is used to detect the relationship between phenotypic variation and genetic polymorphism.
The main objectives of the present study were a) to identify differences in RSA among genetic subpopulations of durum wheat Mediterranean landraces, b) to find correlations of RSA with yield-related traits in different rainfed Mediterranean environments, and c) to identify molecular markers linked to RSA in the old Mediterranean germplasm through a genome-wide association study.
2. Materials and Methods
2.1. Plant Material
The germplasm used in the current study consisted of a set of 160 durum wheat landraces from 21 Mediterranean countries and 18 modern cultivars from a previously structured collection [2,19]. The landraces were classified into four genetic subpopulations (SPs) that matched their geographical origin as follows: the eastern Mediterranean (19 genotypes), the eastern Balkans and Turkey (20 genotypes), the western Balkans and Egypt (31 genotypes), the western Mediterranean (71 genotypes), and 19 genotypes that remained as admixed (Supplementary Materials Table S1).
2.2. Phenotyping
Eight uniform seeds per genotype were cultured following the paper roll method [20,21] in two replicates of four seeds. The seeds were placed at the top of a filter paper (420 × 520 mm) with the embryo facing down and sprayed with a 0.4% sodium hypochlorite solution. Subsequently, the papers were folded in half to obtain a 210 × 520 mm rectangle with the seeds fixed at the top. The papers were misted with deionized water and rolled by hand. The rolls were placed in plastic pots with deionized water at the bottom that was regularly checked to ensure it did not evaporate. The experiment was conducted in a growth chamber at 25 °C and darkness conditions. One week after sowing, the seeds were transferred to a black surface to take digital images that were processed by SmartRoot software [22] (Figure 1). Nine traits for the seminal root system architecture (RSA) were measured: total root number (TRN), primary root length (PRL, cm), total lateral root length (LRL, cm), primary root surface (PRS, cm2), total lateral root surface (LRS, cm2), primary root volume (PRV, cm3), total lateral root volume (LRV, cm3), primary root diameter (PRD, cm), and mean lateral root diameter (LRD, cm).
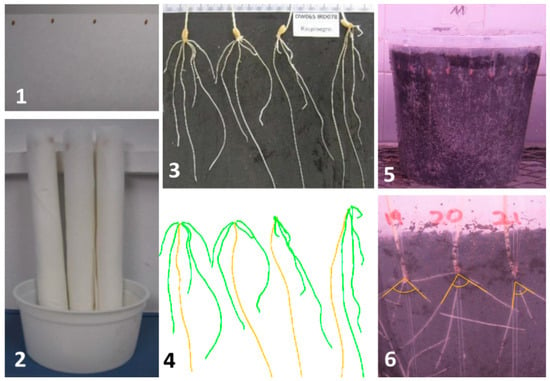
Figure 1.
Experimental setup for root system architecture analysis. First, seeds were placed on humid filter paper (1) and rolled. Paper rolls were placed in plastic pots with deionized water at the bottom for root growth (2). One week after sowing, the seeds were transferred to a black surface for digital imaging (3) that were processed by SmartRoot software [22] (4). The seminal root angle was measured using the clear pots (5,6).
Additionally, the SRA (°) was measured at the facilities of the International Center for Agricultural Research in the Dry Areas (ICARDA) in Rabat (Morocco) using the clear pot method described by Richard et al. [23] (Figure 1). Using a randomized complete block design, eight seeds per genotype were grown in 4 L clear pots filled with peat. The seeds were placed with the embryo facing down and close to the pot wall to facilitate root growth along the transparent wall. The pots were then watered, placed inside 4 L black pots, and kept at 20 °C and darkness conditions in a growth chamber. Five days after sowing, digital images were taken and processed with ImageJ software [24].
Data from field experiments conducted under rainfed conditions during two years of contrasting water input from sowing to physiological maturity (285 mm in 2008 and 104 mm in 2014) in Lleida, North-eastern Spain [25] were used to assess the relationships between RSA traits and yield-related traits.
The experiments were carried out in a non-replicated modified augmented design with three replicated checks (the cultivars ‘Claudio,’ ‘Simeto,’ and ‘Vitron’) and plots of 6 m2 (8 rows, 5 m long with a 0.15 m spacing). Sowing density was adjusted to 250 viable seeds m−2 and the plots were maintained free of weeds and diseases.
2.3. Statistical Analysis
Combined analyses of variance (ANOVA) were performed for the RSA traits of the structured accessions (141 landraces and 18 modern cultivars), considering the accessions and the replicate as random effects. The sum of squares of the cultivar effect was partitioned into differences between SPs and differences within them. The Kenward-Roger correction was used due to the unbalanced number of genotypes within the SPs. Since the experiment was divided into six sets with one check, least squared means were calculated using Simeto as a check and compared using the Tukey test [26] at p < 0.01.
Raw field data were fitted to a linear mixed model with the check cultivars as fixed effects and the row number, column number, and genotype as random effects [27]. Restricted maximum likelihood was used to estimate the variance components and to produce the best linear unbiased predictors (BLUPs) for yield and yield components. The relationships between RSA traits and yield-related traits were assessed through correlation analyses. All calculations were carried out using the SAS statistical package [28].
2.4. Genotyping
DNA isolation was performed from leaf samples following the method reported by Doyle and Doyle [29]. High throughput genotyping was performed at Diversity Arrays Technology Pty Ltd. (Canberra, Australia) (http://www.diversityarrays.com) with the genotyping by sequencing (GBS) DArTseq platform [30]. A total of 46,161 markers were used to genotype the association mapping panel, including 35,837 presence/absence variants (PAVs) and 10,324 single nucleotide polymorphisms (SNPs). Markers were ordered according to the consensus map of wheat v4 available at https://www.diversityarrays.com/.
2.5. Linkage Disequilibrium
Linkage disequilibrium (LD) among markers was calculated for the A and B genomes using markers with a map position on the wheat v4 consensus map, and a minor allele frequency greater than 5%, using TASSEL 5.0 [31]. Pair-wise LD was measured using the squared allele frequency correlations r2 and the values for genomes A and B were plotted against the genetic distance to determine how fast the LD decays. A LOESS curve was fitted to the plot using the JMP v12Pro statistical package (SAS Institute Inc, Cary, NC, USA).
2.6. Genome-Wide Association Study
A genome-wide association study (GWAS) was performed with 160 landraces for the mean of measured traits with TASSEL 5.0 software [31]. A mixed linear model was conducted using the population structure determined by Soriano et al. [19] as the fixed effect and a kinship (K) matrix as the random effect (Q + K) at the optimum compression level. A false discovery rate threshold [32] was established at −log10p > 4.6 (p < 0.05), using 2135 markers according to the results of the LD decay, to consider a marker-trait association (MTA) significant. Moreover, a second, less restrictive threshold was established at −log10p > 3. To simplify the MTA information, those associations located within LD blocks were considered to belong to the same QTL and were named marker-trait association quantitative trait loci (MTA-QTLs). Graphical representation of the genetic position of MTA-QTLs was carried out using MapChart 2.3 [33].
2.7. Gene Annotation
Gene annotation for the target region of significant MTAs was performed using the gene models for high-confidence genes reported for the wheat genome sequence [34] available at https://wheat-urgi.versailles.inra.fr/Seq-Repository/.
3. Results
3.1. Phenotypic Analyses
The ANOVA showed that, for all traits, the phenotypic variability was mainly explained by the cultivar effect, since it accounted for 63.41% (PRD) to 90.57% (LRD) of the total sum of squares (Table 1). A summary of the genetic variation of the RSA traits is shown in Supplementary Materials Table S2. The partitioning of the sum of squares of the cultivar effect into differences between and within SPs revealed that the variability induced by the genotype was mainly explained by differences within SPs on a range from 70.1% for TRN to 91.5 for PRV (Table 1). Differences between SPs were statistically significant for all traits, accounting for 8.5% (PRV) to 30.5% (TRN) of the sum of squares of the genotype effect (Table 1). Western Mediterranean landraces showed the highest number of seminal roots and the narrowest root angle, whereas the eastern Balkans and Turkey SP showed the widest angle (Table 2). The highest values for root size–related traits (length, surface and volume) in both primary and lateral roots were recorded in the eastern Mediterranean landraces. The western Balkans and Egypt subpopulation showed the largest root diameter (Table 2). The comparison of mean values of eastern Balkans and Turkish landraces revealed that the Turkish ones had high values for all traits except TRN, LRL, and root diameter (Supplementary Materials Table S3). The modern cultivars showed intermediate values for all RSA traits (Table 2).

Table 1.
Percentage of the sum of squares of the ANOVA model for the seminal root system architecture traits in a set of 159 Mediterranean durum wheat genotypes structured into five genetic subpopulations by Soriano et al. [19].

Table 2.
Means comparison of seminal root system architecture traits measured in a set of 159 Mediterranean durum wheat genotypes structured into five genetic subpopulations [19]. Means within columns with different letters are significantly different at p < 0.01 following a Tukey test.
Correlation coefficients between RSA traits and yield-related traits were calculated for two field experiments with contrasting water input (285 and 104 mm of rainfall from sowing to physiological maturity). Whereas, for the rainiest environment, only the relationship between SRA and number of spikes per square meter (NSm2) was statistically significant (p = 0.043, r2 = 0.16)). For the driest environment, 14 correlations involving all the yield-related traits and RSA traits except root diameter were statistically significant (Figure 2) (r2 between 0.17 for NSm2 and PRL and PRS to 0.30 for TKW and TRN). Most of the significant correlations were positive. Only the relationship between SRA and thousand kernel weight (TKW) was negative.

Figure 2.
Correlations between seminal root system architecture traits and yield-related traits determined in field experiments receiving high (density ellipse in red, A) and low (density ellipse in green, B) water input from sowing to physiological maturity. Significant correlation coefficients (p < 0.05) are indicated with red and green points. TRN, total root number. SRA, seminal root angle. PRL, primary root length. LRL, total lateral root length. PRS, primary root surface. LRS, total lateral root surface. PRV, primary root volume. LRV, total lateral root volume. PRD, primary root diameter. LRD, mean lateral root diameter. GY, grain yield. NSm2, number of spikes per square meter. NGm2, number of grains per square meter. TKW, thousand kernel weight.
3.2. Marker-Trait Associations
A total of 46,161 DArTseq markers, including PAVs and SNPs, were used to genotype the set of 160 durum wheat landraces. To reduce the risk of false positives, markers and accessions were analyzed for the presence of duplicated patterns and missing values. Of 35,837 PAVs, 24,188 were placed on the wheat v4 consensus map. Of these, those with more than 30% of missing data and those with a minor allele frequency lower than 5% were removed from the analysis, leaving 19,443 PAVs. A total of 6957 SNPs were mapped, leaving a total of 4686 SNPs after marker filtering as before. Additionally, 413 markers were duplicated between PAVs and SNPs, so the corresponding PAVs were eliminated. A total of 23,716 markers remained for the subsequent analysis.
Linkage disequilibrium was estimated for locus pairs in genomes A and B using a sliding window of 50 cM. A total of 471,319 and 681,389 possible pair-wise loci were observed for genomes A and B, respectively. Of these locus pairs, 52% and 43% showed significant linkage disequilibrium at p < 0.01 and p < 0.001, respectively. Mean r2 was 0.12 for genome A and 0.11 for genome B. These means were used as a threshold for estimating the intercept of the LOESS curve to determine the distance at which LD decays in each genome. Markers were in LD in a range from less than 1 cM in genome B to 1 cM in genome A (Supplementary Materials Table S4).
Results of the GWAS are reported in Figure 3 and in Supplementary Materials Table S5. Using a restrictive threshold based on a false discovery rate at p < 0.05 (−log10p > 4.6) and the LD decay, only 12 MTAs corresponding to seven markers were significant. Using a common threshold of −log10p > 3, as previously reported by other authors [35,36,37,38], a total of 233 MTAs involving 176 markers were identified. MTAs were equally distributed in both genomes (50.2% in the A genome and 49.8% in the B genome). Chromosomes 2B and 7A harbored the highest number of MTAs (39 and 32 respectively), carrying 30% of the total number of MTAs, whereas chromosomes 4B and 7B harbored the lowest number of MTAs, 8 and 6 MTAs, respectively (Figure 3A). Root volume was the trait showing the highest number of MTAs (77), followed by root surface (46), root diameter (37), root length and number (26), and SRA (21) (Figure 3B). The mean percentage of phenotypic variance explained (PVE) per MTA was similar for all traits, ranging from 0.09 to 0.11 (Figure 3C). Most of the MTAs showed low PVE, in agreement with the quantitative nature of the analyzed traits. The percentage of MTAs with a PVE lower than 0.1 was 71%, whereas that of MTAs with a PVE lower than 0.15 was 98% (Figure 3D).
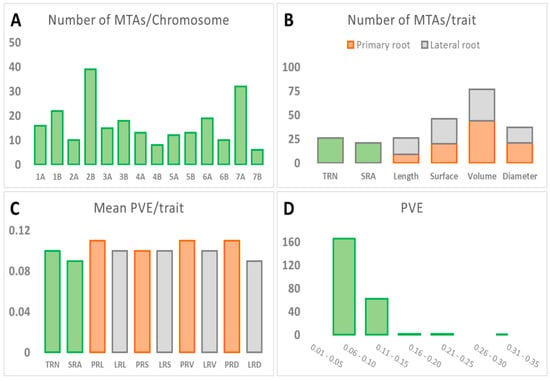
Figure 3.
Summary of marker trait associations (MTA). (A) Number of MTAs per chromosome. (B) Number of MTAs per trait. (C) Mean PVE per trait. (D) PVE. TRN, total root number. SRA, seminal root angle. PRL, primary root length. LRL, total lateral root length. PRS, primary root surface. LRS, total lateral root surface. PRV, primary root volume. LRV, total lateral root volume. PRD, primary root diameter. LRD, mean lateral root diameter.
To simplify the MTA information, those MTAs located within a region of 1 cM, as reported by the LD decay, were considered part of the same QTL. Thus, the 233 associations were restricted to 81 MTA-QTLs (Figure 4 and Table 3). Of the 82 MTA-QTLs, 33 had only one MTA, whereas, for the remaining 49, the number of MTAs per MTA-QTL ranged from 2 in 19 MTA-QTLs to 15 in mtaq-7A.1. When several consecutive pairs of MTAs were separated for a distance of 1 cm, the whole block was considered as the same MTA-QTL. The genomic distribution of MTA-QTLs showed that chromosome 1A, 4A, and 5B harbored 8 MTA-QTLs, chromosomes 1B, 3A, 3B, and 5A 7 MTA-QTLs, 6A 6 MTA-QTLs, 7A 5 MTA-QTLs, 2A, 2B, 4B, and 6B 4 MTA-QTLs and chromosome 7B harbored 3 MTA-QTLs. For the 48 MTA-QTLs with more than one MTA, 10 were related to one trait. Of these, mtaq-1A.5, mtaq-3A.1, mtaq-4A.4, and mtaq-4A.5 carried associations related to root volume, mtaq-2B.1, and mtaq-3B.7 to root diameter, mtaq-3B.1, and mtaq-7A.5 to root number, and mtaq-4A.3 and mtaq-6A.5 to the root angle.
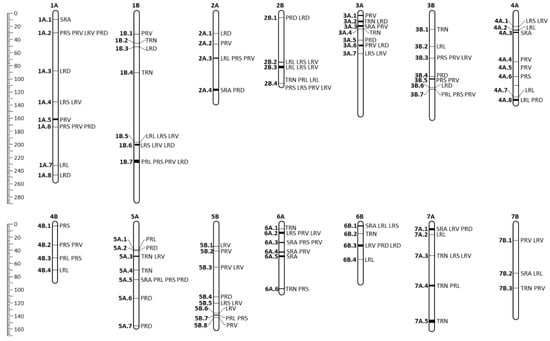
Figure 4.
MTA-QTL map. MTA-QTLs are indicated in bold on the left side of the chromosome and traits involved in each MTA-QTL are on the right side. The rule on the left indicates genetic distance in cM. TRN, total root number. SRA, seminal root angle. PRL, primary root length. LRL, total lateral root length. PRS, primary root surface. LRS, total lateral root surface. PRV, primary root volume. LRV, total lateral root volume. PRD, primary root diameter. LRD, mean lateral root diameter.

Table 3.
MTA-QTLS.
Among all significant MTAs, markers with different alleles between extreme genotypes for each trait (i.e., the upper and lower 10th percentile) were identified except for PRL (Table 4, Figure 5). Frequency of the most common allele among genotypes from the upper 10th percentile ranged from 67% for LRD to 90% for PRV, whereas, for the lower 10th percentile, they ranged from 74% for TRN to 93% for LRD (Figure 5).

Table 4.
Selected significant markers from the GWAS with different allele composition for the upper (UP) and lower (LOW) 10th percentile of genotypes. Different letters on the UP and LOW 10th phenotype indicate that means are significantly different at p < 0.01 following a Tukey test.

Figure 5.
Marker allele frequency means from landraces within the upper and lower 10th percentile for the analyzed traits. All significant markers shown in Table 4 are included. TRN, total root number. SRA, seminal root angle. PRL, primary root length. LRL, total lateral root length. PRS, primary root surface. LRS, total lateral root surface. PRV, primary root volume. LRV, total lateral root volume. PRD, primary root diameter. LRD, mean lateral root diameter.
3.3. Gene Annotation
Of the 176 markers showing significant associations, 31 were identified in the reference sequence of the wheat genome [34] (Table 5). Eight of them were positioned within gene models, whereas, for the rest, the closest gene model to the corresponding marker was taken into consideration. The gene models described in Table 5 included molecules related to abiotic stress resistance, seed formation, carbohydrate remobilization, disease resistance proteins, and other genes involved in different cellular metabolic pathways.

Table 5.
Gene models within MTA-QTL positions. Only MTAs with markers mapped against the genome sequence are included. Genome position of the gene model is indicated in Mb.
4. Discussion
Roots exhibit a high level of morphological plasticity in response to soil conditions, which allows plants to better adapt, particularly under drought conditions. Several authors have reported the role of RSA traits in response to drought stress [39,40]. Wasson et al. [11] suggested that a deep root system with the appropriate density along the soil profile would confer an advantage on wheat grown in rainfed agricultural systems. Therefore, identifying new alleles for improving root architecture under drought conditions and introgressing them into adapted phenotypes is a desirable approach for breeding purposes. The current study analyzed a collection of durum wheat landraces representative of the variability existing within the Mediterranean Basin in an attempt to broaden the genetic background present in commercial cultivars.
Evaluating root architecture in the field is a difficult, expensive, and time-consuming assignment, especially when a large number of plants need to be phenotyped. It has been reported that the root geometry of adult plants is strongly related to the seminal root angle (SRA), with deeply rooted wheat genotypes showing a narrower SRA [10]. Different systems have been adopted to enable early screening of the root system architecture in wheat, assuming that genotypes that differ in root architecture at an early developmental stage would also differ in the field at stages when nutrient and/or water capture become critical for grain yield [12].
4.1. Phenotypic Variation
The germplasm analyzed in the present study, including mostly durum wheat landraces from the Mediterranean Basin, showed wide variability in RSA traits. The variability found was higher than that observed in other studies using elite accessions [12,14] or even landraces, as reported by Ruiz et al. [41] analyzing a collection of Spanish durum wheat landraces. These results, and the intermediate values obtained for all traits in modern cultivars, support the use of ancient local germplasm for widening the genetic background in breeding programs.
Means comparison of phenotypic traits revealed large differences among SPs associated with their geographical origin. Eastern Mediterranean landraces, collected in the area closest to the origin of tetraploid wheat, showed the largest root size in terms of length, surface, and volume, and the widest root angle. The wheat-growing areas of this region, which comprises Syria, Jordan, Israel, and Egypt, are the warmest and driest within the Mediterranean Basin [42]. In addition, when SRA traits were analyzed separately for the two components of the eastern Balkans and Turkey subpopulation, large differences appeared between them, with Turkish landraces being much more similar to the eastern Mediterranean ones than to the eastern Balkan ones, since the latter showed the lowest values for root length, surface, and volume. Turkish landraces also showed a wide root angle, as did the eastern Mediterranean ones. The differences found in SRA between the eastern Balkans and Turkish landraces are sustained by two lines of evidence. One is the contrasting environmental conditions of the wheat-growing areas of northern Balkan countries and Turkey, since the analysis of long-term climate data demonstrated less rainfall and higher temperatures and solar radiation in the latter [42]. The other is that the northern Balkan landraces likely originated in the steps of southern Russia and the Volga region [2,43], which also suggests contrasting environmental conditions in the zones of origin of the eastern Balkan and Turkish landraces. The phenotypic analysis carried out in the current study revealed that landraces from regions where drought stress is prevalent have a larger root size and a wider root angle. This architecture should allow a larger proportion of the soil to be covered for more efficient water capture, and this hypothesis is supported by correlations between RSA and yield traits. Although low, likely due to the very early stage when the root traits were measured, differences in the number of significant correlations were observed between the two environments with the highest and lowest water input reported by Roselló et al. [25]. Root size–related traits were positively correlated with the number of grains and spikes per unit area (primary roots) and with grain yield and grain weight (lateral roots) in the driest environment. SRA was negatively correlated with TKW, as reported previously by Canè et al. [12], who concluded that it was due to the influence of the root angle on the distribution of roots in the soil layers, which affects the water uptake from deeper layers. In our study, the genotypes with the narrowest angle corresponded to those from the western Mediterranean countries, which Royo et al. [42] and Soriano et al. [19] reported to have heavier grains.
4.2. Marker-Trait Associations
The current study attempts to dissect the genetic architecture controlling the seminal root system in a collection of landraces from the Mediterranean Basin by association analysis. A mixed linear model accounting for the genetic relatedness between cultivars (random effect) and their population structure (fixed effect) (K + Q model) was used in order to reduce the number of spurious associations.
A total of 233 significant associations were identified for the 10 RSA traits underlying the complex genetic control of RSA. However, in order to simplify this information and to integrate closely linked MTAs in the same QTL, those MTAs located within LD blocks were considered as belonging to the same MTA-QTL. As a result, the number of genome regions involved in RSA was reduced to 82. The relationships between RSA and yield-related traits was also suggested by the presence of pleiotropic MTA-QTLs. The comparison of the genome regions identified in the current study with those related to yield and yield components by Roselló et al. [44] showed that 45% of the RSA MTA-QTLs were located with yield-related trait MTA-QTLs. These results are in agreement with the findings of Canè et al. [12], who found that 30% of the RSA-QTLs affected agronomic traits, which provided evidence of the implications of RSA in field performance of durum wheat at early growth stages.
In the last few years, GWAS for RSA have been limited in comparison with QTL mapping for root traits based on bi-parental populations (see Soriano and Álvaro [13] for a review). A comparison with previous studies reporting MTAs for RSA resulted in several common regions with the current study. Three common regions were found with the study of Canè et al. [12], but different traits were included for MTAs in those QTLs (mtaq-3A.3, mtaq-3A.5, mtaq-3A.6, and mtaq-6B.2). Two MTAs were in common with those reported by Ayalew et al. [15], who identified five significant associations with root length under stress (2) and non-stress (3) conditions. The MTA reported under stress conditions in chromosome 2B may correspond with mtaq-2B.2, which also shows an association with LRL. However, the association on chromosome 3B, although in a common region with mtaq-3B.4, differed in RSA. When MTA-QTLs were compared with QTLs from bi-parental populations, twelve genomic regions were located within the meta-QTL positions defined by Soriano and Álvaro [13] after the compilation of 754 QTLs from 30 studies.
Candidate genes at the MTA peak were sought using the high-confidence gene annotation from the wheat genome sequence [34]. Among these genes, those involved in plant growth and development as well as tolerance to abiotic stresses may be of special interest. On chromosome 1A, the marker 1210090_SNP in mtaq-1A.7 is located close to a cellulose synthase gene. This type of gene is involved in plant cell growth and structure [45]. A trichome birefringence (TB) protein was identified in mtaq-1B.1. According to Zhu et al. [46], the TB-like27 protein mutants in Arabidopsis increased aluminium accumulation in cell walls, which inhibited root elongation through structural and functional damage. Three peaks corresponded with F-box domains located in mtaq-1B.7, mtaq-3A.1, and mtaq-6A.6. According to Hua et al. [47], this is the protein subunit of E3 ubiquitin ligases involved in the response to abiotic stresses. Li et al. [48] overexpressed the F-box TaFBA1 in transgenic tobacco to improve heat tolerance, and one of the results was increased root length in the transgenic plants. 9-cis-epoxycarotenoid dioxygenase (NCED) is a key enzyme in the biosynthesis of ABA in higher plants, which regulates the response to various environmental stresses [49]. This enzyme is located within mtaq-2A.3. In mtaq-2A.4, the marker 1117775_PAV corresponded with a late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein. These proteins play a role in the response to abiotic stresses. They are mainly accumulated in seeds, but have been found in roots during the whole developmental cycle [50]. The marker 1098568_PAV, in mtaq-6B.4, is located within a gene coding a bZIP transcription factor family protein. This type of transcription factor is involved in abiotic stresses [51]. Zhang et al. [52] observed that the root growth of transgenic plants overexpressing the gene TabZIP14-B was hindered more severely than that of the control plants. Another gene involved in abiotic stress tolerance is the mitochondrial pyruvate carrier (MPC) located in mtaq-7A.1 [53]. This gene is involved in cadmium tolerance in Arabidopsis, which prevents its accumulation. Roots are the predominant plant tissue for cadmium absorption or exclusion. He et al. [53] found that the root length of mutant plants of Arabidopsis for MPC genes was substantially shorter than the wild-type plants. A protein related to WAT1 (WALLS ARE THIN1) involved in secondary cell wall thickness [54] is located in the peak of mtaq-7A.1.
5. Conclusions
Including local landraces in breeding programs is a useful approach to broadening the genetic variability of crops [3]. The variability for root system architecture traits found in Mediterranean landraces and the high number of genome regions controlling them—most of them not reported previously—makes this germplasm a valuable source for root architecture improvement. The identification of extreme genotypes for root architecture traits can help identify parents for the development of new mapping populations to tackle a map-based cloning approach to the genes of interest. In the present study, we identified the molecular markers linked to these genotypes with different allele composition that facilitate the introgression of the corresponding traits through marker-assisted breeding.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4395/9/7/364/s1, Table S1: Accessions included in the study, Table S2: Statistics of the seminal RSA traits, Table S3: Means comparison of seminal root system architecture traits for the eastern Balkans (EB) and Turkish durum wheat landraces. Means within columns with different letters are significantly different at p < 0.05 following a Tukey test, Table S4: Linkage disequilibrium decay plots. (A) Genome A. (B) Genome B. The LOESS curve is represented in blue. The horizontal red line corresponds to the r2 mean for each genome, Table S5: Significant markers associated with seminal root system architecture traits obtained in 160 durum wheat Mediterranean landraces.
Author Contributions
Conceptualization, M.R. and J.M.S.; Methodology, M.R., M.S.-G., and J.M.S.; Formal Analysis, M.R.; Investigation, M.R., M.S.-G., J.M.S.; Resources, C.R.; Data Curation, M.R., C.R.; Writing—Original Draft Preparation, M.R., J.M.S.; Writing—Review & Editing, C.R., M.S.-G., and J.M.S.; Visualization, J.M.S.; Supervision, C.R., M.S.-G., and J.M.S.; Project Administration, C.R. and J.M.S.; Funding Acquisition, C.R. and J.M.S.
Funding
This research was funded by Spanish Ministry of Science, Innovation and Universities (http://www.ciencia.gob.es/), grant numbers AGL-2012-37217 (C.R.) and AGL2015-65351-R (J.M.S. and C.R.).
Acknowledgments
Projects AGL-2012-37217 (C.R.) and AGL2015-65351-R (J.M.S. and C.R.) of the Spanish Ministry of Science, Innovation and Universities (http://www.ciencia.gob.es/) funded this study. The authors acknowledge the contribution of the CERCA Program (Generalitat de Catalunya). M.R. is a recipient of a PhD grant from the Instituto Nacional de Investigación y Tecnología Agraria y Alimentaria (INIA). J.M.S. was hired by the INIA-CCAA program funded by INIA and the Generalitat de Catalunya.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
| BP | Before Present |
| DArTseq | Diversity Arrays Technology sequencing |
| EB + T | Eastern Balkans and Turkey |
| EM | Eastern Mediterranean |
| FDR | False Discovery Rate |
| GWAS | Genome Wide Association Study |
| GY | Grain Yield |
| LRD | Lateral Roots Diameter |
| LRL | Lateral Roots Length |
| LRS | Lateral Roots Surface |
| LRV | Lateral Roots Volume |
| MTA | Marker-Trait Association |
| NGm2 | Number of Grains per square meter |
| NSm2 | Number of Spikes per square meter |
| PAV | Presence/Absence Variants |
| PRD | Primary Root Diameter |
| PRL | Primary Root Length |
| PRS | Primary Root Surface |
| PRV | Primary Root Volume |
| PVE | Phenotypic Variance Explained |
| QTL | Quantitative Trait Loci |
| RSA | Root System Architecture |
| SNP | Single Nucleotide Polymorphism |
| SP | Subpopulation |
| SRA | Seminal Root Angle |
| TKW | Thousand Kernel Weight |
| TRN | Total Root Number |
| WB + E | Western Balkans and Egypt |
| WM | Western Mediterranean |
References
- Feldman, M. Origin of cultivated wheat. In The World Wheat Book: A History of Wheat Breeding; Bonjean, A.P., Angus, W.J., Eds.; Lavoisier Publishing: Paris, France, 2001; pp. 3–56. ISBN 1898298726. [Google Scholar]
- Nazco, R.; Villegas, D.; Ammar, K.; Peña, R.J.; Moragues, M.; Royo, C. Can Mediterranean durum wheat landraces contribute to improved grain quality attributes in modern cultivars? Euphytica 2012, 185, 1–17. [Google Scholar] [CrossRef]
- Lopes, M.S.; El-Basyoni, I.; Baenziger, P.S.; Singh, S.; Royo, C.; Ozbek, K.; Aktas, H.; Ozer, E.; Ozdemir, F.; Manickavelu, A.; et al. Exploiting genetic diversity from landraces in wheat breeding for adaptation to climate change. J. Exp. Bot. 2015, 66, 3477–3486. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, M.P.; Mujeeb-Kazi, A.; Sawkins, M. Prospects for utilising plant-adaptive mechanisms to improve wheat and other crops in drought- and salinity-prone environments. Ann. Appl. Biol. 2005, 146, 239–259. [Google Scholar] [CrossRef]
- Maccaferri, M.; Sanguineti, M.C.; Demontis, A.; El-Ahmed, A.; García del Moral, L.; Maalouf, F.; Nachit, M.; Nserallah, N.; Ouabbou, H.; Rhouma, S.; et al. Association mapping in durum wheat grown across a broad range of water regimes. J. Exp. Bot. 2011, 62, 409–438. [Google Scholar] [CrossRef] [PubMed]
- Royo, C.; García del Moral, L.; Slafer, G.A.; Nachit, M.; Araus, J.L. Selection tools for improving yield-associated physiological traits. In Durum Wheat Breeding: Current Approaches and Future Strategies; Royo, C., Nachit, M.M., Di Fonzo, N., Araus, J., Pfeiffer, W., Slafer, G., Eds.; Food Products Press: New York, NY, USA, 2005; pp. 563–598. [Google Scholar]
- Royo, C.; Villegas, D. Field measurements of canopy spectra for biomass assessment of small-grain cereals. In Biomass—Detection, Production and Usage; Darko, M., Ed.; InTech Open: London, UK, 2011; pp. 27–52. ISBN 978-953-307-492-4. [Google Scholar]
- Tuberosa, R. Phenotyping for drought tolerance of crops in the genomics era. Front. Physiol. 2012, 3, 347. [Google Scholar] [CrossRef] [PubMed]
- Løes, A.-K.; Gahoonia, T.S. Genetic variation in specific root length in Scandinavian wheat and barley accessions. Euphytica 2004, 137, 243–249. [Google Scholar] [CrossRef]
- Manschadi, A.M.; Hammer, G.L.; Christopher, J.T.; Manschadi, A.M.; Hammer, G.L.; Christopher, J.T. Genotypic variation in seedling root architectural traits and implications for drought adaptation in wheat (Triticum aestivum L.). Plant Soil 2008, 303, 115–129. [Google Scholar] [CrossRef]
- Wasson, A.P.; Richards, R.A.; Chatrath, R.; Misra, S.C.; Prasad, S.V.S.; Rebetzke, G.J.; Kirkegaard, J.A.; Christopher, J.; Watt, M. Traits and selection strategies to improve root systems and water uptake in water-limited wheat crops. J. Exp. Bot. 2012, 63, 3485–3498. [Google Scholar] [CrossRef]
- Canè, M.A.; Maccaferri, M.; Nazemi, G.; Salvi, S.; Francia, R.; Colalongo, C.; Tuberosa, R. Association mapping for root architectural traits in durum wheat seedlings as related to agronomic performance. Mol. Breed. 2014, 34, 1629–1645. [Google Scholar] [CrossRef]
- Soriano, J.M.; Álvaro, F. Discovering consensus genomic regions in wheat for root-related traits by QTL meta-analysis. Sci. Rep. 2019, in press. [Google Scholar]
- Sanguineti, M.C.; Li, S.; Maccaferri, M.; Corneti, S.; Rotondo, F.; Chiari, T.; Tuberosa, R. Genetic dissection of seminal root architecture in elite durum wheat germplasm. Ann. Appl. Biol. 2007, 151, 291–305. [Google Scholar] [CrossRef]
- Ayalew, H.; Liu, H.; Börner, A.; Kobiljski, B.; Liu, C.; Yan, G. Genome-wide association mapping of major root length QTLs under PEG induced water stress in wheat. Front. Plant Sci. 2018, 9, 1759. [Google Scholar] [CrossRef] [PubMed]
- Alahmad, S.; El Hassouni, K.; Bassi, F.M.; Dinglasan, E.; Youssef, C.; Quarry, G.; Aksoy, A.; Mazzucotelli, E.; Juhász, A.; Able, J.A.; et al. A major root architecture QTL responding to water limitation in durum wheat. Front. Plant Sci. 2019, 10, 436. [Google Scholar] [CrossRef] [PubMed]
- Beyer, S.; Daba, S.; Tyagi, P.; Bockelman, H.; Brown-Guedira, G.; IWGSC; Mohammadi, M. Loci and candidate genes controlling root traits in wheat seedlings—A wheat root GWAS. Funct. Integr. Genom. 2019, 19, 91–107. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Peng, Z.; Mao, X.; Wang, J.; Chang, X.; Reynolds, M.; Jing, R. Genome-wide association study reveals genomic regions controlling root and shoot traits at late growth stages in wheat. Ann. Bot. 2019, 1–14. [Google Scholar] [CrossRef]
- Soriano, J.; Villegas, D.; Aranzana, M.; García del Moral, L.; Royo, C. Genetic structure of modern durum wheat cultivars and mediterranean landraces matches with their agronomic performance. PLoS ONE 2016, 11, e0160983. [Google Scholar] [CrossRef] [PubMed]
- Rahnama, A.; Munns, R.; Poustini, K.; Watt, M. A screening method to identify genetic variation in root growth response to a salinity gradient. J. Exp. Bot. 2011, 62, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Watt, M.; Moosavi, S.; Cunningham, S.C.; Kirkegaard, J.A.; Rebetzke, G.J.; Richards, R.A. A rapid, controlled-environment seedling root screen for wheat correlates well with rooting depths at vegetative, but not reproductive, stages at two field sites. Ann. Bot. 2013, 112, 447–455. [Google Scholar] [CrossRef] [PubMed]
- Lobet, G.; Pagès, L.; Draye, X. A Novel image-analysis toolbox enabling quantitative analysis of root system architecture. Breakthr. Technol. 2011, 157, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Richard, C.A.I.; Hickey, L.T.; Fletcher, S.; Jennings, R.; Chenu, K.; Christopher, J.T. High-throughput phenotyping of seminal root traits in wheat. Plant Methods 2015, 11, 13. [Google Scholar] [CrossRef] [PubMed]
- Abramoff, M.D.; Magalhaes, P.J.; Ram, S.J. Image Processing with ImageJ. Biophotonics Int. 2004, 11, 36–42. [Google Scholar]
- Roselló, M.; Villegas, D.; Álvaro, F.; Soriano, J.M.; Lopes, M.S.; Nazco, R.; Royo, C. Unravelling the relationship between adaptation pattern and yield formation strategies in Mediterranean durum wheat landraces. Eur. J. Agron. 2019, 107, 43–52. [Google Scholar] [CrossRef]
- Tukey, J.W. Comparing individual means in the analysis of variance. Biometrics 1949, 5, 99–114. [Google Scholar] [CrossRef] [PubMed]
- Little, R.; Milliken, G.; Stroup, W.; Wolfinger, R. SAS system for mixed models. Technometrics 1997, 39, 344. [Google Scholar]
- SAS Institute. SAS Enterprise Guide; SAS Institute: Cary, NC, USA, 2014. [Google Scholar]
- Doyle, J.; Doyle, J. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Sansaloni, C.; Petroli, C.; Jaccoud, D.; Carling, J.; Detering, F.; Grattapaglia, D.; Kilian, A. Diversity arrays technology (DArT) and next-generation sequencing combined: Genome-wide, high throughput, highly informative genotyping for molecular breeding of Eucalyptus. BMC Proc. 2011, 5, 54. [Google Scholar] [CrossRef]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Voorrips, R.E. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef]
- IWGSC. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361, eaar7191. [Google Scholar] [CrossRef]
- Mwadzingeni, L.; Shimelis, H.; Rees, D.J.G.; Tsilo, T.J. Genome-wide association analysis of agronomic traits in wheat under drought-stressed and non-stressed conditions. PLoS ONE 2017, 12, e0171692. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.-X.; Zhu, Y.-L.; Zhang, D.-X.; Shao, H.; Liu, P.; Hu, J.-B.; Zhang, H.; Zhang, H.-P.; Chang, C.; Lu, J.; et al. Genome-wide association study for grain yield and related traits in elite wheat varieties and advanced lines using SNP markers. PLoS ONE 2017, 12, e0188662. [Google Scholar] [CrossRef] [PubMed]
- Mangini, G.; Gadaleta, A.; Colasuonno, P.; Marcotuli, I.; Signorile, A.M.; Simeone, R.; De Vita, P.; Mastrangelo, A.M.; Laidò, G.; Pecchioni, N.; et al. Genetic dissection of the relationships between grain yield components by genome-wide association mapping in a collection of tetraploid wheats. PLoS ONE 2018, 13, e0190162. [Google Scholar] [CrossRef] [PubMed]
- Sukumaran, S.; Reynolds, M.P.; Sansaloni, C. Genome-wide association analyses identify QTL hotspots for yield and component traits in durum wheat grown under yield potential, drought, and heat stress environments. Front. Plant Sci. 2018, 9, 81. [Google Scholar] [CrossRef] [PubMed]
- Christopher, J.; Christopher, M.; Jennings, R.; Jones, S.; Fletcher, S.; Borrell, A.; Manschadi, A.M.; Jordan, D.; Mace, E.; Hammer, G. QTL for root angle and number in a population developed from bread wheats (Triticum aestivum) with contrasting adaptation to water-limited environments. Theor. Appl. Genet. 2013, 126, 1563–1574. [Google Scholar] [CrossRef] [PubMed]
- Paez-García, A.; Motes, C.; Scheible, W.-R.; Chen, R.; Blancaflor, E.; Monteros, M. Root traits and phenotyping strategies for plant improvement. Plants 2015, 4, 334–355. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, M.; Giraldo, P.; González, J.M. Phenotypic variation in root architecture traits and their relationship with eco-geographical and agronomic features in a core collection of tetraploid wheat landraces (Triticum turgidum L.). Euphytica 2018, 214, 54. [Google Scholar] [CrossRef]
- Royo, C.; Nazco, R.; Villegas, D. The climate of the zone of origin of Mediterranean durum wheat (Triticum durum Desf.) landraces affects their agronomic performance. Genet. Resour. Crop Evol. 2014, 61, 1345–1358. [Google Scholar] [CrossRef]
- Dedkova, O.S.; Badaeva, E.D.; Amosova, A.V.; Martynov, S.P.; Ruanet, V.V.; Mitrofanova, O.P.; Pukhal’skiy, V.A. Diversity and the origin of the European population of Triticum dicoccum (Schrank) Schuebl. As revealed by chromosome analysis. Russ. J. Genet. 2009, 45, 1082–1091. [Google Scholar] [CrossRef]
- Roselló, M.; Royo, C.; Sansaloni, C.; Soriano, J.M. GWAS for yield and related traits under rainfed mediterranean environments revealed different genetic architecture in pre- and post-green revolution durum wheat collections. Front. Plant Sci. 2019, in press. [Google Scholar]
- Lei, L.; Li, S.; Gu, Y. Cellulose synthase complexes: Composition and regulation. Front. Plant Sci. 2012, 3, 75. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.F.; Sun, Y.; Zhang, B.C.; Mansoori, N.; Wan, J.X.; Liu, Y.; Wang, Z.W.; Shi, Y.Z.; Zhou, Y.H.; Zheng, S.J. Trichome Birefringence-Like27 affects aluminum sensitivity by modulating the O-acetylation of xyloglucan and aluminum-binding capacity in Arabidopsis. Plant Physiol. 2014, 166, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Hua, Z.; Vierstra, R.D. The cullin-RING ubiquitin-protein ligases. Annu. Rev. Plant Biol. 2011, 62, 299–334. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Wang, W.; Wang, W.; Zhang, G.; Liu, Y.; Wang, Y.; Wang, W. Wheat f-box protein gene TaFBA1 is involved in plant tolerance to heat stress. Front. Plant Sci. 2018, 9, 521. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.J.; Song, G.Q.; Li, Y.L.; Gao, J.; Liu, J.J.; Fan, Q.Q.; Huang, C.Y.; Sui, X.X.; Chu, X.S.; Guo, D.; et al. Cloning of 9-cis-epoxycarotenoid dioxygenase gene (TaNCED1) from wheat and its heterologous expression in tobacco. Biol. Plant. 2014, 58, 89–98. [Google Scholar] [CrossRef]
- Gao, J.; Lan, T. Functional characterization of the late embryogenesis abundant (LEA) protein gene family from Pinus tabuliformis (Pinaceae) in Escherichia coli. Sci. Rep. 2016, 6, 19467. [Google Scholar] [CrossRef]
- Sornaraj, P.; Luang, S.; Lopato, S.; Hrmova, M. Basic leucine zipper (bZIP) transcription factors involved in abiotic stresses: A molecular model of a wheat bZIP factor and implications of its structure in function. Biochim. Biophys. Acta 2016, 1860, 46–56. [Google Scholar] [CrossRef]
- Zhang, W.; Chen, S.; Abate, Z.; Nirmala, J.; Rouse, M.N.; Dubcovsky, J. Identification and characterization of Sr13, a tetraploid wheat gene that confers resistance to the Ug99 stem rust race group. Proc. Natl. Acad. Sci. USA 2017, 114, E9483–E9492. [Google Scholar] [CrossRef]
- He, L.; Jing, Y.; Shen, J.; Li, X.; Liu, H.; Geng, Z.; Wang, M.; Li, Y.; Chen, D.; Gao, J.; et al. Mitochondrial pyruvate carriers prevent cadmium toxicity by sustaining the TCA cycle and glutathione synthesis. Plant Physiol. 2019, 180, 198–211. [Google Scholar] [CrossRef]
- Ranocha, P.; Dima, O.; Nagy, R.; Felten, J.; Corratgé-Faillie, C.; Novák, O.; Morreel, K.; Lacombe, B.; Martinez, Y.; Pfrunder, S.; et al. Arabidopsis WAT1 is a vacuolar auxin transport facilitator required for auxin homoeostasis. Nat. Commun. 2013, 4, 2625. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).