High-Throughput Field-Phenotyping Tools for Plant Breeding and Precision Agriculture
Abstract
1. Introduction
2. High-Throughput Field-Phenotyping (HTPP) Techniques
2.1. Satellite Imaging
2.2. UAVs
2.3. Proximal Phenotyping
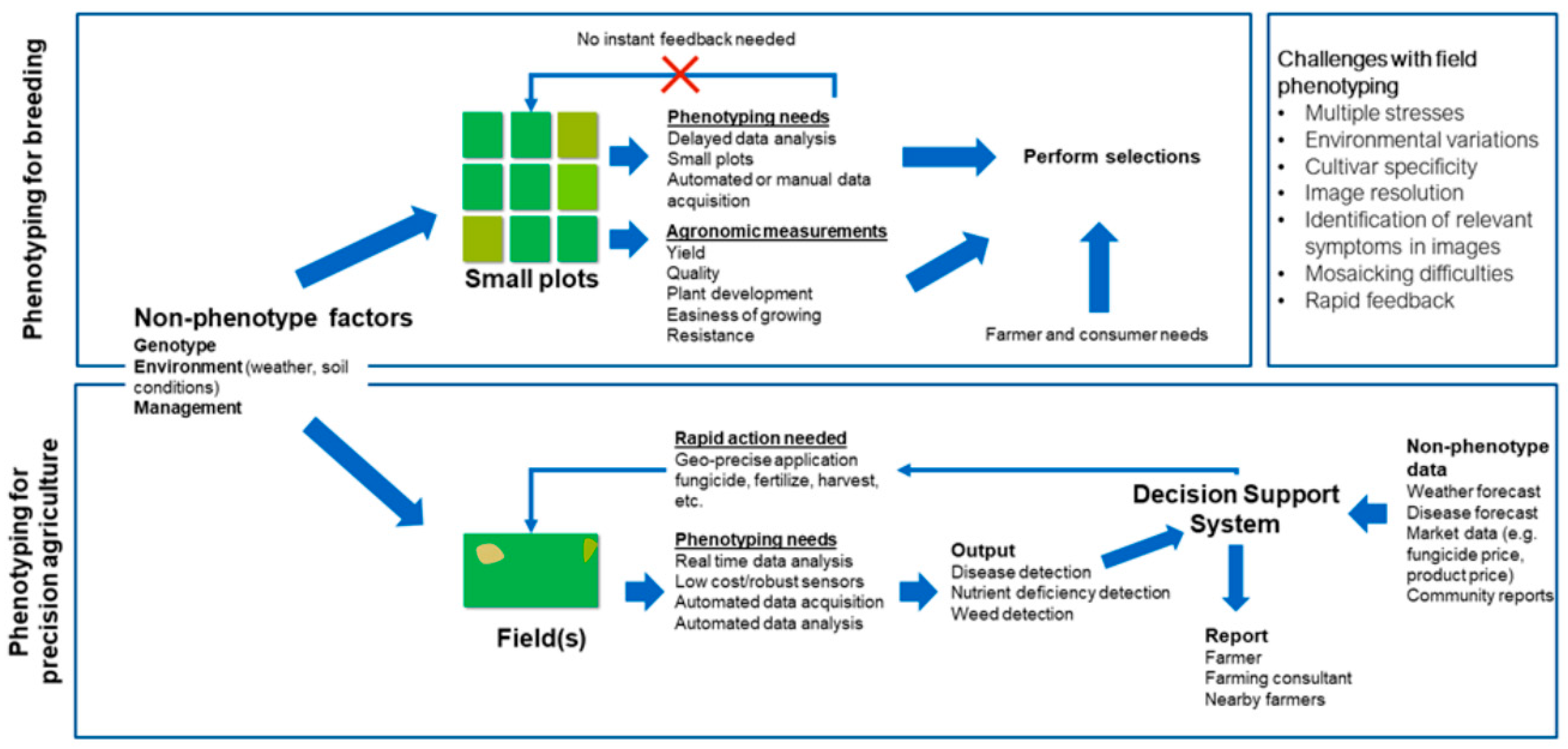
3. High-Throughput Phenotyping for Plant Breeding
4. Phenotyping for Precision Agriculture
4.1. Optimizing Fertilization
4.2. Detecting Diseases and Pests
4.3. Detecting Weeds
4.4. Decision Support Systems
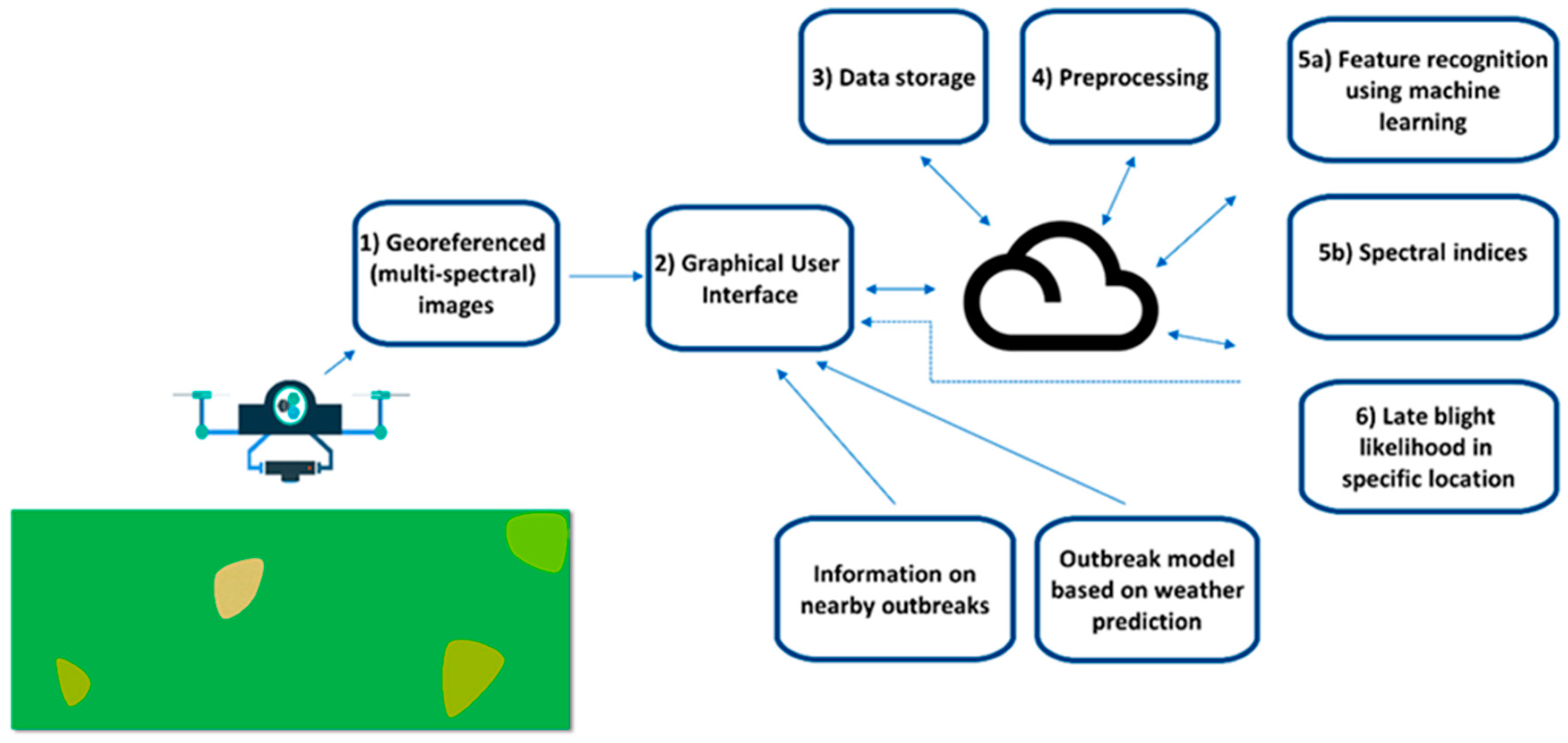
5. Case Study–Developing a DSS App to Combat Potato Late Blight
6. Outlook
7. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hunter, M.C.; Smith, R.G.; Schipanski, M.E.; Atwood, L.W.; Mortensen, D.A. Agriculture in 2050: Recalibrating Targets for Sustainable Intensification. Bioscience 2017, 67, 386–391. [Google Scholar] [CrossRef]
- Altieri, M.A.; Nicholls, C.I. The adaptation and mitigation potential of traditional agriculture in a changing climate. Clim. Chang. 2013, 140, 33–45. [Google Scholar] [CrossRef]
- Chawade, A.; Armoniene, R.; Berg, G.; Brazauskas, G.; Frostgard, G.; Geleta, M.; Gorash, A.; Henriksson, T.; Himanen, K.; Ingver, A.; et al. A transnational and holistic breeding approach is needed for sustainable wheat production in the Baltic Sea region. Physiol. Plant. 2018, 164, 442–451. [Google Scholar] [CrossRef] [PubMed]
- Collard, B.C.Y.; Mackill, D.J. Marker-assisted selection: An approach for precision plant breeding in the twenty-first century. Philos. Trans. R. Soc. B Boil. Sci. 2008, 363, 557–572. [Google Scholar] [CrossRef]
- Ghaffary, S.M.T.; Chawade, A.; Singh, P.K. Practical breeding strategies to improve resistance to Septoria tritici blotch of wheat. Euphytica 2018, 214, 122. [Google Scholar] [CrossRef]
- Bazakos, C.; Hanemian, M.; Trontin, C.; Jiménez-Gómez, J.M.; Loudet, O. New Strategies and Tools in Quantitative Genetics: How to Go from the Phenotype to the Genotype. Annu. Rev. Plant Biol. 2017, 68, 435–455. [Google Scholar] [CrossRef] [PubMed]
- Desta, Z.A.; Ortiz, R. Genomic selection: Genome-wide prediction in plant improvement. Trends Plant Sci. 2014, 19, 592–601. [Google Scholar] [CrossRef]
- Martin, L.B.B.; Fei, Z.; Giovannoni, J.J.; Rose, J.K.C. Catalyzing plant science research with RNA-seq. Front. Plant Sci. 2013, 4, 66. [Google Scholar] [CrossRef] [PubMed]
- Meijón, M.; Satbhai, S.B.; Tsuchimatsu, T.; Busch, W. Genome-wide association study using cellular traits identifies a new regulator of root development in Arabidopsis. Nat. Genet. 2013, 46, 77–81. [Google Scholar] [CrossRef]
- Chawade, A.; Alexandersson, E.; Bengtsson, T.; Andreasson, E.; Levander, F. Targeted proteomics approach for precision plant breeding. J. Proteome Res. 2016, 15, 638–646. [Google Scholar] [CrossRef]
- Acharjee, A.; Chibon, P.-Y.; Kloosterman, B.; America, T.; Renaut, J.; Maliepaard, C.; Visser, R.G.F. Genetical genomics of quality related traits in potato tubers using proteomics. BMC Plant Biol. 2018, 18, 20. [Google Scholar] [CrossRef]
- Alexandersson, E.; Jacobson, D.; Vivier, M.; Weckwerth, W.; Andreasson, E. Field-omics—Understanding large-scale molecular data from field crops. Front. Plant Sci. 2014, 5, 286. [Google Scholar] [CrossRef] [PubMed]
- Mahlein, A.K. Plant Disease Detection by Imaging Sensors—Parallels and Specific Demands for Precision Agriculture and Plant Phenotyping. Plant Dis. 2016, 100, 241–251. [Google Scholar] [CrossRef]
- Minervini, M.; Scharr, H.; Tsaftaris, S.A. Image Analysis: The New Bottleneck in Plant Phenotyping [Applications Corner]. IEEE Signal Process. Mag. 2015, 32, 126–131. [Google Scholar] [CrossRef]
- Reynolds, D.; Baret, F.; Welcker, C.; Bostrom, A.; Ball, J.; Cellini, F.; Lorence, A.; Chawade, A.; Khafif, M.; Noshita, K.; et al. What is cost-efficient phenotyping? Optimizing costs for different scenarios. Plant Sci. 2019, 282, 14–22. [Google Scholar] [CrossRef] [PubMed]
- Pretty, J.N. The sustainable intensification of agriculture. Nat. Resour. Forum 1997, 21, 247–256. [Google Scholar] [CrossRef]
- Elmgren, R.; Larsson, U. Nitrogen and the Baltic Sea: Managing nitrogen in relation to phosphorus. Sci. World J. 2001, 1, 371–377. [Google Scholar] [CrossRef] [PubMed]
- Heick, T.M.; Justesen, A.F.; Jørgensen, L.N. Resistance of wheat pathogen Zymoseptoria tritici to DMI and QoI fungicides in the Nordic-Baltic region—A status. Eur. J. Plant Pathol. 2017, 149, 669–682. [Google Scholar] [CrossRef]
- Kempenaar, C.; Been, T.; Booij, J.; van Evert, F.; Michielsen, J.-M.; Kocks, C. Advances in Variable Rate Technology Application in Potato in The Netherlands. Potato Res. 2018, 60, 295–305. [Google Scholar] [CrossRef] [PubMed]
- Mahlein, A.K.; Steiner, U.; Dehne, H.W.; Oerke, E.C. Spectral signatures of sugar beet leaves for the detection and differentiation of diseases. Precis. Agric. 2010, 11, 413–431. [Google Scholar] [CrossRef]
- Araus, J.L.; Kefauver, S.C.; Zaman-Allah, M.; Olsen, M.S.; Cairns, J.E. Translating High-Throughput Phenotyping into Genetic Gain. Trends Plant Sci. 2018, 23, 451–466. [Google Scholar] [CrossRef] [PubMed]
- Rutkoski, J.; Poland, J.; Mondal, S.; Autrique, E.; Pérez, L.G.; Crossa, J.; Reynolds, M.; Singh, R. Canopy Temperature and Vegetation Indices from High-Throughput Phenotyping Improve Accuracy of Pedigree and Genomic Selection for Grain Yield in Wheat. G3 Genes Genomes Genet. 2016, 6, 2799–2808. [Google Scholar] [CrossRef] [PubMed]
- Tattaris, M.; Reynolds, M.P.; Chapman, S.C. A Direct Comparison of Remote Sensing Approaches for High-Throughput Phenotyping in Plant Breeding. Front. Plant Sci. 2016, 7, 1131. [Google Scholar] [CrossRef]
- Yang, C. High resolution satellite imaging sensors for precision agriculture. Front. Agric. Sci. Eng. 2018, 5, 393–405. [Google Scholar] [CrossRef]
- Seelan, S.K.; Laguette, S.; Casady, G.M.; Seielstad, G.A. Remote sensing applications for precision agriculture: A learning community approach. Remote Sens. Environ. 2003, 88, 157–169. [Google Scholar] [CrossRef]
- Piikki, K.; Söderström, M. Digital soil mapping of arable land in Sweden—Validation of performance at multiple scales. Geoderma 2017. [Google Scholar] [CrossRef]
- Sankaran, S.; Khot, L.R.; Espinoza, C.Z.; Jarolmasjed, S.; Sathuvalli, V.R.; Vandemark, G.J.; Miklas, P.N.; Carter, A.H.; Pumphrey, M.O.; Knowles, N.R.; et al. Low-altitude, high-resolution aerial imaging systems for row and field crop phenotyping: A review. Eur. J. Agron. 2015, 70, 112–123. [Google Scholar] [CrossRef]
- Zaman-Allah, M.; Vergara, O.; Araus, J.L.; Tarekegne, A.; Magorokosho, C.; Zarco-Tejada, P.J.; Hornero, A.; Albà, A.H.; Das, B.; Craufurd, P.; et al. Unmanned aerial platform-based multi-spectral imaging for field phenotyping of maize. Plant Methods 2015, 11, 35. [Google Scholar] [CrossRef]
- De Souza, C.H.W.; Lamparelli, R.A.C.; Rocha, J.V.; Magalhães, P.S.G. Height estimation of sugarcane using an unmanned aerial system (UAS) based on structure from motion (SfM) point clouds. Int. J. Remote Sens. 2017, 38, 2218–2230. [Google Scholar] [CrossRef]
- Lelong, C.; Burger, P.; Jubelin, G.; Roux, B.; Labbé, S.; Baret, F. Assessment of Unmanned Aerial Vehicles Imagery for Quantitative Monitoring of Wheat Crop in Small Plots. Sensors 2008, 8, 3557–3585. [Google Scholar] [CrossRef]
- Xu, R.; Li, C.; Paterson, A.H.; Jiang, Y.; Sun, S.; Robertson, J.S. Aerial Images and Convolutional Neural Network for Cotton Bloom Detection. Front. Plant Sci. 2018, 8, 2235. [Google Scholar] [CrossRef]
- Ludovisi, R.; Tauro, F.; Salvati, R.; Khoury, S.; Mugnozza Scarascia, G.; Harfouche, A. UAV-Based Thermal Imaging for High-Throughput Field Phenotyping of Black Poplar Response to Drought. Front. Plant Sci. 2017, 8, 1681. [Google Scholar] [CrossRef] [PubMed]
- Khan, Z.; Rahimi-Eichi, V.; Haefele, S.; Garnett, T.; Miklavcic, S.J. Estimation of vegetation indices for high-throughput phenotyping of wheat using aerial imaging. Plant Methods 2018, 14, 20. [Google Scholar] [CrossRef]
- Gnädinger, F.; Schmidhalter, U. Digital Counts of Maize Plants by Unmanned Aerial Vehicles (UAVs). Remote Sens. 2017, 9, 544. [Google Scholar] [CrossRef]
- Madec, S.; Baret, F.; de Solan, B.; Thomas, S.; Dutartre, D.; Jezequel, S.; Hemmerlé, M.; Colombeau, G.; Comar, A. High-Throughput Phenotyping of Plant Height: Comparing Unmanned Aerial Vehicles and Ground LiDAR Estimates. Front. Plant Sci. 2017, 8, 2002. [Google Scholar] [CrossRef] [PubMed]
- Geipel, J.; Link, J.; Claupein, W. Combined Spectral and Spatial Modeling of Corn Yield Based on Aerial Images and Crop Surface Models Acquired with an Unmanned Aircraft System. Remote Sens. 2014, 6, 10335–10355. [Google Scholar] [CrossRef]
- Bendig, J.; Yu, K.; Aasen, H.; Bolten, A.; Bennertz, S.; Broscheit, J.; Gnyp, M.L.; Bareth, G. Combining UAV-based plant height from crop surface models, visible, and near infrared vegetation indices for biomass monitoring in barley. Int. J. Appl. Earth Obs. Geoinform. 2015, 39, 79–87. [Google Scholar] [CrossRef]
- Chapman, S.; Merz, T.; Chan, A.; Jackway, P.; Hrabar, S.; Dreccer, M.; Holland, E.; Zheng, B.; Ling, T.; Jimenez-Berni, J. Pheno-Copter: A Low-Altitude, Autonomous Remote-Sensing Robotic Helicopter for High-Throughput Field-Based Phenotyping. Agronomy 2014, 4, 279–301. [Google Scholar] [CrossRef]
- Deery, D.; Jimenez-Berni, J.; Jones, H.; Sirault, X.; Furbank, R. Proximal Remote Sensing Buggies and Potential Applications for Field-Based Phenotyping. Agronomy 2014, 4, 349–379. [Google Scholar] [CrossRef]
- Keener, M.E.; Kircher, P.L. The use of canopy temperature as an indicator of drought stress in humid regions. Agric. Meteorol. 1983, 28, 339–349. [Google Scholar] [CrossRef]
- Koc, A.; Henriksson, T.; Chawade, A. Specalyzer—An interactive online tool to analyze spectral reflectance measurements. PeerJ 2018, 6, e5031. [Google Scholar] [CrossRef] [PubMed]
- Garriga, M.; Romero-Bravo, S.; Estrada, F.; Escobar, A.; Matus, I.A.; del Pozo, A.; Astudillo, C.A.; Lobos, G.A. Assessing Wheat Traits by Spectral Reflectance: Do We Really Need to Focus on Predicted Trait-Values or Directly Identify the Elite Genotypes Group? Front. Plant Sci. 2017, 8, 280. [Google Scholar] [CrossRef]
- Huang, W.; Lamb, D.W.; Niu, Z.; Zhang, Y.; Liu, L.; Wang, J. Identification of yellow rust in wheat using in-situ spectral reflectance measurements and airborne hyperspectral imaging. Precis. Agric. 2007, 8, 187–197. [Google Scholar] [CrossRef]
- Odilbekov, F.; Armoniene, R.; Henriksson, T.; Chawade, A. Proximal Phenotyping and Machine Learning Methods to Identify Septoria Tritici Blotch Disease Symptoms in Wheat. Front. Plant Sci. 2018, 9, 685. [Google Scholar] [CrossRef] [PubMed]
- Debaeke, P.; Rouet, P.; Justes, E. Relationship Between the Normalized SPAD Index and the Nitrogen Nutrition Index: Application to Durum Wheat. J. Plant Nutr. 2006, 29, 75–92. [Google Scholar] [CrossRef]
- Yang, H.; Yang, J.; Lv, Y.; He, J. SPAD Values and Nitrogen Nutrition Index for the Evaluation of Rice Nitrogen Status. Plant Prod. Sci. 2015, 17, 81–92. [Google Scholar] [CrossRef]
- Andrianto, H.; Suhardi; Faizal, A. Measurement of chlorophyll content to determine nutrition deficiency in plants: A systematic literature review. In Proceedings of the 2017 International Conference on Information Technology Systems and Innovation (ICITSI), Bandung, Indonesia, 23–24 October 2017; pp. 392–397. [Google Scholar]
- Chawade, A.; Linden, P.; Brautigam, M.; Jonsson, R.; Jonsson, A.; Moritz, T.; Olsson, O. Development of a model system to identify differences in spring and winter oat. PLoS ONE 2012, 7, e29792. [Google Scholar] [CrossRef] [PubMed]
- Crain, J.L.; Wei, Y.; Barker, J.; Thompson, S.M.; Alderman, P.D.; Reynolds, M.; Zhang, N.; Poland, J. Development and Deployment of a Portable Field Phenotyping Platform. Crop Sci. 2016, 56, 965–975. [Google Scholar] [CrossRef]
- White, J.W.; Conley, M.M. A Flexible, Low-Cost Cart for Proximal Sensing. Crop Sci. 2013, 53, 1646–1649. [Google Scholar] [CrossRef]
- Thompson, A.L.; Thorp, K.R.; Conley, M.; Andrade-Sanchez, P.; Heun, J.T.; Dyer, J.M.; White, J.W. Deploying a Proximal Sensing Cart to Identify Drought-Adaptive Traits in Upland Cotton for High-Throughput Phenotyping. Front. Plant Sci. 2018, 9, 507. [Google Scholar] [CrossRef] [PubMed]
- Bai, G.; Ge, Y.; Hussain, W.; Baenziger, P.S.; Graef, G. A multi-sensor system for high throughput field phenotyping in soybean and wheat breeding. Comput. Electron. Agric. 2016, 128, 181–192. [Google Scholar] [CrossRef]
- Barmeier, G.; Schmidhalter, U. High-Throughput Field Phenotyping of Leaves, Leaf Sheaths, Culms and Ears of Spring Barley Cultivars at Anthesis and Dough Ripeness. Front. Plant Sci. 2017, 8, 1920. [Google Scholar] [CrossRef]
- Rischbeck, P.; Elsayed, S.; Mistele, B.; Barmeier, G.; Heil, K.; Schmidhalter, U. Data fusion of spectral, thermal and canopy height parameters for improved yield prediction of drought stressed spring barley. Eur. J. Agron. 2016, 78, 44–59. [Google Scholar] [CrossRef]
- Barmeier, G.; Schmidhalter, U. High-Throughput Phenotyping of Wheat and Barley Plants Grown in Single or Few Rows in Small Plots Using Active and Passive Spectral Proximal Sensing. Sensors 2016, 16, 1860. [Google Scholar] [CrossRef]
- Andrade-Sanchez, P.; Gore, M.A.; Heun, J.T.; Thorp, K.R.; Carmo-Silva, A.E.; French, A.N.; Salvucci, M.E.; White, J.W. Development and evaluation of a field-based high-throughput phenotyping platform. Funct. Plant Biol. 2014, 41, 68–79. [Google Scholar] [CrossRef]
- Sun, S.; Li, C.; Paterson, A.H.; Jiang, Y.; Xu, R.; Robertson, J.S.; Snider, J.L.; Chee, P.W. In-field High Throughput Phenotyping and Cotton Plant Growth Analysis Using LiDAR. Front. Plant Sci. 2018, 9, 16. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Li, C.; Robertson, J.S.; Sun, S.; Xu, R.; Paterson, A.H. Gphenovision: A Ground Mobile System with Multi-modal Imaging for Field-Based High Throughput Phenotyping of Cotton. Sci. Rep. 2018, 8, 1213. [Google Scholar] [CrossRef] [PubMed]
- Kirchgessner, N.; Liebisch, F.; Yu, K.; Pfeifer, J.; Friedli, M.; Hund, A.; Walter, A. The ETH field phenotyping platform FIP: A cable-suspended multi-sensor system. Funct. Plant Boil. 2017, 44, 154–168. [Google Scholar] [CrossRef]
- Xu, Y.; Li, P.; Zou, C.; Lu, Y.; Xie, C.; Zhang, X.; Prasanna, B.M.; Olsen, M.S. Enhancing genetic gain in the era of molecular breeding. J. Exp. Bot. 2017, 68, 2641–2666. [Google Scholar] [CrossRef]
- Hatfield, J.L.; Walthall, C.L. Meeting Global Food Needs: Realizing the Potential via Genetics × Environment × Management Interactions. Agron. J. 2015, 107, 1215–1226. [Google Scholar] [CrossRef]
- Hunt, E.R.; Daughtry, C.S.T. What good are unmanned aircraft systems for agricultural remote sensing and precision agriculture? Int. J. Remote Sens. 2017, 39, 5345–5376. [Google Scholar] [CrossRef]
- Crain, J.; Mondal, S.; Rutkoski, J.; Singh, R.P.; Poland, J. Combining High-Throughput Phenotyping and Genomic Information to Increase Prediction and Selection Accuracy in Wheat Breeding. Plant Genome 2018, 11. [Google Scholar] [CrossRef]
- Juliana, P.; Montesinos-López, O.A.; Crossa, J.; Mondal, S.; González Pérez, L.; Poland, J.; Huerta-Espino, J.; Crespo-Herrera, L.; Govindan, V.; Dreisigacker, S.; et al. Integrating genomic-enabled prediction and high-throughput phenotyping in breeding for climate-resilient bread wheat. Theor. Appl. Genet. 2018, 132, 177–194. [Google Scholar] [CrossRef] [PubMed]
- Brown, T.B.; Cheng, R.; Sirault, X.R.R.; Rungrat, T.; Murray, K.D.; Trtilek, M.; Furbank, R.T.; Badger, M.; Pogson, B.J.; Borevitz, J.O. TraitCapture: Genomic and environment modelling of plant phenomic data. Curr. Opin. Plant Biol. 2014, 18, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Ganapathysubramanian, B.; Singh, A.K.; Sarkar, S. Machine Learning for High-Throughput Stress Phenotyping in Plants. Trends Plant Sci. 2016, 21, 110–124. [Google Scholar] [CrossRef]
- Shakoor, N.; Lee, S.; Mockler, T.C. High throughput phenotyping to accelerate crop breeding and monitoring of diseases in the field. Curr. Opin. Plant Biol. 2017, 38, 184–192. [Google Scholar] [CrossRef]
- Joalland, S.; Screpanti, C.; Liebisch, F.; Varella, H.V.; Gaume, A.; Walter, A. Comparison of visible imaging, thermography and spectrometry methods to evaluate the effect of Heterodera schachtii inoculation on sugar beets. Plant Methods 2017, 13, 73. [Google Scholar] [CrossRef]
- Joalland, S.; Screpanti, C.; Varella, H.; Reuther, M.; Schwind, M.; Lang, C.; Walter, A.; Liebisch, F. Aerial and Ground Based Sensing of Tolerance to Beet Cyst Nematode in Sugar Beet. Remote Sens. 2018, 10, 787. [Google Scholar] [CrossRef]
- Kuska, M.T.; Mahlein, A.K. Aiming at decision making in plant disease protection and phenotyping by the use of optical sensors. Eur. J. Plant Pathol. 2018, 152, 987–992. [Google Scholar] [CrossRef]
- Wu, D.; Ma, C. The Support Vector Machine (SVM) Based Near-Infrared Spectrum Recognition of Leaves Infected by the Leafminers. In Proceedings of the First International Conference on Innovative Computing, Information and Control, Volume I (ICICIC’06), Beijing, China, 30 August–1 September 2006; pp. 448–451. [Google Scholar]
- Mohanty, S.P.; Hughes, D.P.; Salathé, M. Using Deep Learning for Image-Based Plant Disease Detection. Front. Plant Sci. 2016, 7, 1419. [Google Scholar] [CrossRef]
- Sa, I.; Popović, M.; Khanna, R.; Chen, Z.; Lottes, P.; Liebisch, F.; Nieto, J.; Stachniss, C.; Walter, A.; Siegwart, R. WeedMap: A Large-Scale Semantic Weed Mapping Framework Using Aerial Multispectral Imaging and Deep Neural Network for Precision Farming. Remote Sens. 2018, 10, 1423. [Google Scholar] [CrossRef]
- Sawyer, J.E. Concepts of Variable Rate Technology with Considerations for Fertilizer Application. J. Prod. Agric. 1994, 7, 195–201. [Google Scholar] [CrossRef]
- Reyes, J.F.; Esquivel, W.; Cifuentes, D.; Ortega, R. Field testing of an automatic control system for variable rate fertilizer application. Comput. Electron. Agric. 2015, 113, 260–265. [Google Scholar] [CrossRef]
- Naik, H.S.; Zhang, J.; Lofquist, A.; Assefa, T.; Sarkar, S.; Ackerman, D.; Singh, A.; Singh, A.K.; Ganapathysubramanian, B. A real-time phenotyping framework using machine learning for plant stress severity rating in soybean. Plant Methods 2017, 13, 23. [Google Scholar] [CrossRef]
- Bravo, C.; Moshou, D.; West, J.; McCartney, A.; Ramon, H. Early Disease Detection in Wheat Fields using Spectral Reflectance. Biosyst. Eng. 2003, 84, 137–145. [Google Scholar] [CrossRef]
- Griffel, L.M.; Delparte, D.; Edwards, J. Using Support Vector Machines classification to differentiate spectral signatures of potato plants infected with Potato Virus Y. Comput. Electron. Agric. 2018, 153, 318–324. [Google Scholar] [CrossRef]
- Sugiura, R.; Tsuda, S.; Tamiya, S.; Itoh, A.; Nishiwaki, K.; Murakami, N.; Shibuya, Y.; Hirafuji, M.; Nuske, S. Field phenotyping system for the assessment of potato late blight resistance using RGB imagery from an unmanned aerial vehicle. Biosystems Engineering 2016, 148, 1–10. [Google Scholar] [CrossRef]
- Franceschini, M.H.D.; Bartholomeus, H.; van Apeldoorn, D.F.; Suomalainen, J.; Kooistra, L. Feasibility of Unmanned Aerial Vehicle Optical Imagery for Early Detection and Severity Assessment of Late Blight in Potato. Remote Sens. 2019, 11, 224. [Google Scholar] [CrossRef]
- Oerke, E.C.; Dehne, H.W. Safeguarding production—Losses in major crops and the role of crop protection. Crop Prot. 2004, 23, 275–285. [Google Scholar] [CrossRef]
- Harker, K.N.; O’Donovan, J.T. Recent Weed Control, Weed Management, and Integrated Weed Management. Weed Technol. 2017, 27, 1–11. [Google Scholar] [CrossRef]
- Gonzalez-Andujar, J.L.; Huang, H.; Deng, J.; Lan, Y.; Yang, A.; Deng, X.; Zhang, L. A fully convolutional network for weed mapping of unmanned aerial vehicle (UAV) imagery. PLoS ONE 2018, 13, e0196302. [Google Scholar] [CrossRef]
- Peña, J.; Torres-Sánchez, J.; Serrano-Pérez, A.; de Castro, A.; López-Granados, F. Quantifying Efficacy and Limits of Unmanned Aerial Vehicle (UAV) Technology for Weed Seedling Detection as Affected by Sensor Resolution. Sensors 2015, 15, 5609–5626. [Google Scholar] [CrossRef] [PubMed]
- Rose, D.C.; Sutherland, W.J.; Parker, C.; Lobley, M.; Winter, M.; Morris, C.; Twining, S.; Ffoulkes, C.; Amano, T.; Dicks, L.V. Decision support tools for agriculture: Towards effective design and delivery. Agric. Syst. 2016, 149, 165–174. [Google Scholar] [CrossRef]
- Johannes, A.; Picon, A.; Alvarez-Gila, A.; Echazarra, J.; Rodriguez-Vaamonde, S.; Navajas, A.D.; Ortiz-Barredo, A. Automatic plant disease diagnosis using mobile capture devices, applied on a wheat use case. Comput. Electron. Agric. 2017, 138, 200–209. [Google Scholar] [CrossRef]
- Ramcharan, A.; Baranowski, K.; McCloskey, P.; Ahmed, B.; Legg, J.; Hughes, D.P. Deep Learning for Image-Based Cassava Disease Detection. Front. Plant Sci. 2017, 8, 1852. [Google Scholar] [CrossRef]
- Hallau, L.; Neumann, M.; Klatt, B.; Kleinhenz, B.; Klein, T.; Kuhn, C.; Röhrig, M.; Bauckhage, C.; Kersting, K.; Mahlein, A.K.; et al. Automated identification of sugar beet diseases using smartphones. Plant Pathol. 2018, 67, 399–410. [Google Scholar] [CrossRef]
- Devaux, A.; Kromann, P.; Ortiz, O. Potatoes for sustainable global food security. Potato Res. 2014, 57, 185–199. [Google Scholar] [CrossRef]
- Eriksson, D.; Carlson-Nilsson, U.; Ortíz, R.; Andreasson, E. Overview and Breeding Strategies of Table Potato Production in Sweden and the Fennoscandian Region. Potato Res. 2016, 59, 279–294. [Google Scholar] [CrossRef]
- Gijzen, M.; Lehsten, V.; Wiik, L.; Hannukkala, A.; Andreasson, E.; Chen, D.; Ou, T.; Liljeroth, E.; Lankinen, Å.; Grenville-Briggs, L. Earlier occurrence and increased explanatory power of climate for the first incidence of potato late blight caused by Phytophthora infestans in Fennoscandia. PLoS ONE 2017, 12, e0177580. [Google Scholar] [CrossRef]
- Fry, W.E. Evaluation of Potato Late Blight Forecasts Modified to Incorporate Host Resistance and Fungicide Weathering. Phytopathology 1983, 73, 1054–1059. [Google Scholar] [CrossRef]
- Alexandersson, E.; Antkowiak, P.; Holmberg, M.; Piikki, K.; Söderström, M.; Liljeroth, E. The possibilities and challenges of UAV-borne remote sensing for detection of potato late blight in the field. In Abstract Book for the Plant Biology Europe Conference in Copenhagen; Department of Plant and Environmental Sciences, University of Copenhagen: Copenhagen, Denmark, 2018; p. 10. ISBN 978-87-996274-1-7. [Google Scholar]
- Brown, B. The Total Economic Impact™ of IBM’s Design Thinking Practice; Forrester Consulting: Massachusetts, MA, USA, 2018; pp. 1–45. [Google Scholar]
- Ćwiek-Kupczyńska, H.; Altmann, T.; Arend, D.; Arnaud, E.; Chen, D.; Cornut, G.; Fiorani, F.; Frohmberg, W.; Junker, A.; Klukas, C.; et al. Measures for interoperability of phenotypic data: Minimum information requirements and formatting. Plant Methods 2016, 12, 44. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chawade, A.; van Ham, J.; Blomquist, H.; Bagge, O.; Alexandersson, E.; Ortiz, R. High-Throughput Field-Phenotyping Tools for Plant Breeding and Precision Agriculture. Agronomy 2019, 9, 258. https://doi.org/10.3390/agronomy9050258
Chawade A, van Ham J, Blomquist H, Bagge O, Alexandersson E, Ortiz R. High-Throughput Field-Phenotyping Tools for Plant Breeding and Precision Agriculture. Agronomy. 2019; 9(5):258. https://doi.org/10.3390/agronomy9050258
Chicago/Turabian StyleChawade, Aakash, Joost van Ham, Hanna Blomquist, Oscar Bagge, Erik Alexandersson, and Rodomiro Ortiz. 2019. "High-Throughput Field-Phenotyping Tools for Plant Breeding and Precision Agriculture" Agronomy 9, no. 5: 258. https://doi.org/10.3390/agronomy9050258
APA StyleChawade, A., van Ham, J., Blomquist, H., Bagge, O., Alexandersson, E., & Ortiz, R. (2019). High-Throughput Field-Phenotyping Tools for Plant Breeding and Precision Agriculture. Agronomy, 9(5), 258. https://doi.org/10.3390/agronomy9050258






