Transcriptome Analysis of Banana (Musa acuminate L.) in Response to Low-Potassium Stress
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Low −K+ Stress Treatment
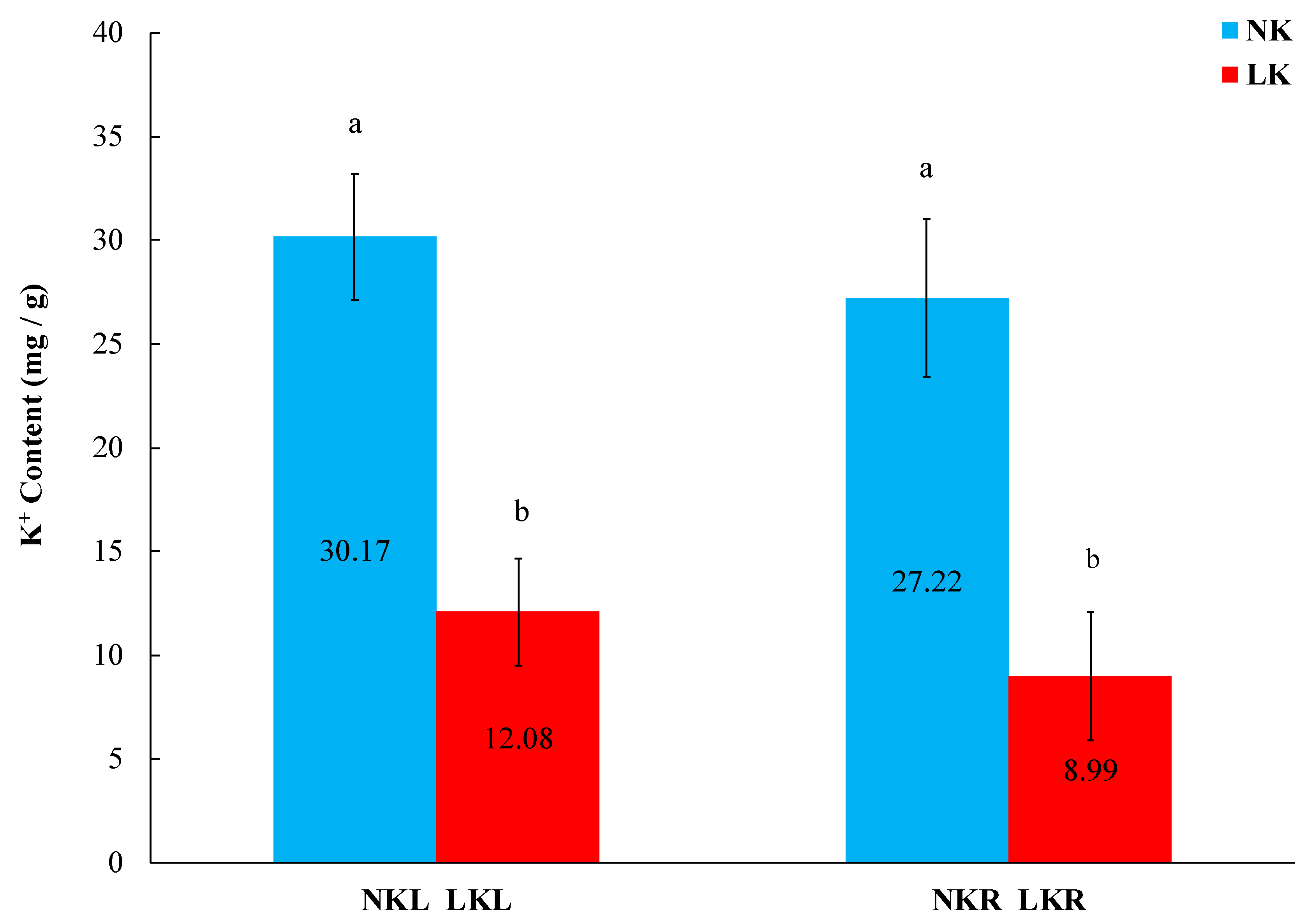
2.2. Determination of Phenotypic Traits and Measurement of K+ Content
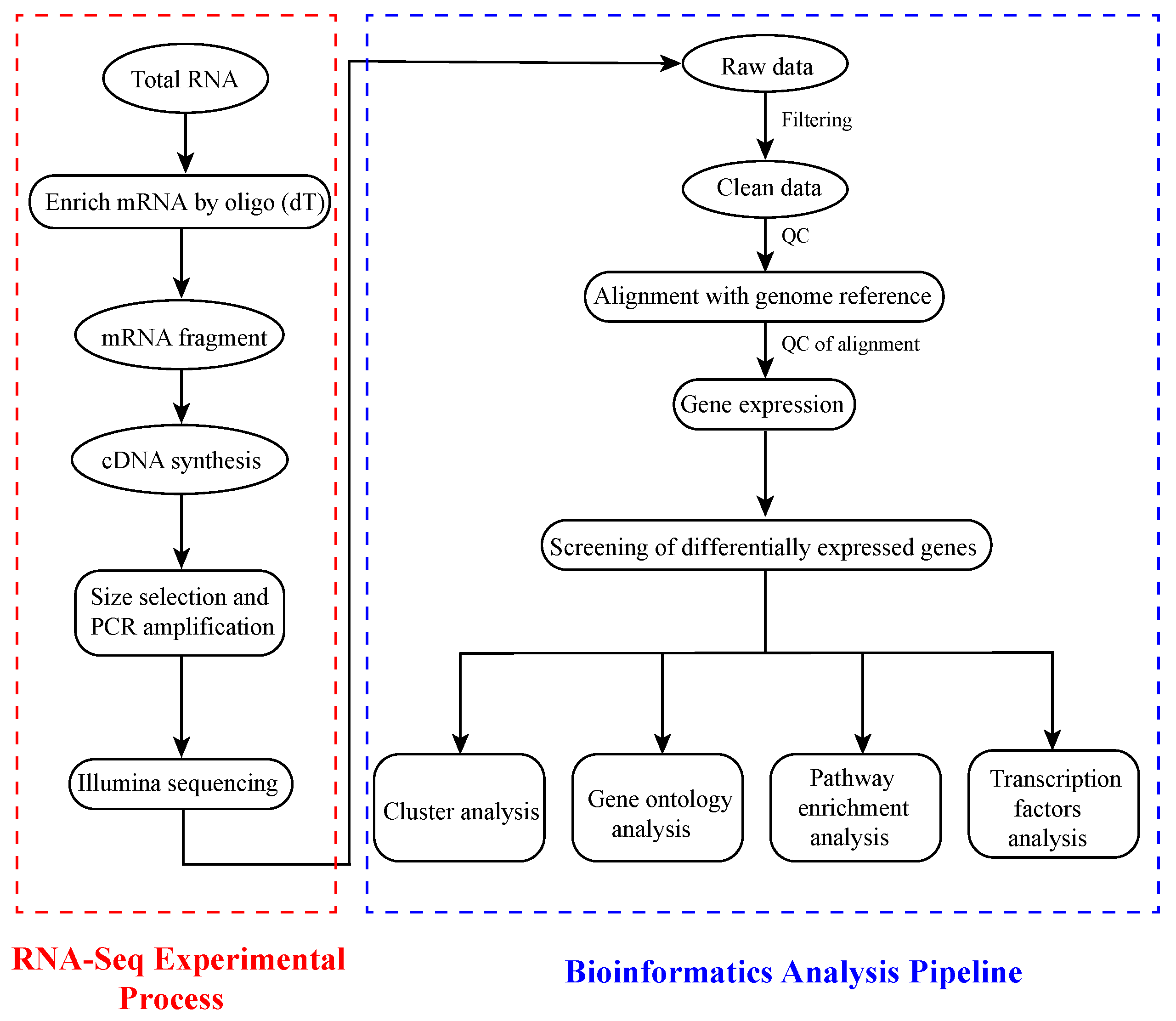
2.3. RNA-Seq and Bioinformatics Analysis
2.4. Differentially Expressed Genes (DEGs) Screening
2.5. Quantitative Real-Time PCR (qRT-PCR) Analysis
3. Results
3.1. K+ Deficiency Inhibits the Growth of Banana Seedlings
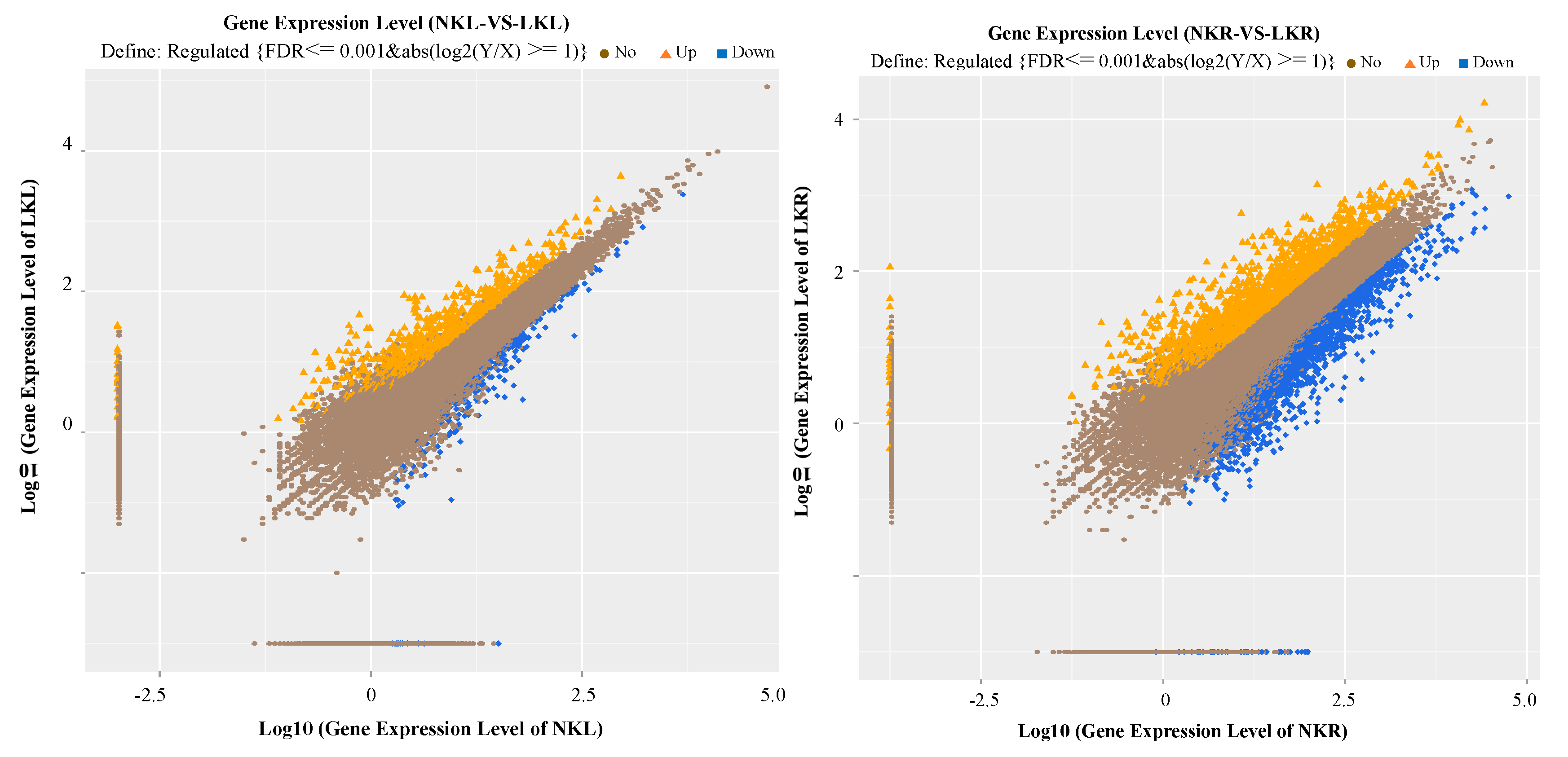
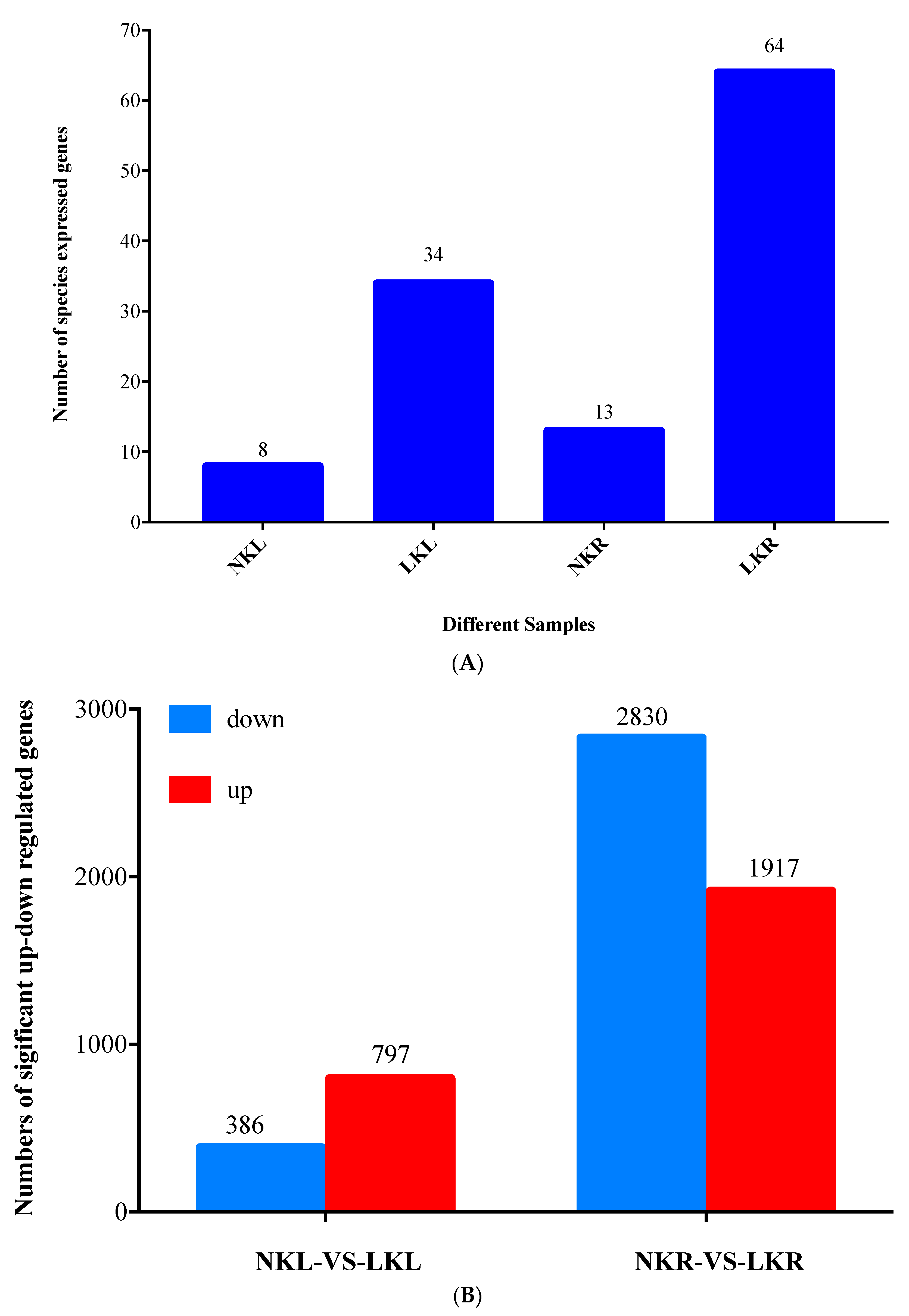
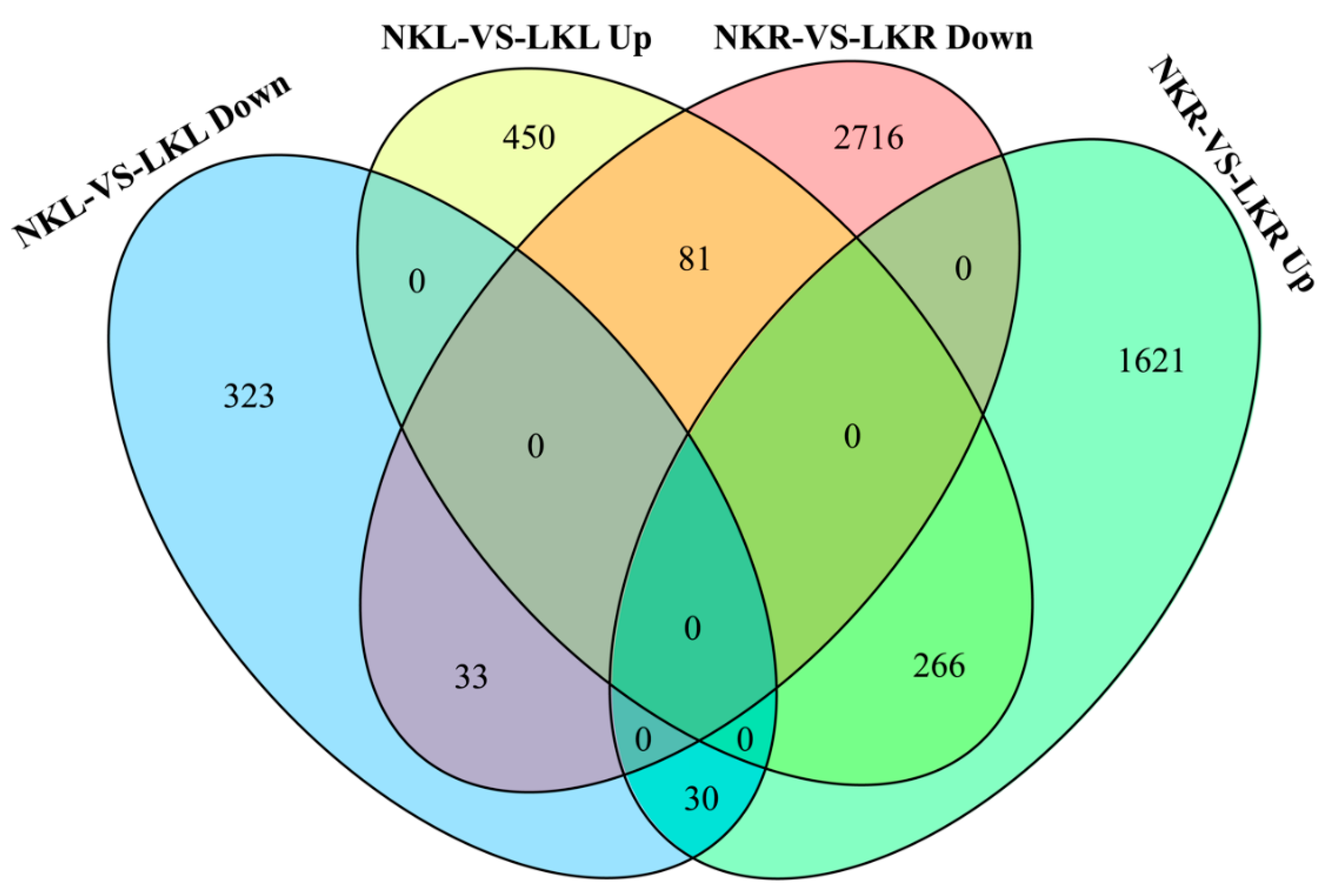
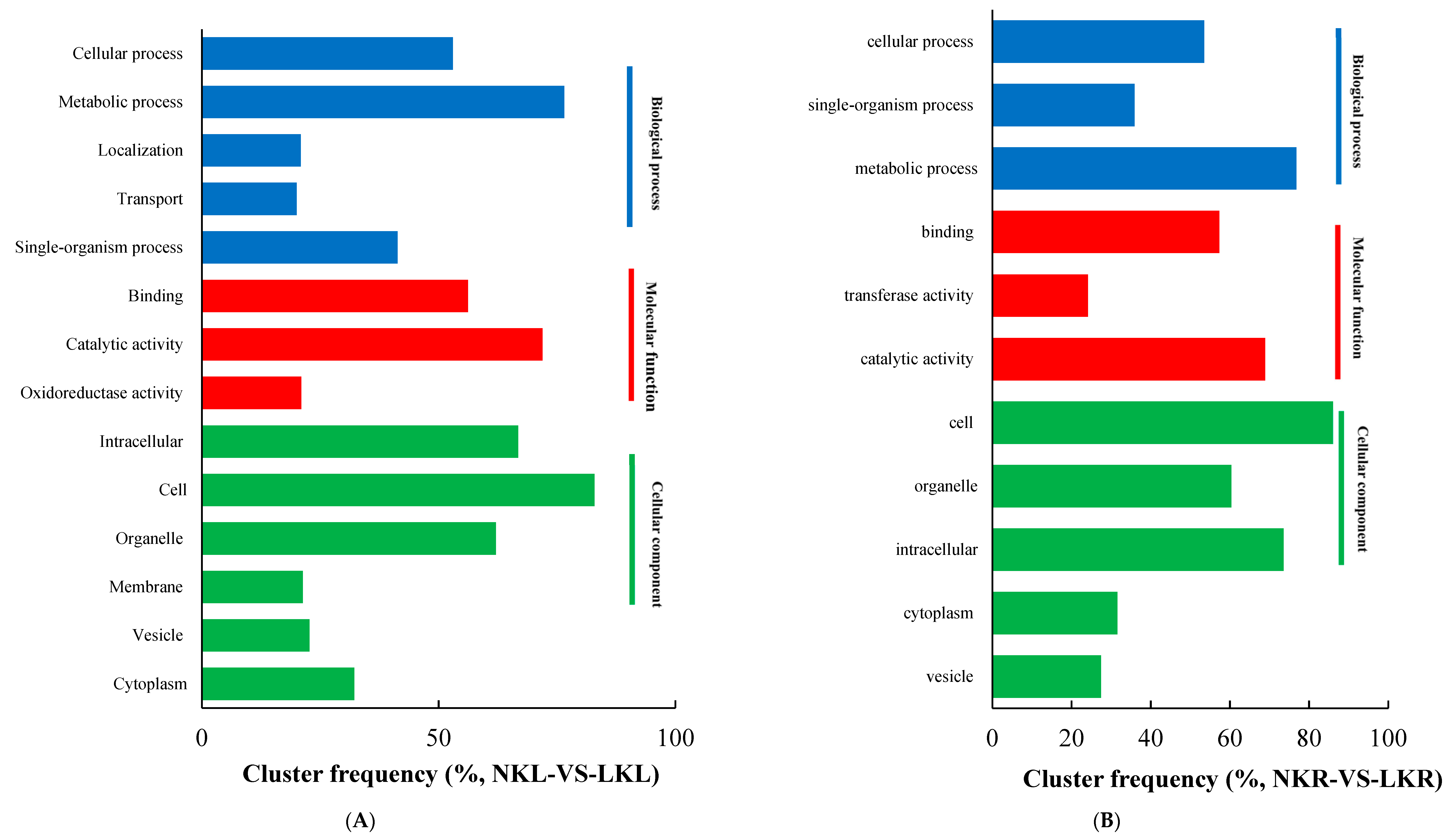
3.2. RNA Sequencing, Alignment, and Bioinformatics Analysis in Banana Leaves and Roots
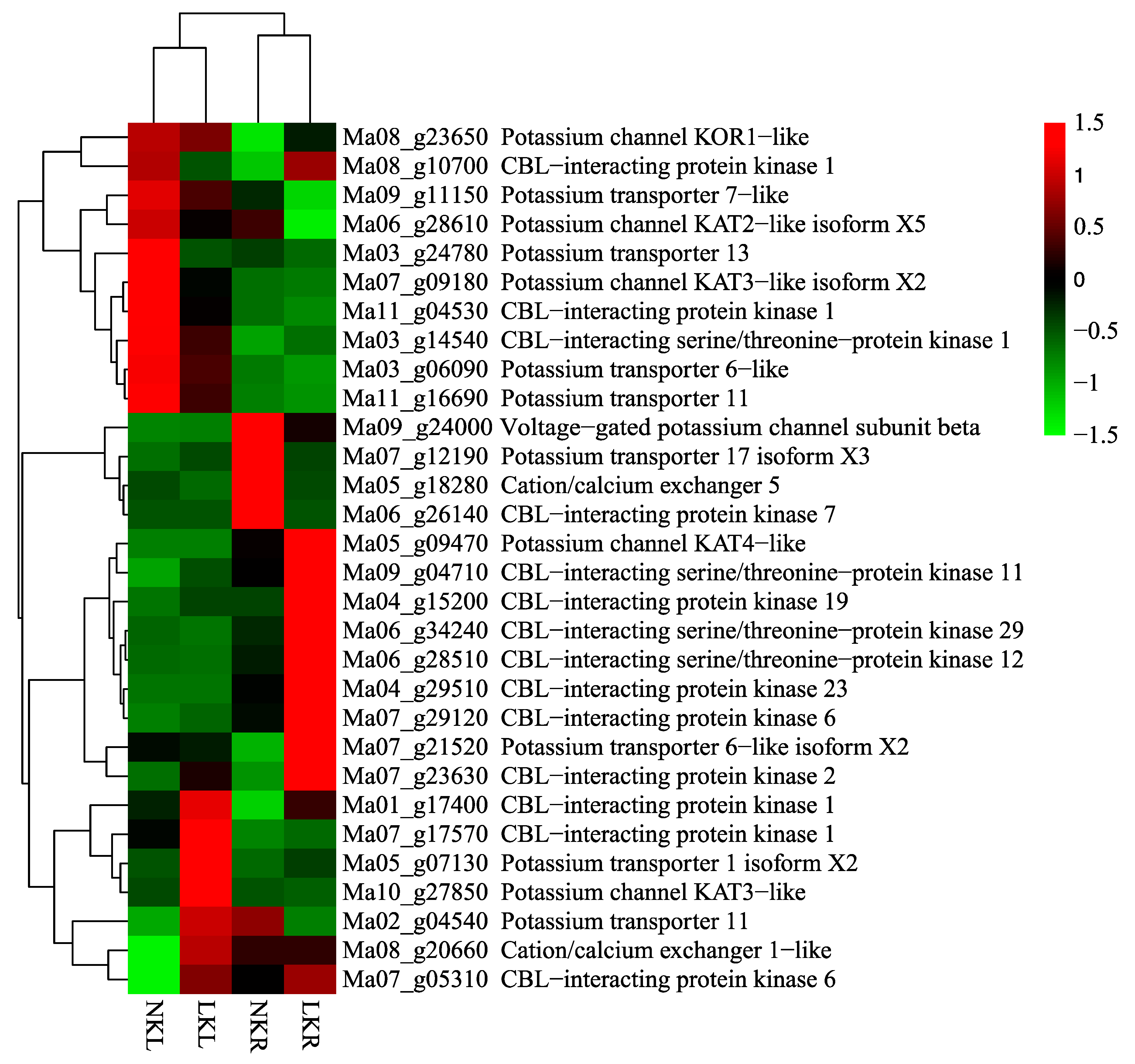
3.3. DEGs Related to K+ Transport and Uptake in Roots and Leaves
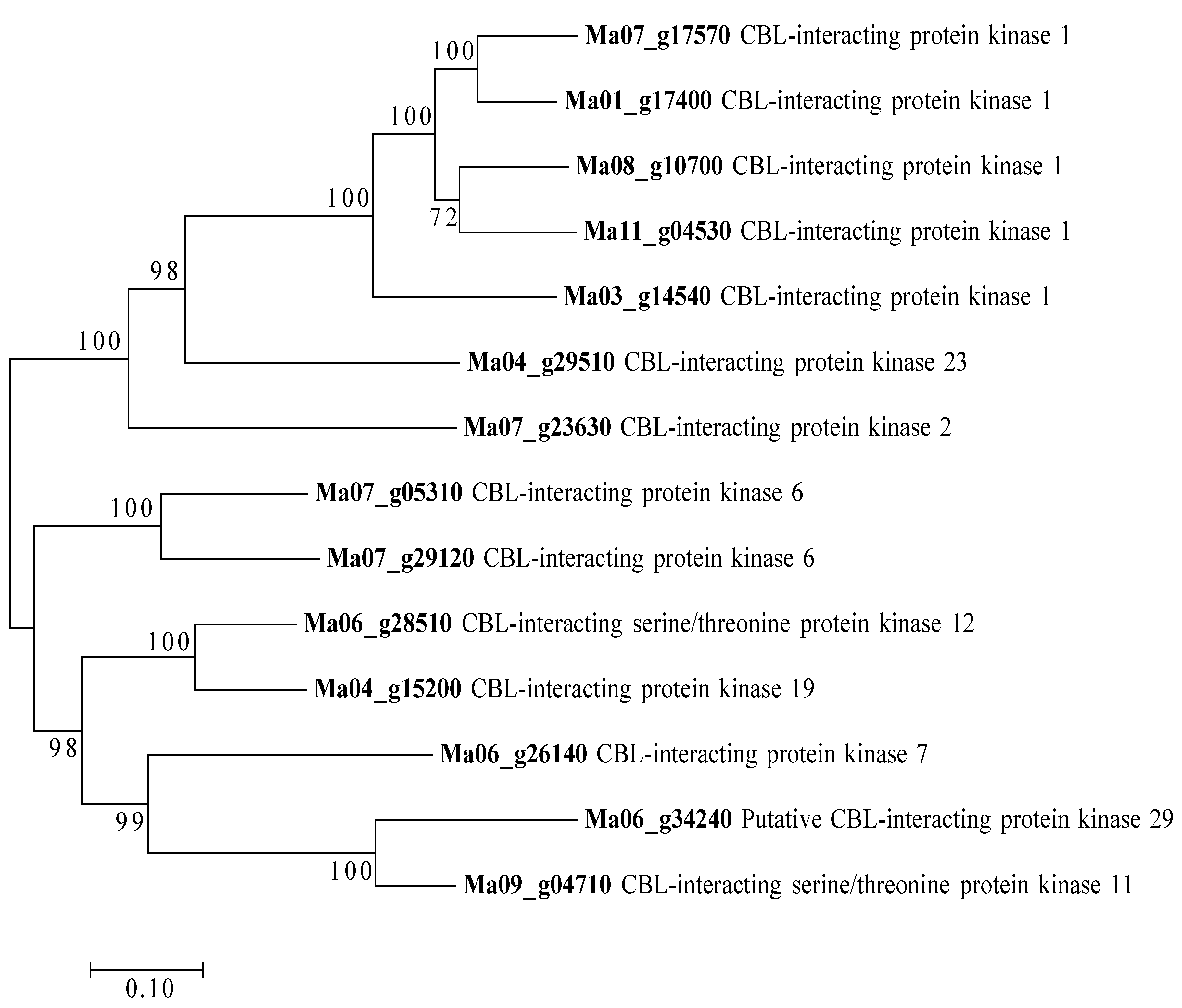
3.4. Transcription Factors (TFs) Analysis in Banana Leaves and Roots
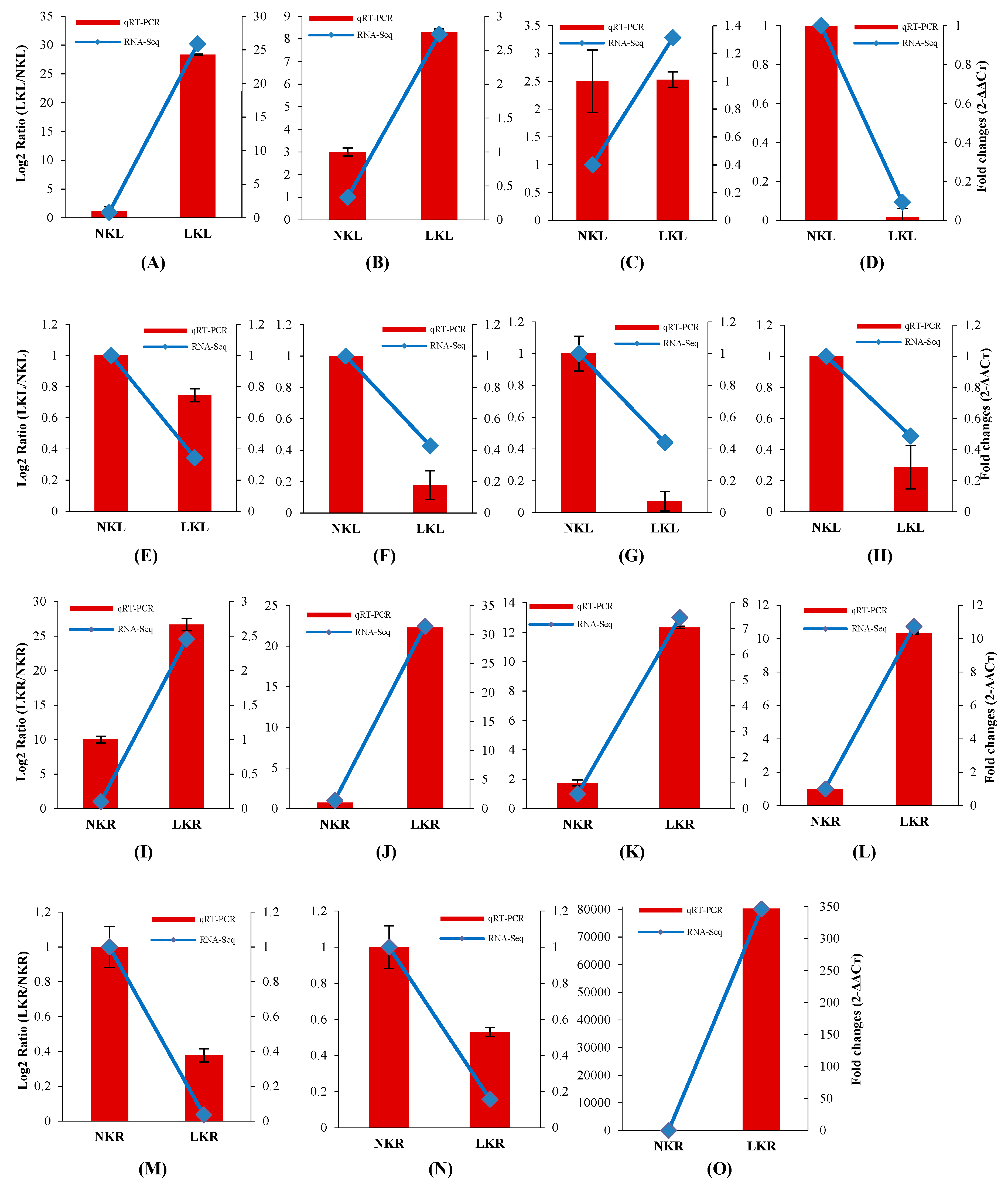
3.5. Validation of Gene Expression by qRT-PCR
4. Discussion
5. Conclusions
Supplementary Materials
Availability of Supporting Data
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Doran, I.; Sen, B.; Kaya, Z. The effects of compost prepared from waste material of banana plants on the nutrient contents of banana leaves. J. Environ. Biol. 2003, 24, 437–444. [Google Scholar]
- And, D.T.C.; Hanson, J.B. The Mineral Nutrition of Higher Plants. Annu. Rev. Plant Physiol. 1980, 31, 239–298. [Google Scholar]
- Leigh, R.A. A Hypothesis Relating Critical Potassium Concentrations for Growth to the Distribution and Functions of this Ion in the Plant Cell. New Phytol. 2010, 97, 1–13. [Google Scholar] [CrossRef]
- Jin, S.H.; Huang, J.Q.; Li, X.Q.; Zheng, B.S.; Wu, J.S.; Wang, Z.J.; Liu, G.H.; Chen, M. Effects of potassium supply on limitations of photosynthesis by mesophyll diffusion conductance in Carya cathayensis. Tree Physiol. 2011, 31, 1142–1151. [Google Scholar] [CrossRef]
- Luan, S. The CBL-CIPK network in plant calcium signaling. Trends Plant Sci. 2009, 14, 37–42. [Google Scholar] [CrossRef]
- Hedrich, R.; Kudla, J. Calcium signaling networks channel plant K+ uptake. Cell 2006, 125, 1221–1223. [Google Scholar] [CrossRef]
- Ma, T.L.; Wu, W.H.; Wang, Y. Transcriptome analysis of rice root responses to potassium deficiency. BMC Plant Biol. 2012, 12, 161. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, W.H. Regulation of potassium transport and signaling in plants. Curr. Opin. Plant Biol. 2017, 39, 123–128. [Google Scholar] [CrossRef]
- Zhang, X.; Jiang, H.; Wang, H.; Cui, J.; Wang, J.; Hu, J.; Guo, L.; Qian, Q.; Xue, D. Transcriptome Analysis of Rice Seedling Roots in Response to Potassium Deficiency. Sci. Rep. 2017, 7, 5523. [Google Scholar] [CrossRef]
- Ruan, L.; Zhang, J.; Xin, X.; Zhang, C.; Ma, D.; Chen, L.; Zhao, B. Comparative analysis of potassium deficiency-responsive transcriptomes in low potassium susceptible and tolerant wheat (Triticum aestivum L.). Sci. Rep. 2015, 5, 10090. [Google Scholar] [CrossRef]
- Wang, Q.; Guan, C.; Wang, P.; Lv, M.L.; Ma, Q.; Wu, G.Q.; Bao, A.K.; Zhang, J.L.; Wang, S.M. AtHKT1;1 and AtHAK5 mediate low-affinity Na+ uptake in Arabidopsis thaliana under mild salt stress. Plant Growth Regul. 2015, 75, 615–623. [Google Scholar] [CrossRef]
- Haro, R.; Fraile-Escanciano, A.; Gonzalez-Melendi, P.; Rodriguez-Navarro, A. The potassium transporters HAK2 and HAK3 localize to endomembranes in Physcomitrella patens. HAK2 is required in some stress conditions. Plant Cell Physiol. 2013, 54, 1441–1454. [Google Scholar] [CrossRef]
- Kim, E.J.; Kwak, J.M.; Uozumi, N.; Schroeder, J.I. AtKUP1: An Arabidopsis gene encoding high-affinity potassium transport activity. Plant Cell 1998, 10, 51–62. [Google Scholar] [CrossRef]
- Horie, T.; Costa, A.; Kim, T.H.; Han, M.J.; Horie, R.; Leung, H.Y.; Miyao, A.; Hirochika, H.; An, G.; Schroeder, J.I. Rice OsHKT2;1 transporter mediates large Na+ influx component into K+-starved roots for growth. EMBO J. 2007, 26, 3003–3014. [Google Scholar] [CrossRef]
- Horie, T.; Brodsky, D.E.; Costa, A.; Kaneko, T.; Lo Schiavo, F.; Katsuhara, M.; Schroeder, J.I. K+ transport by the OsHKT2;4 transporter from rice with atypical Na+ transport properties and competition in permeation of K+ over Mg2+ and Ca2+ ions. Plant Physiol. 2011, 156, 1493–1507. [Google Scholar] [CrossRef]
- Caballero, F.; Botella, M.A.; Rubio, L.; Fernandez, J.A.; Martinez, V.; Rubio, F. A Ca(2+)-sensitive system mediates low-affinity K(+) uptake in the absence of AKT1 in Arabidopsis plants. Plant Cell Physiol. 2012, 53, 2047–2059. [Google Scholar] [CrossRef]
- Fuchs, I.; Stolzle, S.; Ivashikina, N.; Hedrich, R. Rice K+ uptake channel OsAKT1 is sensitive to salt stress. Planta 2005, 221, 212–221. [Google Scholar] [CrossRef]
- Li, J.; Long, Y.; Qi, G.N.; Li, J.; Xu, Z.J.; Wu, W.H.; Wang, Y. The Os-AKT1 channel is critical for K+ uptake in rice roots and is modulated by the rice CBL1-CIPK23 complex. Plant Cell 2014, 26, 3387–3402. [Google Scholar] [CrossRef]
- Ashley, M.K.; Grant, M.; Grabov, A. Plant responses to potassium deficiencies: A role for potassium transport proteins. J. Exp. Bot. 2006, 57, 425–436. [Google Scholar] [CrossRef]
- Amtmann, A.; Troufflard, S.; Armengaud, P. The effect of potassium nutrition on pest and disease resistance in plants. Physiol. Plant. 2008, 133, 682–691. [Google Scholar] [CrossRef]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef]
- D’Hont, A.; Denoeud, F.; Aury, J.M.; Baurens, F.C.; Carreel, F.; Garsmeur, O.; Noel, B.; Bocs, S.; Droc, G.; Rouard, M.; et al. The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature 2012, 488, 213–217. [Google Scholar] [CrossRef]
- Martin, G.; Baurens, F.C.; Droc, G.; Rouard, M.; Cenci, A.; Kilian, A.; Hastie, A.; Dolezel, J.; Aury, J.M.; Alberti, A.; et al. Improvement of the banana “Musa acuminata” reference sequence using NGS data and semi-automated bioinformatics methods. BMC Genom. 2016, 17, 243. [Google Scholar] [CrossRef]
- White, E.J.; Venter, M.; Hiten, N.F.; Burger, J.T. Modified cetyltrimethylammonium bromide method improves robustness and versatility: The benchmark for plant RNA extraction. Biotechnol. J. 2008, 3, 1424–1428. [Google Scholar] [CrossRef]
- Cock, P.J.; Fields, C.J.; Goto, N.; Heuer, M.L.; Rice, P.M. The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic Acids Res. 2010, 38, 1767–1771. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Gul, H.; Tong, Z.; Han, X.; Nawaz, I.; Wahocho, S.; Khan, S.; Zhang, C.; Tian, Y.; Cong, P.; Zhang, L. Comparative Transcriptome Analysis between Ornamental Apple Species Provides Insights into Mechanism of Double Flowering. Agronomy 2019, 9, 112. [Google Scholar] [CrossRef]
- Shivani; Awasthi, P.; Sharma, V.; Kaur, N.; Kaur, N.; Pandey, P.; Tiwari, S. Genome-wide analysis of transcription factors during somatic embryogenesis in banana (Musa spp.) cv. Grand Naine. PLoS ONE 2017, 12, e0182242. [Google Scholar] [CrossRef]
- Luan, S.; Lan, W.; Chul Lee, S. Potassium nutrition, sodium toxicity, and calcium signaling: Connections through the CBL-CIPK network. Curr. Opin. Plant Biol. 2009, 12, 339–346. [Google Scholar] [CrossRef] [PubMed]
- Barros, F.; Dominguez, P.; de la Pena, P. Cytoplasmic domains and voltage-dependent potassium channel gating. Front. Pharmacol. 2012, 3, 49. [Google Scholar] [CrossRef] [PubMed]
- Emery, L.; Whelan, S.; Hirschi, K.D.; Pittman, J.K. Protein Phylogenetic Analysis of Ca(2+)/cation Antiporters and Insights into their Evolution in Plants. Front. Plant Sci. 2012, 3, 1. [Google Scholar] [CrossRef] [PubMed]
- He, C.; O’Halloran, D.M. Analysis of the Na+/Ca2+ exchanger gene family within the phylum Nematoda. PLoS ONE 2014, 9, e112841. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, R.E.; Lewis, B.D.; Spalding, E.P.; Sussman, M.R. A role for the AKT1 potassium channel in plant nutrition. Sciences 1998, 280, 918–921. [Google Scholar] [CrossRef]
- Yang, T.; Zhang, S.; Hu, Y.; Wu, F.; Hu, Q.; Chen, G.; Cai, J.; Wu, T.; Moran, N.; Yu, L.; et al. The role of a potassium transporter OsHAK5 in potassium acquisition and transport from roots to shoots in rice at low potassium supply levels. Plant Physiol. 2014, 166, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Verkhratsky, A.; Parpura, V. Calcium signalling and calcium channels: Evolution and general principles. Eur. J. Pharmacol. 2014, 739, 1–3. [Google Scholar] [CrossRef]
- Li, Z.; Matsuoka, S.; Hryshko, L.V.; Nicoll, D.A.; Bersohn, M.M.; Burke, E.P.; Lifton, R.P.; Philipson, K.D. Cloning of the NCX2 isoform of the plasma membrane Na(+)-Ca2+ exchanger. J. Biol. Chem. 1994, 269, 17434–17439. [Google Scholar]
- Sheen, J. Ca2+-dependent protein kinases and stress signal transduction in plants. Sciences 1996, 274, 1900–1902. [Google Scholar] [CrossRef]
- Sun, T.; Wang, Y.; Wang, M.; Li, T.; Zhou, Y.; Wang, X.; Wei, S.; He, G.; Yang, G. Identification and comprehensive analyses of the CBL and CIPK gene families in wheat (Triticum aestivum L.). BMC Plant Biol. 2015, 15, 269. [Google Scholar] [CrossRef] [PubMed]
- Thoday−Kennedy, E.L.; Jacobs, A.K.; Roy, S.J. The role of the CBL–CIPK calcium signalling network in regulating ion transport in response to abiotic stress. Plant Growth Regul. 2015, 76, 3–12. [Google Scholar] [CrossRef]
- Cuellar, T.; Azeem, F.; Andrianteranagna, M.; Pascaud, F.; Verdeil, J.L.; Sentenac, H.; Zimmermann, S.; Gaillard, I. Potassium transport in developing fleshy fruits: The grapevine inward K(+) channel VvK1.2 is activated by CIPK-CBL complexes and induced in ripening berry flesh cells. Plant J. Cell Mol. Biol. 2013, 73, 1006–1018. [Google Scholar] [CrossRef]
- He, Y.; Li, W.; Lv, J.; Jia, Y.; Wang, M.; Xia, G. Ectopic expression of a wheat MYB transcription factor gene, TaMYB73, improves salinity stress tolerance in Arabidopsis thaliana. J. Exp. Bot. 2012, 63, 1511–1522. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.J.; Ruzicka, D.; Shin, R.; Schachtman, D.P. The Arabidopsis AP2/ERF transcription factor RAP2.11 modulates plant response to low-potassium conditions. Mol. Plant 2012, 5, 1042–1057. [Google Scholar] [CrossRef] [PubMed]
- Anastacio, M.M.; Kanter, E.M.; Makepeace, C.M.; Keith, A.D.; Zhang, H.; Schuessler, R.B.; Nichols, C.G.; Lawton, J.S. Relationship between mitochondrial matrix volume and cellular volume in response to stress and the role of ATP-sensitive potassium channel. Circulation 2013, 128, S130–S135. [Google Scholar] [CrossRef]
- Baek, D.; Kim, M.C.; Chun, H.J.; Kang, S.; Park, H.C.; Shin, G.; Park, J.; Shen, M.; Hong, H.; Kim, W.Y.; et al. Regulation of miR399f transcription by AtMYB2 affects phosphate starvation responses in Arabidopsis. Plant Physiol. 2013, 161, 362–373. [Google Scholar] [CrossRef]
- Chen, Y.; Li, F.; Ma, Y.; Chong, K.; Xu, Y. Overexpression of OrbHLH001, a putative helix-loop-helix transcription factor, causes increased expression of AKT1 and maintains ionic balance under salt stress in rice. J. Plant Physiol. 2013, 170, 93–100. [Google Scholar] [CrossRef]
- Hong, J.P.; Takeshi, Y.; Kondou, Y.; Schachtman, D.P.; Matsui, M.; Shin, R. Identification and characterization of transcription factors regulating Arabidopsis HAK5. Plant Cell Physiol. 2013, 54, 1478–1490. [Google Scholar] [CrossRef]
- Hichri, I.; Muhovski, Y.; Zizkova, E.; Dobrev, P.I.; Franco-Zorrilla, J.M.; Solano, R.; Lopez-Vidriero, I.; Motyka, V.; Lutts, S. The Solanum lycopersicum Zinc Finger2 cysteine-2/histidine-2 repressor-like transcription factor regulates development and tolerance to salinity in tomato and Arabidopsis. Plant Physiol. 2014, 164, 1967–1990. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Pan, Y.; Hu, W.; Cong, R.; Ren, T.; Guo, S.; Lu, J. The photosynthetic and structural differences between leaves and siliques of Brassica napus exposed to potassium deficiency. BMC Plant Biol. 2017, 17, 240. [Google Scholar] [CrossRef] [PubMed]
- Cherel, I.; Lefoulon, C.; Boeglin, M.; Sentenac, H. Molecular mechanisms involved in plant adaptation to low K(+) availability. J. Exp. Bot. 2014, 65, 833–848. [Google Scholar] [CrossRef]
- Zeng, Q.; Ling, Q.; Fan, L.; Li, Y.; Hu, F.; Chen, J.; Huang, Z.; Deng, H.; Li, Q.; Qi, Y. Transcriptome profiling of sugarcane roots in response to low potassium stress. PLoS ONE 2015, 10, e0126306. [Google Scholar] [CrossRef]
- Nieves-Cordones, M.; Aleman, F.; Martinez, V.; Rubio, F. K+ uptake in plant roots. The systems involved, their regulation and parallels in other organisms. J. Plant Physiol. 2014, 171, 688–695. [Google Scholar] [CrossRef]
- Yu, Q.; An, L.; Li, W. The CBL-CIPK network mediates different signaling pathways in plants. Plant Cell Rep. 2014, 33, 203–214. [Google Scholar] [CrossRef]
- Shin, R.; Schachtman, D.P. Hydrogen peroxide mediates plant root cell response to nutrient deprivation. Proc. Natl. Acad. Sci. USA 2004, 101, 8827–8832. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.Y.; Shin, R.; Schachtman, D.P. Ethylene mediates response and tolerance to potassium deprivation in Arabidopsis. Plant Cell 2009, 21, 607–621. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Kim, B.G.; Cheong, Y.H.; Pandey, G.K.; Luan, S. A Ca(2+) signaling pathway regulates a K(+) channel for low−K response in Arabidopsis. Proc. Natl. Acad. Sci. USA 2006, 103, 12625–12630. [Google Scholar] [CrossRef] [PubMed]
- Lebaudy, A.; Very, A.A.; Sentenac, H. K+ channel activity in plants: Genes, regulations and functions. FEBS Lett. 2007, 581, 2357–2366. [Google Scholar] [CrossRef]
- Liu, L.L.; Ren, H.M.; Chen, L.Q.; Wang, Y.; Wu, W.H. A protein kinase, calcineurin B-like protein-interacting protein Kinase9, interacts with calcium sensor calcineurin B-like Protein3 and regulates potassium homeostasis under low-potassium stress in Arabidopsis. Plant Physiol. 2013, 161, 266–277. [Google Scholar] [CrossRef] [PubMed]
- Nieves-Cordones, M.; Al Shiblawi, F.R.; Sentenac, H. Roles and Transport of Sodium and Potassium in Plants. Met. Ions Life Sci. 2016, 16, 291–324. [Google Scholar]
- Ragel, P.; Rodenas, R.; Garcia-Martin, E.; Andres, Z.; Villalta, I.; Nieves-Cordones, M.; Rivero, R.M.; Martinez, V.; Pardo, J.M.; Quintero, F.J.; et al. The CBL-Interacting Protein Kinase CIPK23 Regulates HAK5-Mediated High-Affinity K+ Uptake in Arabidopsis Roots. Plant Physiol. 2015, 169, 2863–2873. [Google Scholar] [PubMed]
- Liu, W.; Stewart, C.N., Jr. Plant synthetic promoters and transcription factors. Curr. Opin. Biotechnol. 2016, 37, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Dale, J.; James, A.; Paul, J.Y.; Khanna, H.; Smith, M.; Peraza-Echeverria, S.; Garcia-Bastidas, F.; Kema, G.; Waterhouse, P.; Mengersen, K.; et al. Transgenic Cavendish bananas with resistance to Fusarium wilt tropical race 4. Nat. Commun. 2017, 8, 1496. [Google Scholar] [CrossRef] [PubMed]

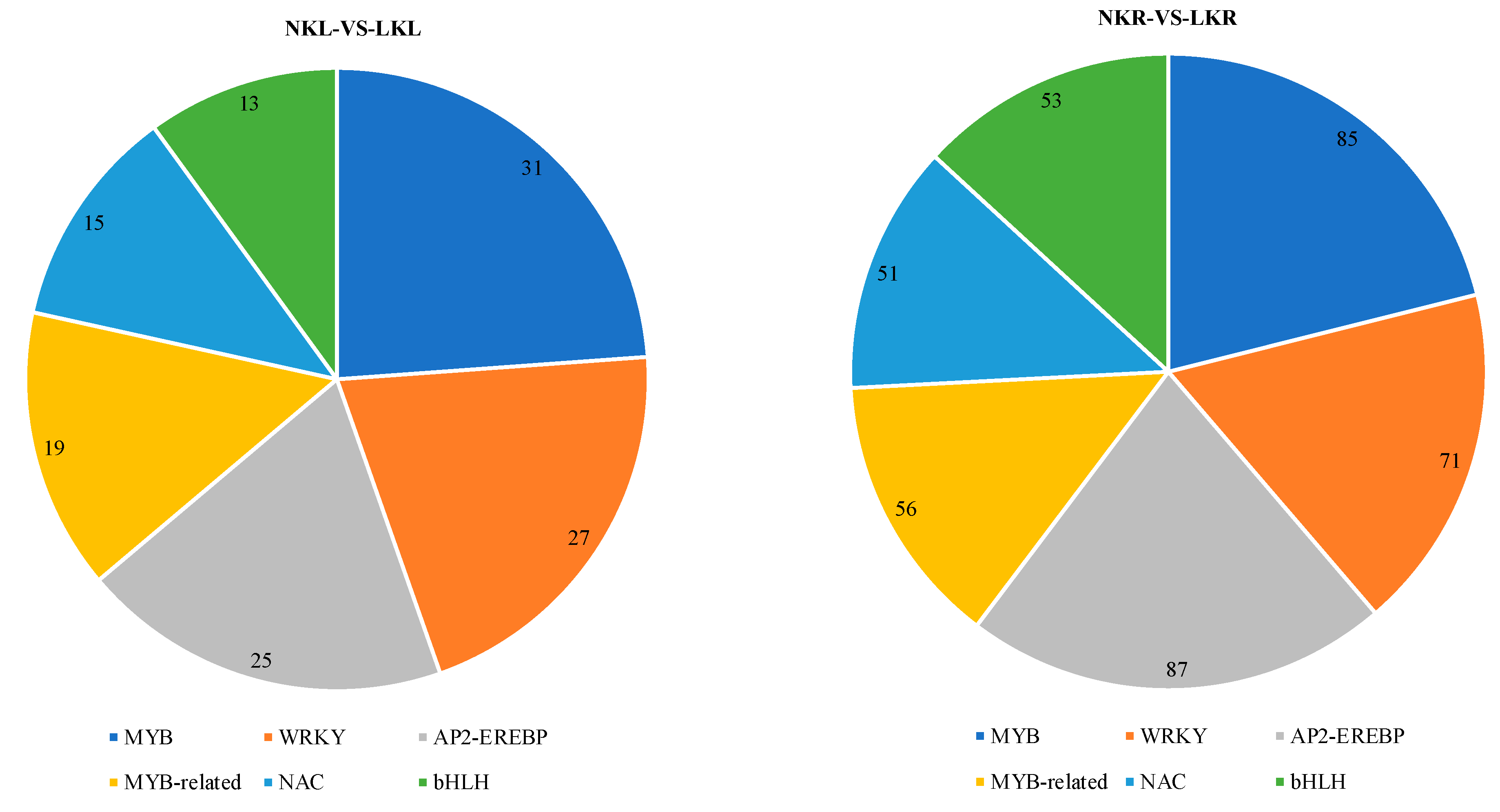
| Total Leaf Number | Number of Leaves Showing Symptoms | Plant Height/cm | Fresh weight of Aerial Part/g | Fresh Weight of Underground Part/g | Dry Weight of Aerial Part/g | Dry Weight of Underground Part/g | |
|---|---|---|---|---|---|---|---|
| NK | 7.40 ± 0.82 | 1.55 ± 0.68 | 15.12 ± 0.82 | 44.69 ± 4.56 | 21.38 ± 2.96 | 6.02 ± 1.31 | 3.35 ± 0.47 |
| LK | 6.20 ± 0.89 | 2.00 ± 0.72 | 11.77 ± 1.13 | 26.38 ± 4.54 | 11.67 ± 2.37 | 3.08 ± 0.76 | 1.67 ± 0.49 |
| Significance level | ab | ab | ab | ab | ab | ab | ab |
| Sample | Raw Reads | Clean Reads | Genome Map Rate | Expressed Genes | Expressed Transcripts |
|---|---|---|---|---|---|
| NKL | 12,191,789 | 12,121,980 | 84.01% | 26,796 | 33,388 |
| LKL | 12,191,898 | 12,108,969 | 84.81% | 27,014 | 33,469 |
| NKR | 12,191,778 | 12,121,219 | 82.63% | 29,158 | 35,896 |
| LKR | 12,191,844 | 12,087,980 | 81.73% | 28,748 | 35,380 |
| Total | 48,767,309 | 24,209,199 | - | 111,716 | 138,133 |
| (A) Differentially Expressed Genes Related to Potassium in Leaves | |||||
| Gene ID | NKL-FPKM | LKL-FPKM | log2 Ratio (LKL/NKL) | Up–Down-Regulation (LKL/NKL) | Gene Function Annotation |
| Ma03_g24780 | 2.66 | 0.44 | −2.60 | Down | Potassium transporter 13 [Musa acuminata subsp. malaccensis] |
| Ma07_g09180 | 9.3 | 2.77 | −1.75 | Down | Potassium channel KAT3-like isoform X2 [Musa acuminata subsp. malaccensis] |
| Ma11_g04530 | 77.06 | 36.94 | −1.06 | Down | CBL-interacting protein kinase 1 [Musa acuminata subsp. malaccensis] |
| Ma05_g07130 | 1.66 | 20.65 | 3.64 | Up | Potassium transporter 1 isoform X2 [Musa acuminata subsp. Malaccensis] |
| Ma08_g20660 | 3.97 | 21.49 | 2.44 | Up | Cation/calcium exchanger 1–like [Musa acuminata subsp. malaccensis] |
| Ma02_g04540 | 4.92 | 11.5 | 1.22 | Up | Potassium transporter 11 [Musa acuminata subsp. malaccensis] |
| Ma10_g27850 | 0.2 | 3.22 | 4.01 | Up | Potassium channel KAT3–like [Musa acuminata subsp. malaccensis] |
| Ma07_g17570 | 11.26 | 34.39 | 1.61 | Up | CBL-interacting protein kinase 1 [Musa acuminata subsp. malaccensis] |
| Ma07_g05310 | 7.85 | 20.19 | 1.36 | Up | CBL-interacting protein kinase 6 [Musa acuminata subsp. malaccensis] |
| Ma09_g04710 | 38.84 | 86 | 1.15 | Up | CBL-interacting serine/threonine-protein kinase 11 [Musa acuminata subsp. malaccensis] |
| (B) Differentially Expressed Genes Related to Potassium in Root | |||||
| Gene ID | NKR-FPKM | LKR-FPKM | log2 Ratio (LKR/NKR) | Up–Down-Regulation (LKL/NKL) | Gene Function Annotation |
| Ma03_g06090 | 5.79 | 1 | −2.53 | Down | Potassium transporter 6-like [Musa acuminata subsp. malaccensis] |
| Ma07_g12190 | 7.8 | 1.74 | −2.16 | Down | Potassium transporter 17 isoform X3 [Musa acuminata subsp. malaccensis] |
| Ma09_g24000 | 163.17 | 71.21 | −1.20 | Down | Voltage-gated potassium channel subunit beta [Musa acuminata subsp. malaccensis] |
| Ma06_g28610 | 6.23 | 2.73 | −1.19 | Down | Potassium channel KAT2-like isoform X5 [Musa acuminata subsp. malaccensis] |
| Ma05_g18280 | 11.8 | 5.44 | −1.12 | Down | Cation/calcium exchanger 5 [Musa acuminata subsp. malaccensis] |
| Ma09_g11150 | 12.07 | 5.59 | −1.11 | Down | Potassium transporter 7-like [Musa acuminata subsp. malaccensis] |
| Ma11_g16690 | 8.55 | 4.02 | −1.09 | Down | Probable potassium transporter 11 [Musa acuminata subsp. malaccensis] |
| Ma06_g26140 | 4.41 | 0.001 | −12.11 | Down | CBL-interacting protein kinase 7 [Musa acuminata subsp. malaccensis] |
| Ma05_g07130 | 0.37 | 2.73 | 2.88 | Up | Potassium transporter 1 isoform X2 [Musa acuminata subsp. malaccensis] |
| Ma08_g23650 | 1.65 | 7.88 | 2.26 | Up | Potassium channel KOR1-like [Musa acuminata subsp. malaccensis] |
| Ma07_g21520 | 6.71 | 29.6 | 2.14 | Up | Potassium transporter 6-like isoformX2 [Musa acuminata subsp. malaccensis] |
| Ma05_g09470 | 22.34 | 59.24 | 1.41 | Up | Potassium channel KAT4-like [Musa acuminata subsp. malaccensis] |
| Ma06_g34240 | 0.88 | 4.47 | 2.34 | Up | CBL-interacting serine/threonine-protein kinase 29 [Musa acuminata subsp. malaccensis] |
| Ma06_g28510 | 3.15 | 14.66 | 2.22 | Up | CBL-interacting serine/threonine-protein kinase 12 [Musa acuminata subsp. malaccensis] |
| Ma07_g23630 | 51.84 | 228.13 | 2.14 | Up | CBL-interacting protein kinase 2 [Musa acuminata subsp. malaccensis] |
| Ma01_g17400 | 12 | 39.95 | 1.74 | Up | CBL-interacting protein kinase 1 [Musa acuminata subsp. malaccensis] |
| Ma04_g29510 | 24.94 | 74.58 | 1.58 | Up | CBL-interacting protein kinase 23 [Musa acuminata subsp. malaccensis] |
| Ma07_g29120 | 91.26 | 249.13 | 1.45 | Up | CBL-interacting protein kinase 6 [Musa acuminata subsp. malaccensis] |
| Ma03_g14540 | 10.2 | 26.64 | 1.39 | Up | CBL-interacting serine/threonine-protein kinase 1 [Musa acuminata subsp. malaccensis] |
| Ma04_g15200 | 26.57 | 66.81 | 1.33 | Up | CBL-interacting protein kinase 19 [Musa acuminata subsp. malaccensis] |
| Ma08_g10700 | 5.38 | 12.27 | 1.19 | Up | CBL-interacting protein kinase 1 [Musa acuminata subsp. malaccensis] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, M.; Zeng, C.-B.; He, R.; Yan, Z.; Qi, Z.; Xiong, R.; Cheng, Y.; Wei, S.-S.; Tang, H. Transcriptome Analysis of Banana (Musa acuminate L.) in Response to Low-Potassium Stress. Agronomy 2019, 9, 169. https://doi.org/10.3390/agronomy9040169
Xu M, Zeng C-B, He R, Yan Z, Qi Z, Xiong R, Cheng Y, Wei S-S, Tang H. Transcriptome Analysis of Banana (Musa acuminate L.) in Response to Low-Potassium Stress. Agronomy. 2019; 9(4):169. https://doi.org/10.3390/agronomy9040169
Chicago/Turabian StyleXu, Min, Can-Bin Zeng, Rui He, Zhen Yan, Zhao Qi, Rui Xiong, Yu Cheng, Shuang-Shuang Wei, and Hua Tang. 2019. "Transcriptome Analysis of Banana (Musa acuminate L.) in Response to Low-Potassium Stress" Agronomy 9, no. 4: 169. https://doi.org/10.3390/agronomy9040169
APA StyleXu, M., Zeng, C.-B., He, R., Yan, Z., Qi, Z., Xiong, R., Cheng, Y., Wei, S.-S., & Tang, H. (2019). Transcriptome Analysis of Banana (Musa acuminate L.) in Response to Low-Potassium Stress. Agronomy, 9(4), 169. https://doi.org/10.3390/agronomy9040169




