Multi-Parental Advances Generation Inter-Cross Population, to Develop Organic Tomato Genotypes by Participatory Plant Breeding
Abstract
1. Introduction
2. Materials and Methods
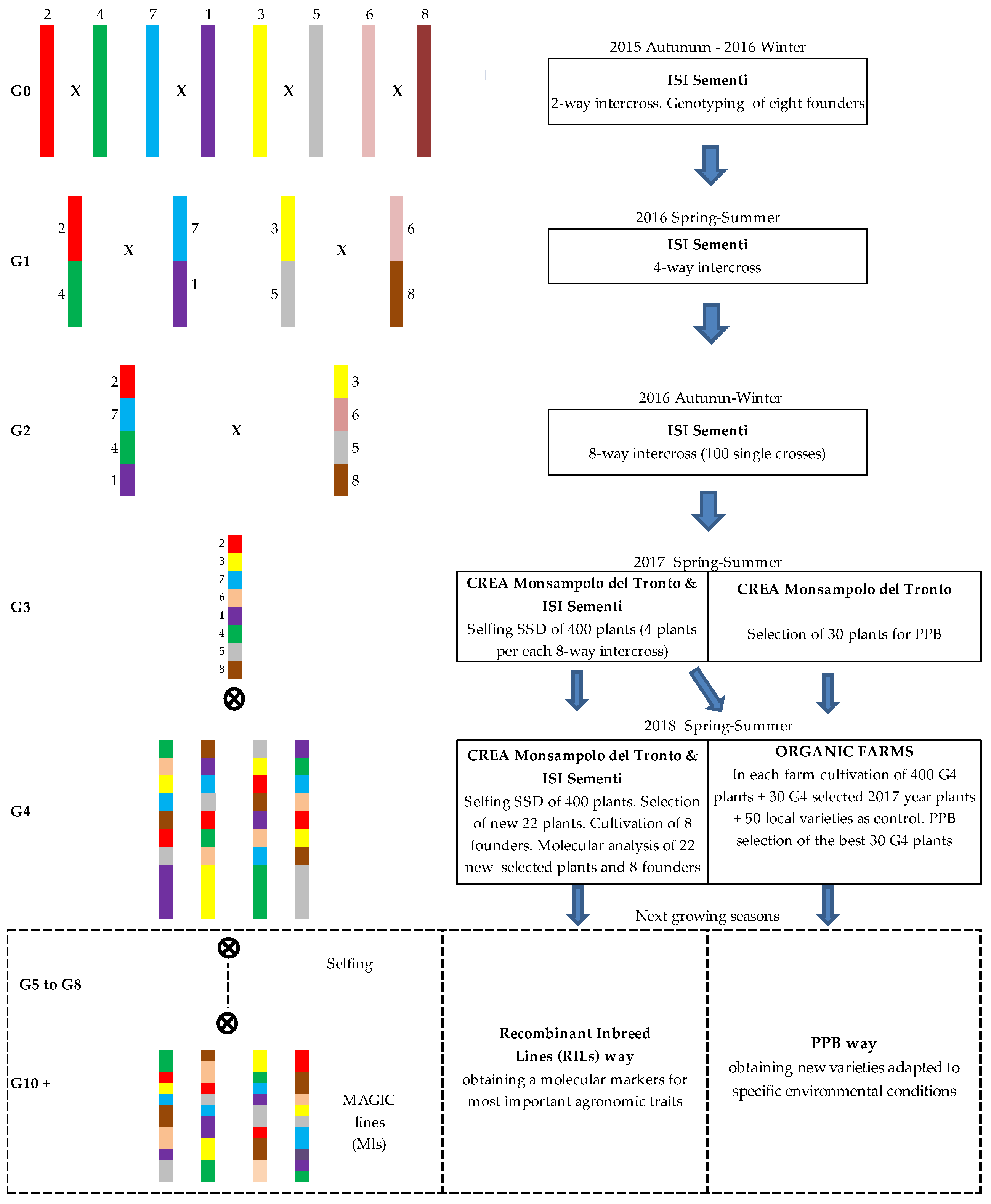
2.1. The MAGIC Population Construction
2.2. Cultivation and Selection of MAGIC Population
2.3. Participatory Plant Breeding Approach and Description
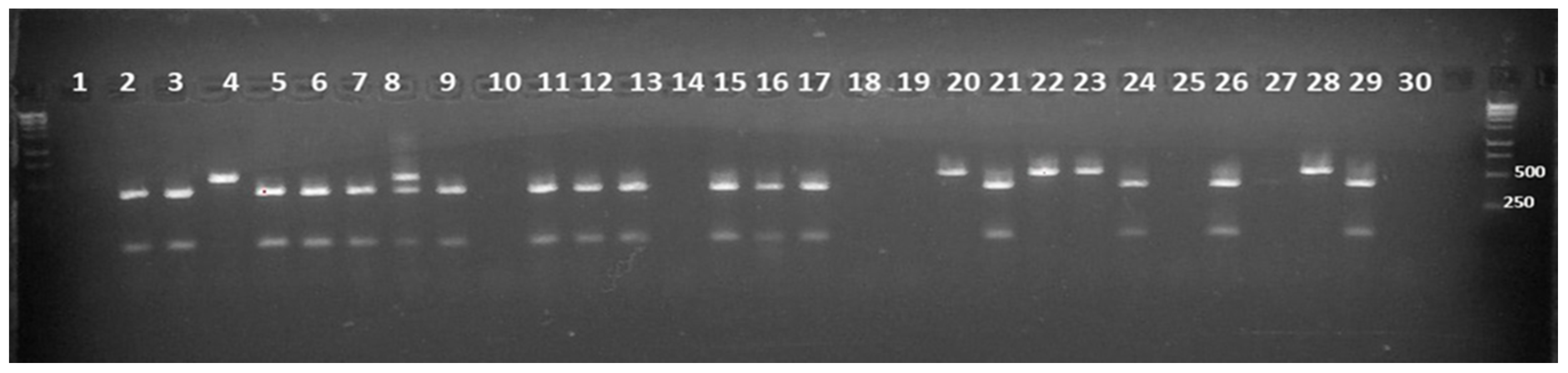
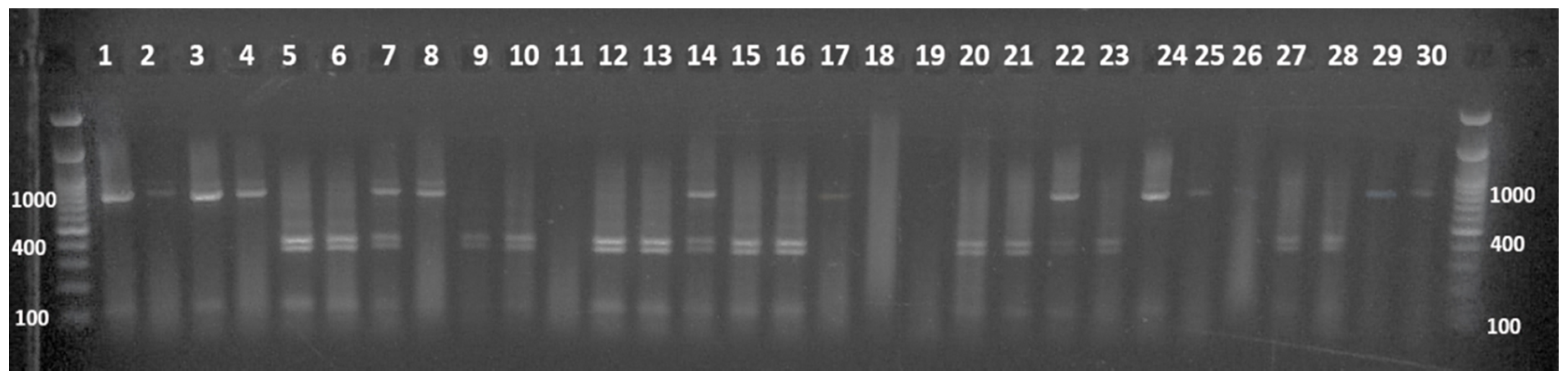
2.4. Molecular Analysis of the Selected G4 Plants
3. Results and Discussion
3.1. MAGIC Development and Tomato Selection of G3 Population
3.2. Evaluation and Selection within the MAGIC Population
3.3. Molecular Characterization of the G4 Selected Plants
3.4. Tomato Genotype Selection by Participatory Plant Breeding Program
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References and Notes
- Gafsi, M.; Legagneux, B.; Nguyen, G.; Robin, P. Towards sustainable farming systems: Effectiveness and deficiency of the French procedure of sustainable agriculture. Agric. Syst. 2006, 90, 226–242. [Google Scholar] [CrossRef]
- Montemurro, F.; Persiani, A.; Diacono, M. Environmental sustainability assessment of horticultural systems: A multi-criteria evaluation approach applied in a case study in Mediterranean conditions. Agronomy 2018, 8, 98. [Google Scholar] [CrossRef]
- De Benedetto, D.; Montemurro, F.; Diacono, M. Impacts of Agro-Ecological Practices on Soil Losses and Cash Crop Yield. Agriculture 2017, 7, 103. [Google Scholar] [CrossRef]
- Campanelli, G.; Acciarri, N.; Campion, B.; Delvecchio, S.; Leteo, F.; Fusari, F.; Angelini, P.; Ceccarelli, S. Participatory tomato breeding for organic conditions in Italy. Euphytica 2015, 204, 179–197. [Google Scholar] [CrossRef]
- Ceccarelli, S. Efficiency of plant breeding. Crop Sci. 2015, 55, 87–97. [Google Scholar] [CrossRef]
- Barabaschi, D.; Tondelli, A.; Desiderio, F.; Volante, A.; Vaccino, P.; Valè, G.; Cattivelli, L. Next generation breeding. Plant Sci. 2016, 242, 3–13. [Google Scholar] [CrossRef] [PubMed]
- D’Agostino, N.; Tripodi, P. NGS-Based Genotyping, High-Throughput Phenotyping and Genome-Wide Association Studies Laid the Foundations for Next-Generation Breeding in Horticultural Crops. Diversity 2017, 9, 38. [Google Scholar] [CrossRef]
- Pascual, L.; Desplat, N.; Huang, B.E.; Desgroux, A.; Bruguier, L.; Bouchet, J.P.; Le, Q.H.; Chauchard, B.; Verschave, P.; Causse, M. Potential of a tomato magic population to decipher the genetic control of quantitative traits and detect causal variants in the resequencing era. Plant Biotechnol. J. 2015, 13, 565–577. [Google Scholar] [CrossRef] [PubMed]
- Huynh, B.L.; Ehlers, J.D.; Huang, B.E.; Muñoz-Amatriaín, M.; Lonardi, S.; Santos, J.R.P.; Ndeve, A.; Batieno, B.J.; Boukar, O.; Cisse, N.; et al. A multi-parent advanced generation inter-cross (MAGIC) population for genetic analysis and improvement of cowpea (Vigna unguiculata L. Walp.). Plant J. 2018, 93, 1129–1142. [Google Scholar] [CrossRef] [PubMed]
- Gaur, P.M.; Thudi, M.; Srinivasan, S.; Varshney, R.K. Advances in chickpea genomics. In Legumes in the Omic Era; Nadarajan, N., Gupta, D.S., Eds.; Springer: New York, NY, USA, 2013; pp. 73–94. [Google Scholar]
- Bandillo, N.; Raghavan, C.; Muyco, P.A.; Sevilla, M.A.; Lobina, I.T.; Dilla-Ermita, C.J.; Tung, C.W.; McCouch, S.; Thomson, M.; Mauleon, R.; et al. Multi-parent advanced generation inter-cross (MAGIC) populations in rice: Progress and potential for genetics research and breeding. Rice 2013, 6, 11. [Google Scholar] [CrossRef] [PubMed]
- Sannemann, W.; Huang, B.E.; Mathew, B.; Leon, J. Multi-parent advanced generation inter-cross in barley: High-resolution quantitative trait locus mapping for flowering time as a proof of concept. Mol. Breed. 2015, 35, 86. [Google Scholar] [CrossRef]
- Mackay, I.J.; Bansept-Basler, P.; Barber, T.; Bentley, A.R.; Cockram, J.; Gosman, N.; Greenland, A.J.; Horsnell, R.; Howells, R.; O’sullivan, D.M.; et al. An eight-parent multiparent advanced generation inter-cross population for winter-sown wheat: Creation, properties, and validation. G3 2014, 4, 1603–1610. [Google Scholar] [CrossRef] [PubMed]
- Hoagland, L.; Navazio, J.; Zystro, J.; Vargas, J.G.; Gibson, K. Key Traits and Promising Germplasm for an Organic Participatory Tomato Breeding Program in the U.S. Midwest. Hortscience 2015, 50, 1301–1308. [Google Scholar] [CrossRef]
- Nesbitt, T.C.; Tanksley, S.D. Comparative sequencing in the genus lycopersicon: Implications for the evolution of fruit size in the domestication of cultivated tomatoes. Genetics 2002, 162, 365–379. [Google Scholar] [PubMed]
- Campanelli, G.; Canali, S. Crop production and environmental effects in conventional and organic vegetable farming systems: The case of a long-term experiment in Mediterranean conditions (Central Italy). J. Sustain. Agric. 2012, 36, 599–619. [Google Scholar] [CrossRef]
- Cullis, B.R.; Smith, A.B.; Coombes, N.E. On the Design of Early Generation Variety Trials with Correlated Data. J. Agric. Biol. Environ. Stat. 2006, 11, 381–393. [Google Scholar] [CrossRef]
- Coombes, N.E. DiGGer, a Design Generator. 2006. Available online: http://www.austatgen.org/files/software/downloads/ (accessed on 14 February 2019).
- Commission Regulation (EC) No 889/2008. Official Journal of The European Union 18 September 2008.
- Doyle, J.J.; Doyle, J.L. Isolation of plant DNA from fresh tissue. Focus 1990, 12, 13–15. [Google Scholar]
- Staniaszek, M.; Kozic, E.U.; Marczewski, W. A CAPS marker TAO1902 diagnostic for the I-2 gene conferring resistance to Fusarium Oxysporum f. sp. Lycopersici race 2 in tomato. Plant Breed. 2007, 126, 331–333. [Google Scholar] [CrossRef]
- Vakalounakis, D.J.; Laterrot, H.; Moretti, A.; Ligoxigaki, E.K.; Smardas, K. Linkage between Frl (Fusarium oxysporum f. sp. radicislycopersici resistance) and Tm-2 (tobacco mosaic virus resistance-2) loci in tomato (Lycopersicon esculentum). Ann. Appl. Biol. 1997, 130, 319–323. [Google Scholar] [CrossRef]
- Pérez de Castro, A.; Blanca, J.M.; Dìez, M.J.; Nuez Viñals, F. Identification of a CAPS marker tightly linked to the Tomato yellow leaf curl disease resistance gene Ty-1 in tomato. Eur. J. Plant Pathol. 2007, 117, 347–356. [Google Scholar] [CrossRef]
- Shi, A.; Vierling, R.; Grazzini, R.; Chen, P.; Caton, H.; Panthee, D. Identification of molecular markers for Sw-5 gene of tomato spotted wilt virus resistance. Am. J. Biotechnol. Mol. Sci. 2011, 1, 8–16. [Google Scholar] [CrossRef]
- Yang, W.; Francis, D.M. Marker-assisted Selection for Combining Resistance to Bacterial Spot and Bacterial Speck in Tomato. J. Am. Soc. Hortic. Sci. 2005, 130, 716–721. [Google Scholar] [CrossRef]
- Sabatini, E.; Rotino, G.; Voltattorni, S.; Nazzareno, A. A novel CAPS marker derived from the Ovate gene in tomato (L. esculentum Mill.) is useful to distinguish two Italian ecotypes and to recover pear shape in marker assisted selection. Eur. J. Hortic. Sci. 2006, 71, 193–198. [Google Scholar]
- Huang, B.E.; Verbyla, K.L.; Verbyla, A.P.; Raghavan, C.; Singh, V.K. MAGIC populations in crops: Current status and future prospects. Appl. Genet. 2015, 128, 999–1017. [Google Scholar] [CrossRef] [PubMed]
- The Tomato Genome Consortium. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 2012, 485, 635–641. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.C.; Tanksley, S.D. RFLP analysis of phylogenetic relationships and genetic variation in the genus. Lycopersicon. Theor. Appl. Genet. 1990, 80, 437–448. [Google Scholar] [CrossRef] [PubMed]
- Zamir, D. Improving plant breeding with exotic genetic libraries. Nat. Rev. Genet. 2001, 2, 983–989. [Google Scholar] [CrossRef] [PubMed]
- Blanca, J.; Montero-Pau, J.; Sauvage, C.; Bauchet, G.; Illa, E.; Díez, M.J. Genomic variation in tomato, from wild ancestors to contemporary breeding accessions. BMC Genom. 2015, 16, 257. [Google Scholar] [CrossRef] [PubMed]
- Bailey, D.W. Recombinant-inbred strains. An aid to finding identity, linkage, and function of histocompatibility and other genes. Transplantation 1991, 11, 325–327. [Google Scholar] [CrossRef]
- Maluszynski, M.; Kasha, K.J.; Forster, B.P.; Szarejko, I. Doubled Haploid Production in Crop Plants: A Manual; Kluwer Academic Publishers: Dordrecht, The Netherlands, 2013. [Google Scholar]
- Scheben, A.; Batley, J.; Edwards, D. Genotype-by-sequencing approaches to characterize crop genomes: Choosing the right tool for the right application. Plant Biotechnol. J. 2017, 15, 149–161. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Zhao, X.; Laroche, A.; Lu, Z.X.; Liu, H.; Li, Z. Genotyping-by-sequencing (GBS), An ultimate marker-assisted selection (MAS) tool to accelerate plant breeding. Front. Plant Sci. 2014, 5, 484. [Google Scholar] [CrossRef] [PubMed]
- Diacono, M.; Persiani, A.; Fiore, A.; Montemurro, F.; Canali, S. Agro-ecology for adaptation of horticultural systems to climate change: Agronomic and energetic performance evaluation. Agronomy 2017, 7, 35. [Google Scholar] [CrossRef]
- Diacono, M.; Persiani, A.; Canali, S.; Montemurro, F. Agronomic performance and sustainability indicators in organic tomato combining different agro-ecological practices. Nutr. Cycl. Agroecosyst. 2018, 112, 101–117. [Google Scholar] [CrossRef]
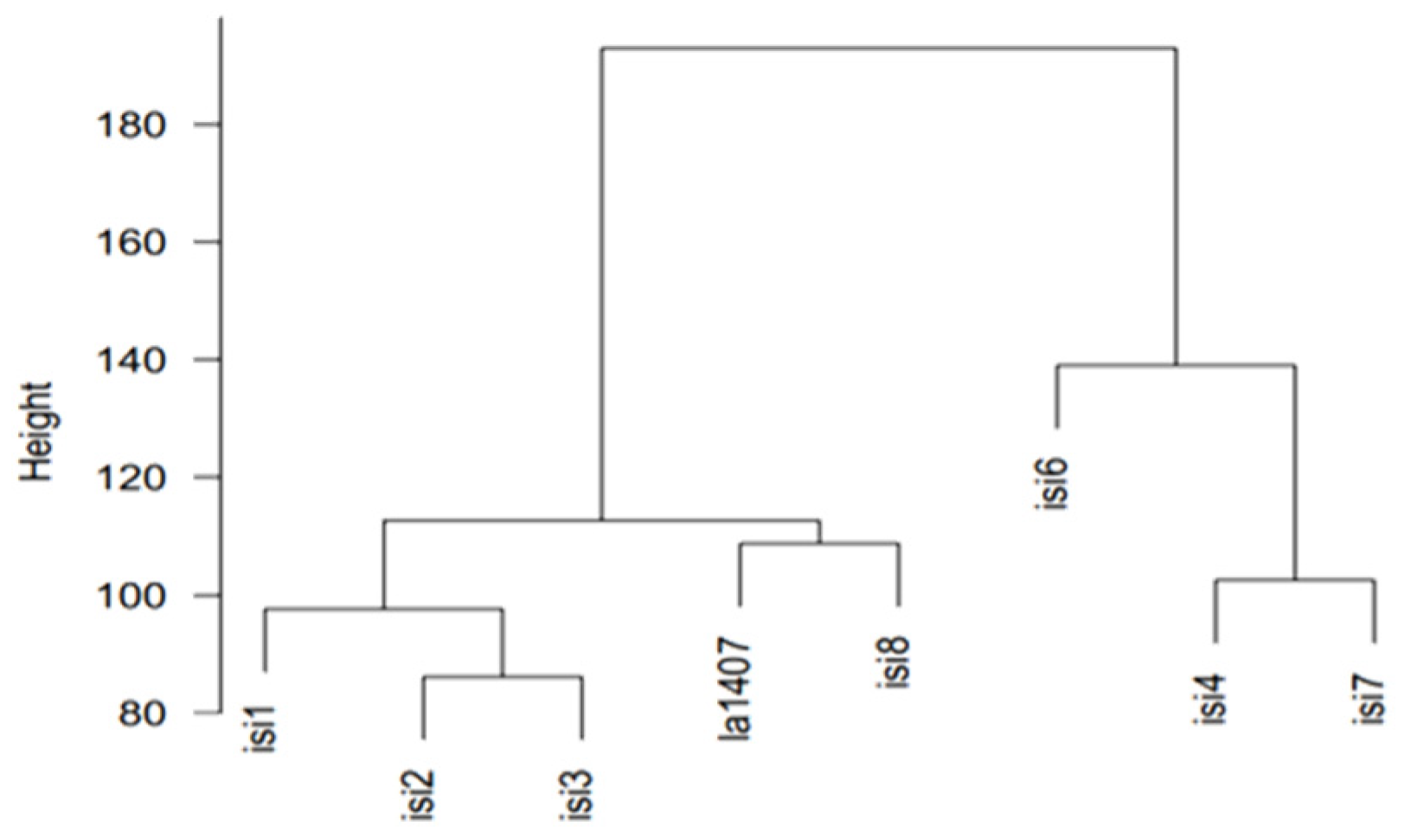
| Founders | Shape | Weight | DTF 1 | Sp 2 | H3 3 | Fruit Color | Peduncle | Green Shoulder | Hypocotil | OBV 4 |
|---|---|---|---|---|---|---|---|---|---|---|
| ISI 1 | Blocky | 70 | 50 | sp/sp | Present | Red | Jointless | Absent | Light green | Not transparent |
| ISI 2 | Round | 160 | 50 | sp/sp | Absent | Red | Jointed | Absent | Violet | Not transparent |
| ISI 3 | Oval | 90 | 47 | sp/sp | Absent | Red | Jointed | Absent | Violet | Not transparent |
| ISI 4 | Blocky | 80 | 43 | sp/sp | Absent | Red | Jointless | Absent | Violet | Transparent |
| LA1407 | Round | 5 | 73 | SP/SP | Absent | Orange | Jointed | Present | Green | Not transparent |
| ISI 6 | Blocky | 80 | 36 | sp/sp | Absent | Red | Jointless | Absent | Violet | Transparent |
| ISI 7 | Elongated | 75 | 46 | sp/sp | Absent | Red | Jointless | Absent | Violet | Not transparent |
| ISI 8 | Round | 50 | 55 | sp/sp | Absent | Red | Jointless | Present | Violet | Not transparent |
| Gene | Marker | Sequences Primer (5′–3′) | References | |
|---|---|---|---|---|
| Forward “F” | Reverse “R” | |||
| Resistance | ||||
| I-2 (FOL) | TAO1 | GGGCTCCTAATCCGTGCTTCA | GGTGGAGGATCGGGTTTGTTTC | [21] |
| Frl (FORL) | T1212 | AAGTGCTCTAGACAAAAAGACTCC | CCAATGTACAATGGAACTCGTTGATG | [22] |
| Mi-1 (M) | Mi23 | TGGAAAAATGTTGAATTTCTTTTG | GCATACTATATGGCTTGTTTACCC | [23] |
| Sw-5 (TSWV) | Sw5 | CGGAACCTGTAACTTGACTG | GAGCTCTCATCCATTTTCCG | [24] |
| Pto | Pto | ATCTACCCACAATGACCATGAGCTG | GTGCATACTCCAGTTTCCAC | [25] |
| Fruit Shape | ||||
| Ovate (Pear shape) | OvaNest | AATGCCAACACCAAGAGGAG | TCTCCCAAATGTCTGAGAACG | [26] |
| Chromosome | Number of Founder Alleles |
|---|---|
| Chr 1 | 224 |
| Chr 2 | 340 |
| Chr 3 | 475 |
| Chr 4 | 1221 |
| Chr 5 | 1309 |
| Chr 6 | 1509 |
| Chr 7 | 157 |
| Chr 8 | 249 |
| Chr 9 | 325 |
| Chr 10 | 205 |
| Chr 11 | 1161 |
| Chr 12 | 392 |
| Total | 7567 |
| Plant | Resistance | Fruit Shape | |
|---|---|---|---|
| Selected | 1 | Meloidogyne; FORL; TSWV | |
| 2 | Meloidogyne; TSWV | ||
| 3 | Meloidogyne; FORL; TSWV | ||
| 4 | Meloidogyne; P. syringae; TSWV | ||
| 5 | Meloidogyne; FOL; FORL; TSWV | Pear shape | |
| 6 | Meloidogyne; FOL; FORL; TSWV | Pear shape | |
| 7 | Meloidogyne; FOL; FORL; TSWV | Pear shape | |
| 8 | Meloidogyne; FORL; P. syringae; TSWV | Pear shape | |
| 9 | Meloidogyne; FOL; P. syringae; TSWV | ||
| 10 | Meloidogyne; FOL; FORL; TSWV | Pear shape | |
| 11 | Meloidogyne; P. syringae; TSWV | Pear shape | |
| 12 | Meloidogyne; FOL; FORL; P. syringae; TSWV | ||
| 13 | Meloidogyne; FOL; FORL; P. syringae; TSWV | Pear shape | |
| 14 | Meloidogyne; FOL; TSWV | Pear shape | |
| 15 | Meloidogyne; FOL; FORL; P. syringae; TSWV | Pear shape | |
| 16 | Meloidogyne; FOL; FORL; P. syringae; TSWV | Pear shape | |
| 17 | Meloidogyne; P. syringae; TSWV | Pear shape | |
| 18 | Meloidogyne; TSWV | ||
| 19 | Meloidogyne; TSWV | ||
| 20 | Meloidogyne; FOL; FORL; TSWV | Pear shape | |
| 21 | Meloidogyne; FOL; FORL; P. syringae; TSWV | ||
| 22 | Meloidogyne; FOL; FORL; TSWV | Pear shape | |
| Parents | 23 | Meloidogyne; FOL; FORL; P. syringae; TSWV | |
| 24 | Meloidogyne; FORL; TSWV | ||
| 25 | TSWV | Pear shape | |
| 26 | Meloidogyne; TSWV | ||
| 27 | Meloidogyne; FOL; FORL; TSWV | ||
| 28 | Meloidogyne; FOL; FORL; P. syringae; TSWV | Pear shape | |
| 29 | Meloidogyne; TSWV | Pear shape | |
| 30 | Meloidogyne; FORL; TSWV | Pear shape | |
| Plant | Column | Row | Researcher and/or Technicians Evaluation | Farmer Evaluation |
|---|---|---|---|---|
| 1 | 1 | 1 | 1 to 4 1 | 1 to 4 1 |
| to 480 | to 12 | to 40 | 1 to 4 1 | 1 to 4 1 |
| Plant Develop | Plant Vigor | Plant Health | Production Rate | Fruit Shape | Fruit Size | Fruit Color | Homogeneity Ripe Fruit | Fruit Solidity | Internal Fruit Solidity | Fruit Taste | °Brix |
|---|---|---|---|---|---|---|---|---|---|---|---|
| VIS 1 | VIS | VIS | VIS | VIS | VIS | VIS | VIS | MEA | VIS | TAS | MEA |
| Organic Farm | Plant Vigor | Plant Health | Abundance Fruiting | Fruit Size | Homogeneity Fruit Ripe | Fruit Firmness | Puffiness | Solid Soluble |
|---|---|---|---|---|---|---|---|---|
| North | 4.13 ± 0.57 | 3.53 ± 0.68 | 4.20 ± 0.85 | 2.97 ± 0.81 | 3.37 ± 0.89 | 6.20 ± 0.93 | 2.83 ± 0.75 | 5.22 ± 0.75 |
| Center | 3.10 ± 0.71 | 2.80 ± 0.85 | 3.50 ± 0.63 | 2.80 ± 0.61 | 3.20 ± 0.85 | 6.17 ± 1.61 | 2.97 ± 0.89 | 5.47 ± 0.99 |
| South | 3.03 ± 0.85 | 2.83 ± 0.79 | 3.30 ± 0.75 | 3.10 ± 0.85 | 3.73 ± 0.74 | 6.04 ± 1.77 | 3.23 ± 0.77 | 5.71 ± 0.86 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Campanelli, G.; Sestili, S.; Acciarri, N.; Montemurro, F.; Palma, D.; Leteo, F.; Beretta, M. Multi-Parental Advances Generation Inter-Cross Population, to Develop Organic Tomato Genotypes by Participatory Plant Breeding. Agronomy 2019, 9, 119. https://doi.org/10.3390/agronomy9030119
Campanelli G, Sestili S, Acciarri N, Montemurro F, Palma D, Leteo F, Beretta M. Multi-Parental Advances Generation Inter-Cross Population, to Develop Organic Tomato Genotypes by Participatory Plant Breeding. Agronomy. 2019; 9(3):119. https://doi.org/10.3390/agronomy9030119
Chicago/Turabian StyleCampanelli, Gabriele, Sara Sestili, Nazzareno Acciarri, Francesco Montemurro, Daniela Palma, Fabrizio Leteo, and Massimiliano Beretta. 2019. "Multi-Parental Advances Generation Inter-Cross Population, to Develop Organic Tomato Genotypes by Participatory Plant Breeding" Agronomy 9, no. 3: 119. https://doi.org/10.3390/agronomy9030119
APA StyleCampanelli, G., Sestili, S., Acciarri, N., Montemurro, F., Palma, D., Leteo, F., & Beretta, M. (2019). Multi-Parental Advances Generation Inter-Cross Population, to Develop Organic Tomato Genotypes by Participatory Plant Breeding. Agronomy, 9(3), 119. https://doi.org/10.3390/agronomy9030119







