Envirotypes Based on Seed Yield Limiting Factors Allow to Tackle G × E Interactions
Abstract
1. Introduction
2. Materials and Methods
2.1. Field Network and Crop Management Description
2.2. Plant Material and Seed Yield Assessment
2.3. Key Periods of the Winter Rapeseed Crop Cycle
2.4. Indicators Used for Environmental Description
2.4.1. Descriptor Definition: Raw Climatic Data and Soil Water Status Evaluation
2.4.2. Set up of Informative Indicators
2.5. Statistical Analyses
2.5.1. Partial Least Squares (PLS) Regression
2.5.2. Envirotyping Based on the PLS Regression Results
2.5.3. Test of the Environment and Genotype by Environment Effects Using Linear Models
3. Results
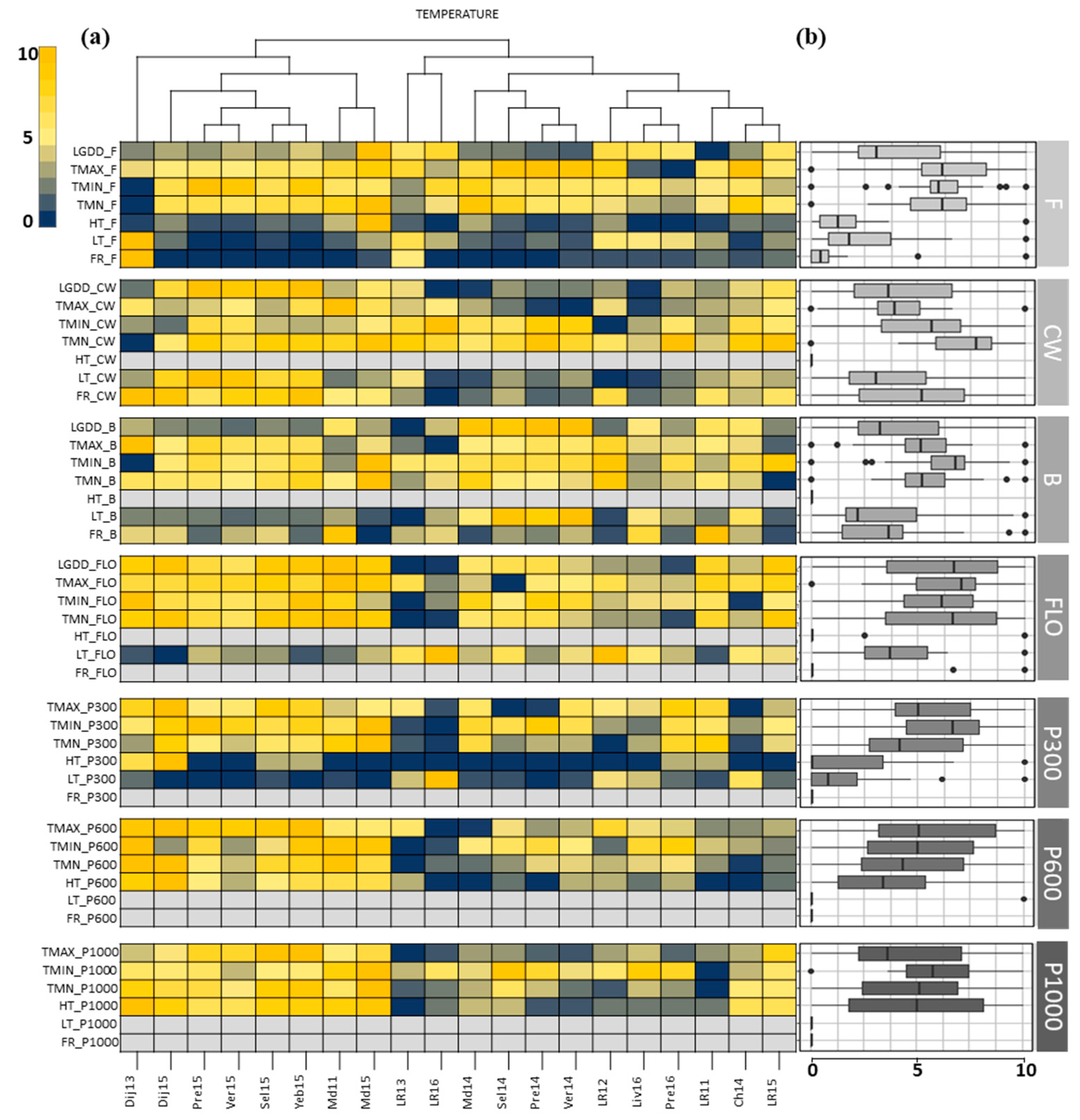
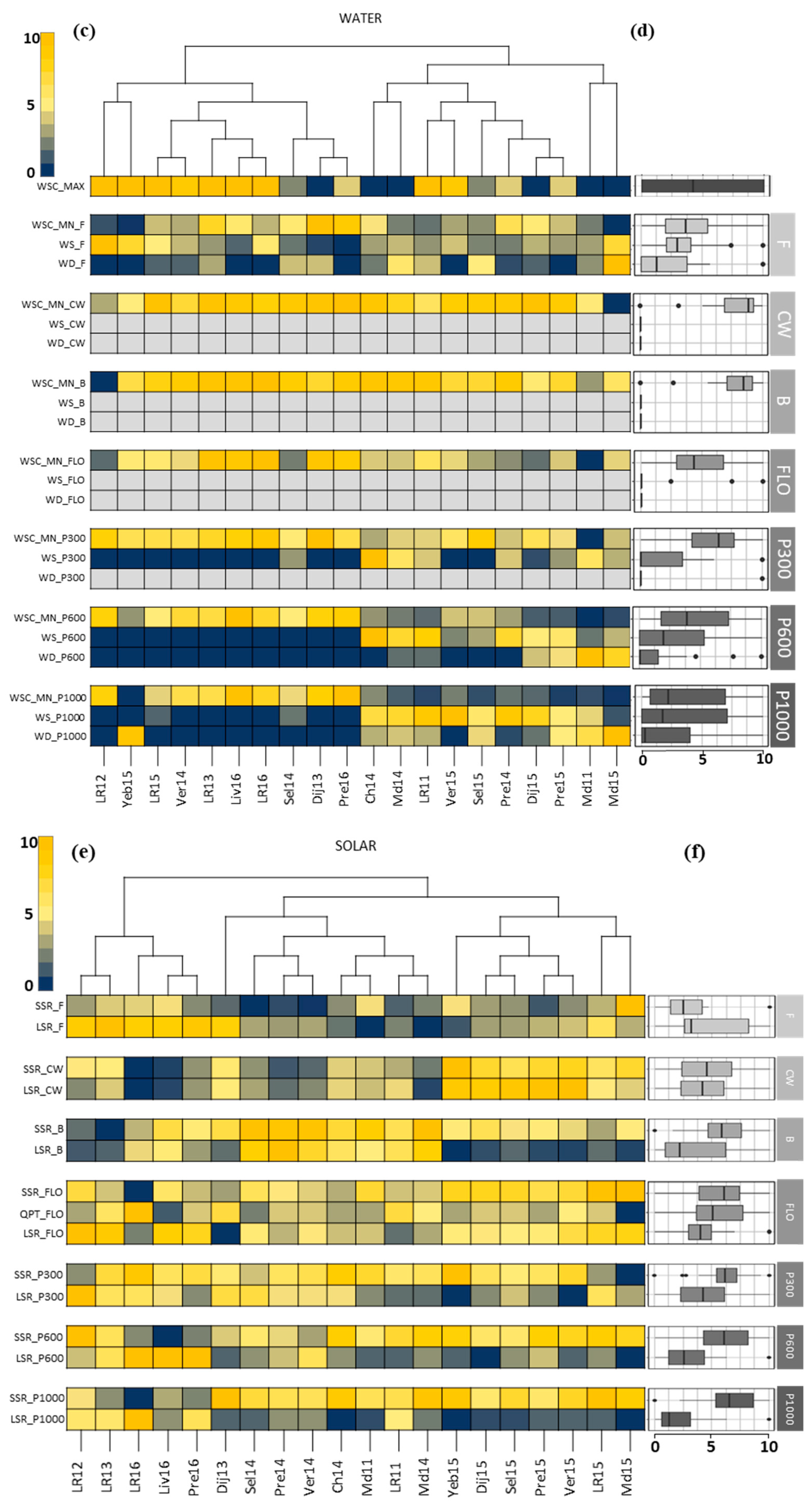
3.1. Description of the Field Network and Pedoclimatic Indicators
3.2. Identification of the Critical Indicators for Seed Yield
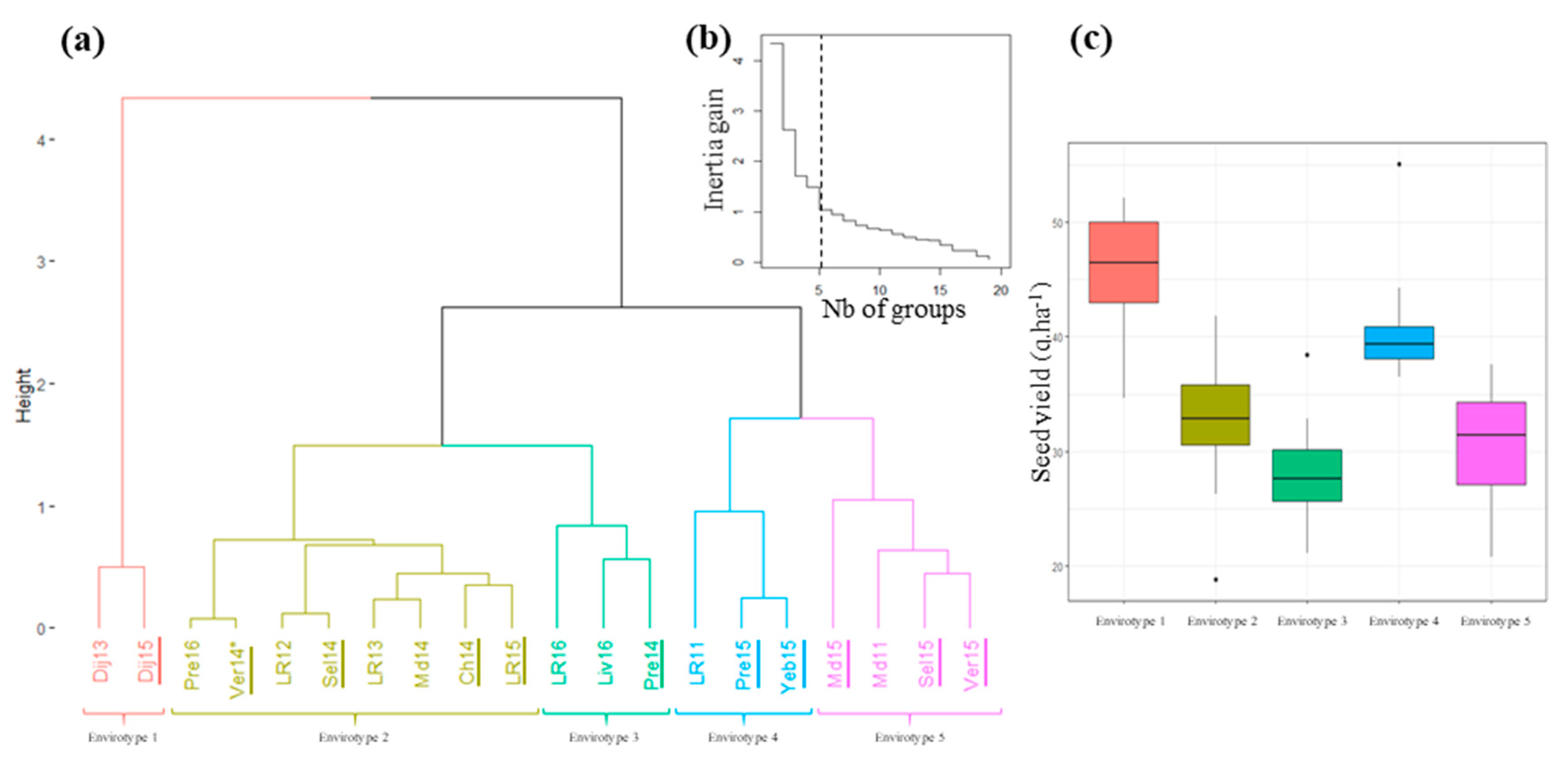
3.3. Definition of the Envirotypes
3.4. Evaluation and Decomposition of the G × E Interaction at the Network Level
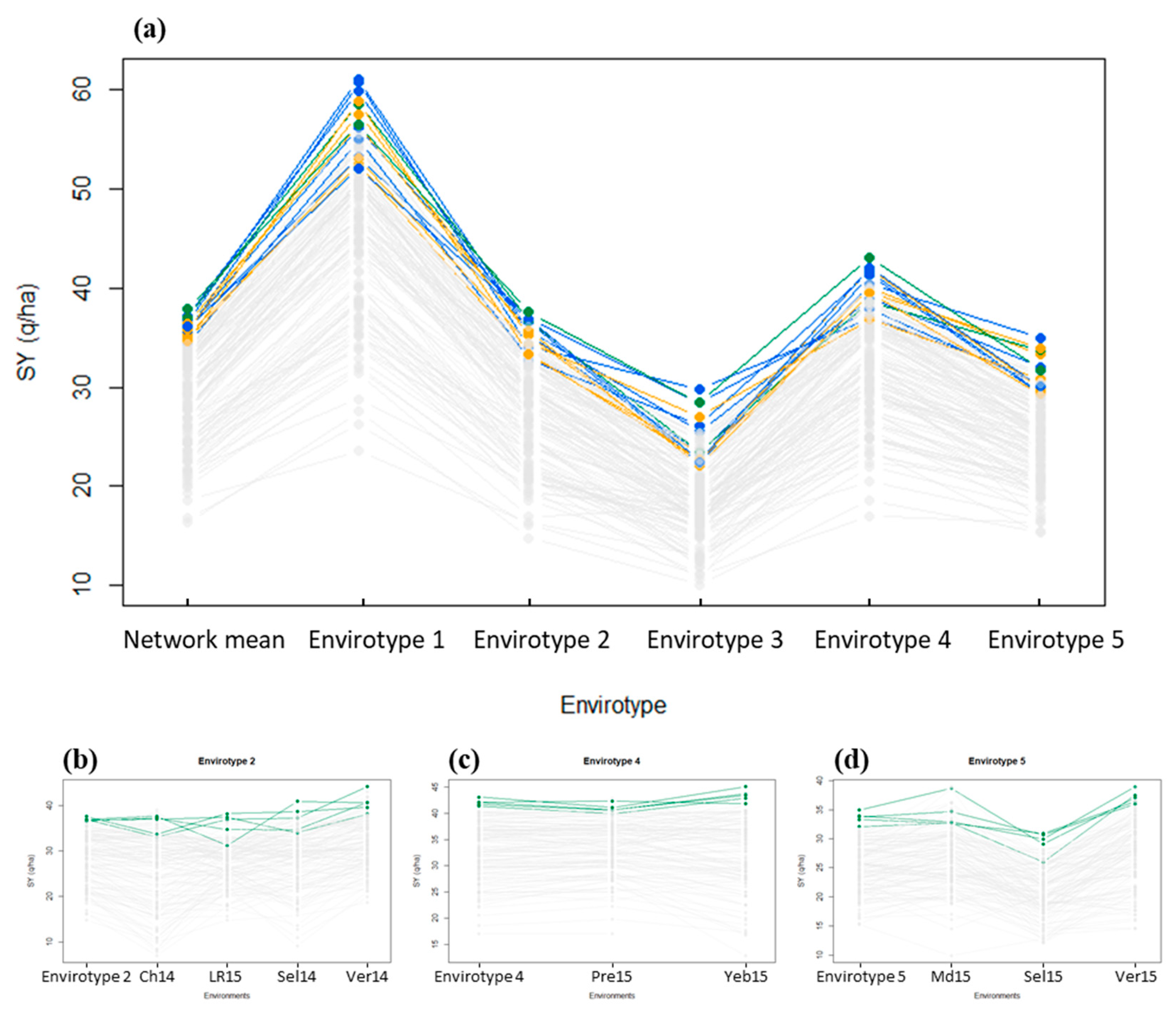
3.4.1. Effect of the Envirotyping on the G × E Decomposition
3.4.2. Ranking of the Genotypes Per Envirotype
4. Discussion
4.1. When Used in A Multi-Constraining Network the PLS Regression Selected Key Indicators That Were Critical for Seed Yield
4.2. Critical Factors for Seed Yield in WOSR Were Mostly Related to Heat Stress, Radiation Deficit, and Water Shortage during Vernalization and Reproductive Periods
4.3. An Approach to Capture the Components of the Environmental Effect
4.4. An Approach that Provided First Clues to Tackle the G × E Interaction
4.5. Get Further into the Genetic Determinant of the G × E Interaction and Its Interest for Breeding Programs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- von Wricke, G. Über eine Methode zur Erfassung der ökologischen Streubreite in Feldversuchen. Z. Planznezücht 1962, 47, 92–96. [Google Scholar]
- Finlay, K.W.; Wilkinson, G.N. The analysis of adaptation in a plant-breeding programme. Aust. J. Agric. Res. 1963, 14, 742–754. [Google Scholar] [CrossRef]
- Brancourt-Hulmel, M.; Biarnès-Dumoulin, V.; Denis, J. Points de repère dans l’analyse de la stabilité et de l’interaction génotype-milieu en amélioration des plantes. Agronomie 1997, 17, 219–246. [Google Scholar] [CrossRef]
- Denis, J.B. Two way analysis using covariates1. Stat. A J. Theor. Appl. Stat. 1988, 19, 123–132. [Google Scholar]
- Vargas, J.; van Eeuwijk, F.; Sayre, K.D.; Reynolds, P.M.C. Interpreting treatment x environment interactin in agronomy trials. Agron. J. 2001, 93, 949–960. [Google Scholar] [CrossRef]
- Van Ittersum, M.K.; Cassman, K.G.; Grassini, P.; Wolf, J.; Tittonell, P.; Hochman, Z. Yield gap analysis with local to global relevance-A review. Filed Crop. Res. 2013, 143, 4–17. [Google Scholar] [CrossRef]
- FAOSTAT Production/Yield Quantities of Rapeseed in World. Available online: http://www.fao.org/faostat/en/?#data/QC/visualize (accessed on 20 April 2019).
- Bouchet, A.S.; Laperche, A.; Bissuel-Belaygue, C.; Baron, C.; Morice, J.; Rousseau-Gueutin, M.; Dheu, J.E.; George, P.; Pinochet, X.; Foubert, T.; et al. Genetic basis of nitrogen use efficiency and yield stability across environments in winter rapeseed. BMC Genet. 2016, 17, 131. [Google Scholar] [CrossRef]
- He, D.; Wang, E.; Wang, J.; Lilley, J.M. Genotype × environment × management interactions of canola across China: A simulation study. Agric. For. Meteorol. 2017, 247, 424–433. [Google Scholar] [CrossRef]
- Moghaddam, M.J.; Pourdad, S.S. Genotype × environment interactions and simultaneous selection for high oil yield and stability in rainfed warm areas rapeseed (Brassica napus L.) from Iran. Euphytica 2011, 180, 321–335. [Google Scholar] [CrossRef]
- Zhang, H.; Berger, J.D.; Milroy, S.P. Genotype×environment interaction studies highlight the role of phenology in specific adaptation of canola (Brassica napus) to contrasting Mediterranean climates. Filed Crop. Res. 2013, 144, 77–88. [Google Scholar] [CrossRef]
- Metzger, M.J.; Bunce, R.G.H.; Jongman, R.H.G.; Mücher, C.A.; Watkins, J.W. A climatic stratification of the environment of Europe. Glob. Ecol. Biogeogr. 2005, 14, 549–563. [Google Scholar] [CrossRef]
- Habekotté, B. Evaluation of seed yield determining factors of winter oilseed rape (Brassica napus L.) by means of crop growth modelling. Filed Crop. Res. 1997, 54, 137–151. [Google Scholar] [CrossRef]
- Champolivier, L.; Merrien, A. Effects of water stress applied at different growth stages to Brassica napus L. var. oleifera on yield, yield components and seed quality. Eur. J. Agron. 1996, 5, 153–160. [Google Scholar] [CrossRef]
- Parnaudeau, V.; Jeuffroy, M.; Machet, J.; Reau, R.; Bissuel, C. Methods for determining the nitrogen fertiliser requirements of some major arable crops in France. In Proceedings of the International Fertiliser Society, Cambridge, UK, 11 December 2009; pp. 1–26. [Google Scholar]
- Rémy, J.; Hébert, J. Le devenir des engrais azotés dans le sol. Acad. l’Agric. Fr. 1977, 63, 700–714. [Google Scholar]
- Colnenne, C.; Meynard, J.; Reau, R.; Justes, E.; Merrien, A. Determination of a Critical Nitrogen Dilution Curve for Winter Oilseed Rape. Ann. Bot. 1998, 81, 311–317. [Google Scholar] [CrossRef]
- Lancashire, P.D.; Bleiholder, H.; Van Den BOOM, T.; Langelüddeke, P.; Stauss, R.; Weber, E.; Witzenberger, A. A uniform decimal code for growth stages of crops and weeds. Ann. Appl. Biol. 1991, 119, 561–601. [Google Scholar] [CrossRef]
- Weber, E.; Bleiholder, H. Explanations of the BBCH decimal codes for the growth stages of maize, rape, faba beans, sunflowers and peas-with illustrations. Gesunde Pflanz. 1990, 42, 308–321. [Google Scholar]
- Gabrielle, B.; Denoroy, P.; Gosse, G.; Justes, E.; Andersen, M.N. Development and evaluation of a CERES-type model for winter oilseed rape. Filed Crop. Res. 1998, 57, 95–111. [Google Scholar] [CrossRef]
- Hebinger, H. Le Colza; Editions France Agricole: Paris, France, 2013; ISBN 978-2-85557-241-3. [Google Scholar]
- Leterme, P. Modelisation De La Croissance Et De La Production Des Siliques Chez Le Colza D’hiver (Brassica napus L.); INA Paris-Grignon: Paris, France, 1985. [Google Scholar]
- Jullien, A.; Mathieu, A.; Allirand, J.M.; Pinet, A.; De Reffye, P.; Cournède, P.H.; Ney, B. Characterization of the interactions between architecture and sourcesink relationships in winter oilseed rape (Brassica napus) using the GreenLab model. Ann. Bot. 2011, 107, 765–779. [Google Scholar] [CrossRef]
- Météo France Données Quotidiennes Du Modèle De Simulation Des Schémas De Surface. Available online: https://donneespubliques.meteofrance.fr/?fond=produit&id_produit=230&id_rubrique=40 (accessed on 1 January 2016).
- Bruand, A.; Duval, O.; Cousin, I. Estimation des propriétés de rétention en eau des sols à partir de la base de données SOLHYDRO: Une première proposition combinant le type d’horizon, sa texture et sa densité apparente. Étude Gest. Des Sols 2004, 11, 323–334. [Google Scholar]
- Fan, J.; McConkey, B.; Wang, H.; Janzen, H. Root distribution by depth for temperate agricultural crops. Filed Crop. Res. 2016, 189, 68–74. [Google Scholar] [CrossRef]
- Lacoste, M.; Mulder, V.L.; Richer-De-Forges, A.C.; Martin, M.P.; Arrouays, D. Evaluating large-extent spatial modeling approaches: A case study for soil depth for France. Geoderma Reg. 2016, 7, 137–152. [Google Scholar] [CrossRef]
- Allen, R.G.; Pereira, L.S.; Raes, D.; Smith, M. Crop evapotranspiration—Guidelines for computing crop water requirements - FAO Irrigation and drainage paper 56. Irrig. Drain. 1998, 1–15. [Google Scholar]
- Weymann, W.; Böttcher, U.; Sieling, K.; Kage, H. Effects of weather conditions during different growth phases on yield formation of winter oilseed rape. Filed Crop. Res. 2015, 173, 41–48. [Google Scholar] [CrossRef]
- Fischer, R.A. Number of kernels in wheat crops and the influence of solar radiation and temperature. J. Agric. Sci. 1985, 105, 447–461. [Google Scholar] [CrossRef]
- Baux, A.; Wegmüller, J.; Holzkämper, A. Exploring Climatic Impact on Oilseed Rape Yield in Switzerland. Procedia Environ. Sci. 2015, 29, 123. [Google Scholar] [CrossRef]
- Ward, J.H. Hierarchical grouping to optimize an objective function. J. Am. Stat. Assoc. 1963, 58, 236–244. [Google Scholar] [CrossRef]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2017. [Google Scholar]
- Wold, S.; Albano, C.; Dunn, W.J., III; Esbensen, K.; Hellberg, S.; Johansson, E.; Sjöström, H. Pattern Recognition: Finding and Using Regularities in Multivariate Data. In “Food Research and Data Analysis”, Proceedings of the IUFoST Conference, Oslo, Norway, 20–23 September 1982; Martens, J., Ed.; Applied Science Publications: London, UK, 1983; pp. 147–188. [Google Scholar]
- Palermo, G.; Piraino, P.; Zucht, H.D. Performance of PLS regression coefficients in selecting variables for each response of a multivariate PLS for omics-type data. Adv. Appl. Bioinform. Chem. 2009, 2, 57–70. [Google Scholar] [CrossRef]
- Tenenhaus, M. La Régression PLS: Théorie Et Pratique; Editions Technip: Paris, France, 1998; ISBN 2-7108-0735-1. [Google Scholar]
- Schüürmann, G.; Ebert, R.U.; Chen, J.; Wang, B.; Kühne, R. External validation and prediction employing the predictive squared correlation coefficient—Test set activity mean vs. training set activity mean. J. Chem. Inf. Model. 2008, 48, 2140–2145. [Google Scholar] [CrossRef]
- Gauchi, J.P.; Chagnon, P. Comparison of selection methods of explanatory variables in PLS regression with application to manufacturing process data. Chemom. Intell. Lab. Syst. 2001, 58, 171–193. [Google Scholar] [CrossRef]
- Sanchez, G. Plsdepot: Partial Least Squares (PLS) Data Analysis Methods, R package version 0.1.17; 2012; Available online: https://CRAN.R-project.org/package=plsdepot (accessed on 22 November 2019).
- Krzanowski, W.J.; Lai, Y.T. A Criterion for Determining the Number of Groups in a Data Set Using Sum-of-Squares Clustering. Biometrics 1988, 44, 23. [Google Scholar] [CrossRef]
- Lê, S.; Josse, J.; Husson, F. FactoMineR: A Package for Multivariate Analysis. J. Stat. Softw. 2008, 25, 1–18. [Google Scholar] [CrossRef]
- Husson, F.; Lê, S.; Pagès, J. Computer Science and Data Analysis Series Exploratory Multivariate Analysis by Example Using R; Chapman and Hall/CRC: Boca Raton, FL, USA, 2017; ISBN 9781138196346. [Google Scholar]
- Chandler, J.; Corbesier, L.; Spielmann, P.; Dettendorfer, J.; Stahl, D.; Apel, K.; Melzer, S. Modulating flowering time and prevention of pod shatter in oilseed rape. Mol. Breed. 2005, 15, 87–94. [Google Scholar] [CrossRef]
- Ferreira, M.E.; Satagopan, J.; Yandell, B.S.; Williams, P.H.; Osborn, T.C. Mapping loci controlling vernalization requirement and flowering time in Brassica napus. Theor. Appl. Genet. 1995, 90, 727–732. [Google Scholar] [CrossRef] [PubMed]
- Sheldon, C.C.; Finnegan, E.J.; Rouse, D.T.; Tadege, M.; Bagnall, D.J.; Helliwell, C.A.; Peacock, W.J.; Dennis, E.S. Control of Flowering By Vernalization. Curr. Opin. Plant Biol. 2000, 3, 418–422. [Google Scholar] [CrossRef]
- Tadege, M.; Sheldon, C.C.; Helliwell, C.A.; Stoutjesdijk, P.; Dennis, E.S.; Peacock, W.J. Control of flowering time by FLC orthologues in Brassica napus. Plant J. 2001, 28, 545–553. [Google Scholar] [CrossRef]
- Morrison, M.J. Heat stress during reproduction in summer rape. Can. J. Bot. 1993, 71, 303–308. [Google Scholar] [CrossRef]
- Angadi, S.V.; Cutforth, H.W.; Miller, P.R.; Mcconkey, B.G.; Entz, M.H.; Brandt, S.A. Response of three Brassica species to high temperature stress during reproductive growth. Can. J. Plant Sci. 2000, 80, 693–702. [Google Scholar] [CrossRef]
- Young, L.W.; Wilen, R.W.; Bonham-smith, P.C. High temperature stress of Brassica napus during fowering reduces micro- and megagametophyte fertility, induces fruit abortion, and disrupts seed production. J. Exp. Bot. 2004, 55, 485–495. [Google Scholar] [CrossRef]
- Nuttal, W.F.; Moulin, A.P.; Townley-Smith, L.J. Yield Response of Canola to Nitrogen, Phosphorus, Precipitation, and Temperature. Agron. J. 1992, 84, 765–768. [Google Scholar] [CrossRef]
- Parisot-Baril, C. Etude De La Stabilite Du Rendement Chez Le Ble Tendre D’hiver (Triticum Aestivum L. THELL.); Université Paris-Sud: Orsay, France, 1992. [Google Scholar]
- Yan, W. Analysis and Handling of G × E in a Practical Breeding Program. Crop Sci. 2016, 56, 2106–2118. [Google Scholar] [CrossRef]


| Descriptor | Description | Category | Unit |
|---|---|---|---|
| LGDD* | Length of a crop period expressed in growing degree-days. Sum of the daily Tmean for the period (base temperature 0 °C) | Temperature | °C.d |
| TMN | Mean of the daily Tmean recorded over a given crop period | Temperature | °C |
| TMIN | Minimal temperature over a given crop period | Temperature | °C |
| TMAX | Maximal temperature over a given crop period | Temperature | °C |
| WSC_MAX* | Maximal water soil content for a given environment | Water | mm |
| WSC_MN | Mean of the water soil content over a given crop period (in proportion of WSC_MAX) | Water | % |
| SSR | Sum of the daily solar radiation over a given crop period | Solar | J cm−2 |
| QPT* | Photothermal quotient of SSR by LGDD (SSR/LGDD) | Solar | MJ m−2 °C−1 |
| Descriptor | Description | Category |
| HT | Number of days with high temperature (Tmax > 25 °C) | Temperature |
| LT | Number of days with low temperature (0 °C < Tmin < 5 °C) | Temperature |
| FR | Number of freezing days (Tmin < 0 °C) | Temperature |
| WS | Number of days when WSC < 1/3 WSC_MAX | Water |
| WD | Number of days when WSC = 0 mm | Water |
| LSR | Number of days with a lack of solar radiation (SR < 900 J cm−2) | Solar |
| VERN_OPT* | Number of days with an optimal vernalization treatment (Tmean < 5 °C and day length < 9 h) | Plant |
| Pedoclimatic Indicator | Confidence Index | Correlated Indicators |
|---|---|---|
| VERN_OPT | 1 | LT_CW |
| TMAX_FLO | 0.95 | - |
| HT_P600 | 0.9 | FR_CW (0.72); TMAX_P600 (0.83); TMN_P600 (0.87); HT_P1000 (0.70) |
| SSR_P600 | 0.85 | LSR_P600 (−0.70) |
| TMN_CW | 0.85 | TMIN_B (0.73) |
| HT_P300 | 0.8 | TMAX_P300 (0.70) |
| LSR_FLO | 0.8 | SSR_FLO (−0.70) |
| WS_P1000 | 0.75 | WS_P600 (0.77); WSC_MN_P600 (−0.68); WSC_MN_P1000 (−0.70) |
| TMIN_P300 | 0.65 | TMN_FLO (0.70); LT_FLO (−0.68); LGDD_FLO (0.70); TMN_P300 (0.71); LT_P300 (−0.88) |
| TMN_P1000 | 0.6 | TMN_FLO (0.70); LGDD_FLO (0.70); TMAX_P1000 (0.76); HT_P1000 (0.89); LSR_P1000 (−0.76) |
| G | E | G × E | E × R | Residuals | |
|---|---|---|---|---|---|
| Sum Sq | 70821 | 125131 | 28620 | 6069 | 14190 |
| %Sum Sq | 28.9 | 51.1 | 11.7 | 2.5 | 5.8 |
| Pvalue | *** | *** | *** | *** |
| G | C | C × E | G × C | G × C × E | C × E × R | Residuals | |
|---|---|---|---|---|---|---|---|
| Sum Sq | 70821 | 113195 | 11936 | 12195 | 16425 | 6069 | 14190 |
| %Sum Sq | 28.9 | 46.2 | 4.9 | 5.0 | 6.7 | 2.5 | 5.8 |
| Pvalue | *** | *** | *** | *** | *** | *** |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Corlouer, E.; Gauffreteau, A.; Bouchet, A.-S.; Bissuel-Bélaygue, C.; Nesi, N.; Laperche, A. Envirotypes Based on Seed Yield Limiting Factors Allow to Tackle G × E Interactions. Agronomy 2019, 9, 798. https://doi.org/10.3390/agronomy9120798
Corlouer E, Gauffreteau A, Bouchet A-S, Bissuel-Bélaygue C, Nesi N, Laperche A. Envirotypes Based on Seed Yield Limiting Factors Allow to Tackle G × E Interactions. Agronomy. 2019; 9(12):798. https://doi.org/10.3390/agronomy9120798
Chicago/Turabian StyleCorlouer, Erwan, Arnaud Gauffreteau, Anne-Sophie Bouchet, Christine Bissuel-Bélaygue, Nathalie Nesi, and Anne Laperche. 2019. "Envirotypes Based on Seed Yield Limiting Factors Allow to Tackle G × E Interactions" Agronomy 9, no. 12: 798. https://doi.org/10.3390/agronomy9120798
APA StyleCorlouer, E., Gauffreteau, A., Bouchet, A.-S., Bissuel-Bélaygue, C., Nesi, N., & Laperche, A. (2019). Envirotypes Based on Seed Yield Limiting Factors Allow to Tackle G × E Interactions. Agronomy, 9(12), 798. https://doi.org/10.3390/agronomy9120798





