Agricultural and Other Biotechnological Applications Resulting from Trophic Plant-Endophyte Interactions
Abstract
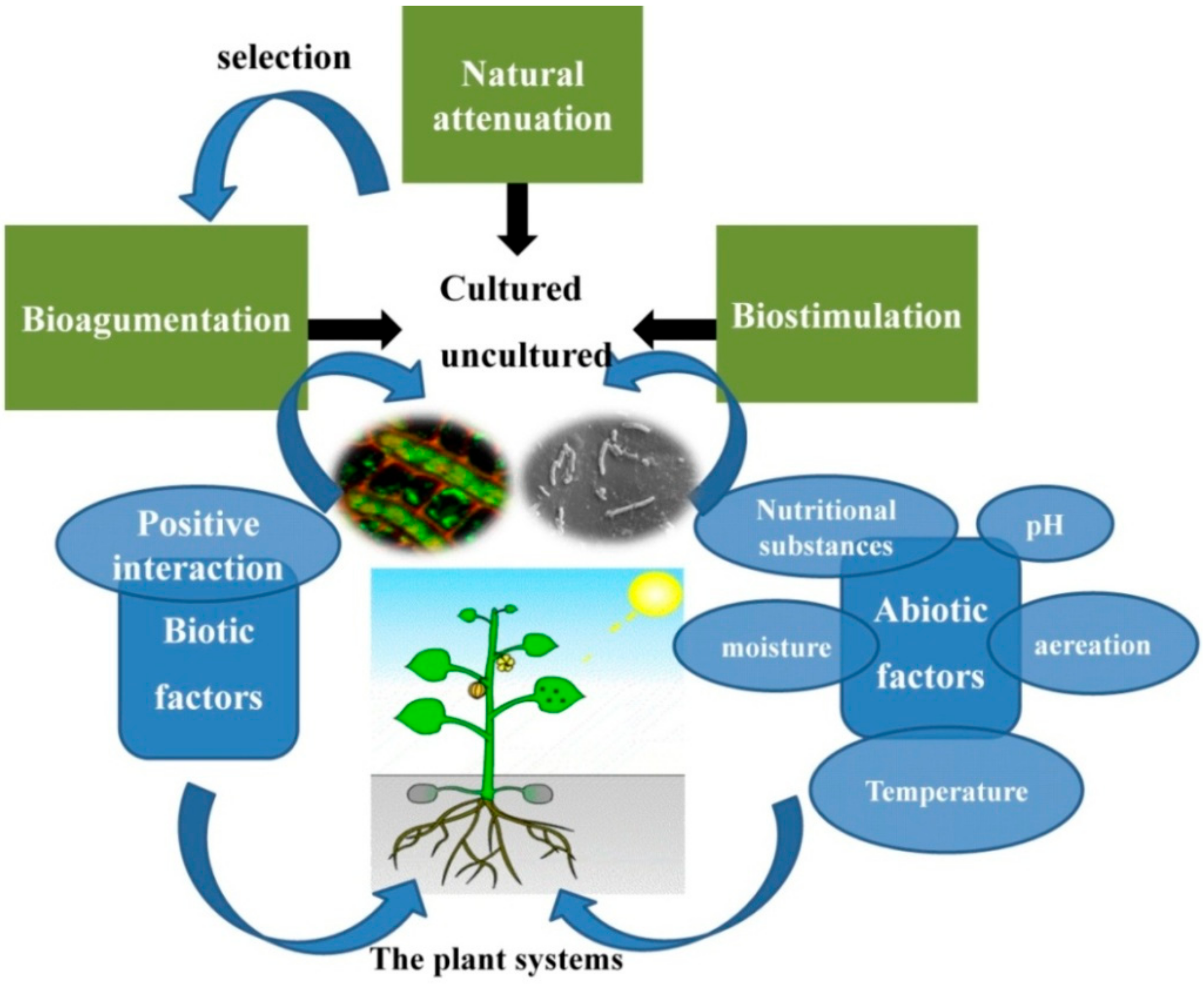
1. Introduction
2. Next Generation Sequencing Platforms as an Effective Tool in Identification of Endophytes
3. Biosynthesis of Factors Stimulating Plant Growth
4. Endophytic Strains Applied for Commercial-Scale Production as Biofertilizers and Biopreparations
5. Studies of Improvement of Ecological Adaptation of the Host in Salinity and Drought Conditions
6. Application of Endophytes for Bioremediation, Biotransformation, and Production of Bioactive Metabolites
7. The Role of Endophytes in Support of Defense Mechanisms in Plants
8. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Tenguria, R.K.; Khan, F.N.; Quereshi, S. Endophytes–mines of pharmacological therapeutics. World J. Sci. Technol. 2011, 1, 127–149. [Google Scholar]
- Stępniewska, Z.; Kuźniar, A. Endophytic microorganisms–promising applications in bioremediation of greenhouse gases. Appl. Microbiol. Biotechnol. 2013, 97, 9589–9596. [Google Scholar] [CrossRef] [PubMed]
- Pylak, M.; Oszust, K.; Frąc, M. Review report on the role of bioproducts, biopreperations, biostimulants and microbial inoculants in organic production of fruit. Rev. Environ. Sci. Biotechnol. 2019. [Google Scholar] [CrossRef]
- Hardoim, P.R.; van Overbeek, L.S.; Berg, G.; Pirttila, A.M.; Compant, S.; Campisano, A.; Doring, M.; Sessitsch, A. The hidden world within plants: Ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol. Mol. Biol. Rev. 2015, 79, 293–320. [Google Scholar] [CrossRef]
- Tan, R.X.; Zou, W.X. Endophytes: A rich source of functional metabolites. Nad. Prod. Rep. 2001, 18, 448–459. [Google Scholar] [CrossRef]
- Frank, A.C.; Saldierna, G.J.P.; Shay, J.E. Transmission of bacterial endophytes. Microorganisms 2017, 5, 70. [Google Scholar] [CrossRef]
- Toubal, S.; Bouchenak, O.; Elhaddad, D.; Yahiaoui, K.; Boumaza, S.; Arab, K. MALDI-TOF MS detection of endophytic bacteria associated with great nettle (Urtica dioica L.), grown in Algeria. Pol. J. Microbiol. 2018, 67, 67–72. [Google Scholar] [CrossRef]
- Yaish, M.W.; Al-Lawati, A.; Jana, G.A.; Patankar, V.H.; Glick, B.R. Impact of soil salinity on the structure of the bacterial endophytic community identified from the roots of Caliph Medic (Medicago truncatula). PLoS ONE 2016, 11, e0179007. [Google Scholar] [CrossRef]
- Le Cocq, K.; Gurr, S.J.; Hirscg, P.R.; Mauchline, T.H. Exploitation of endophytes for sustainable agricultural intensification. Mol. Plant Pathol. 2017, 18, 469–473. [Google Scholar] [CrossRef]
- Liu, H.; Carvalhais, L.C.; Crawford, M.; Singh, E.; Dennis, P.G.; Pieterse, C.M.J.; Schenk, P.M. Inner plant values: Diversity, colonization and benefits from endophytic bacteria. Front. Microbiol. 2017, 8, 2552. [Google Scholar] [CrossRef]
- Akinsanya, M.A.; Goh, J.K.; Lim, S.P.; Ting, A.S. Metagenomics study of endophytic bacteria in Aloe vera using next-generation technology. Genom. Data 2015, 6, 159–163. [Google Scholar] [CrossRef] [PubMed]
- Eichmeier, A.; Pećenka, J.; Peňázová, E.; Baránek, M.; Catalá-García, S.; León, M.; Armengol, J.; Gramaje, D. High-throughput amplicon sequencing-based analysis of active fungal communities inhabiting grapevine after hot-water treatments reveals unexpectedly high fungal diversity. Fungal Ecol. 2018, 36, 26–38. [Google Scholar] [CrossRef]
- Kunda, P.; Dhal, P.K.; Mukherjee, A. Endophytic bacterial community of rice (Oryza sativa L.) from coastal saline zone of West Bengal: 16S rRNA gene based metagenomics approach. Meta Gene 2018, 18, 79–86. [Google Scholar] [CrossRef]
- Nguyen, H.Q.; Vu, N.T.H.; Chu, H.H.; Chu, S.K.; Hoang, H.; Tran, T.T.; Nguyen, C.; Dinh, L.T.M.; Trinh, H.T.T.; Phib, T.Q. Draft genome sequence of Streptomyces cavourensis YBQ59, an endophytic producer of antibiotics bafilomycin D, Nonactic Acid, Prelactone B, and 5,11-Epoxy-10-Cadinanol. Microbiol. Res. Ann. 2018, 7, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Battu, L.; Reddy, M.M.; Goud, B.S.; Ulaganathan, K.; Kandasamy, U. Genome inside genome: NGS based identification and assembly of endophytic Sphingopyxis granuli and Pseudomonas aeruginosa genomes from rice genomic reads. Genomics 2017, 109, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Tanjung, Z.A.; Aditama, R.; Sudania, W.M.; Utomo, C.; Liwang, T. Metagenomics workflow analysis of endophytic bacteria from oil palm fruits. J. Phys. Conf. Ser. 2017, 835, 1–6. [Google Scholar] [CrossRef]
- Mareque, C.; Freitas da Silva, T.; Vollú, R.E.; Beracochea, M.; Seldin, L.; Battistoni, F. The endophytic bacterial microbiota associated with sweet sorghum (Sorghum bicolor) is modulated by the application of chemical N fertilizer to the field. Int. J. Genom. 2018, 2018, 1–10. [Google Scholar] [CrossRef]
- Wolińska, A. Metagenomic achievements in microbial diversity determination in croplands: A review. In Microbial Diversity in Genomic Era; Das, S., Dash, H.R., Eds.; Academic Press: Cambridge, MA, USA, 2019; Chapter 2; pp. 15–35. [Google Scholar]
- Goodwin, S.; McPherson, J.D.; McCombie, W.R. Coming of age: Ten years of next-generation sequencing technologies. Nat. Rev. Genet. 2016, 17, 333–351. [Google Scholar] [CrossRef]
- Dijk, W.; Jaszczyszyn, Y.; Naquin, D.; Thermes, C. The third revolution in sequencing technology. Trends Genet. 2018, 34, 666–681. [Google Scholar] [CrossRef]
- Wolińska, A.; Kuźniar, A.; Zielenkiewicz, U.; Banach, A.; Izak, D.; Stępniewska, Z.; Błaszczyk, M. Metagenomic analysis of some potential nitrogen-fixing bacteria in arable soils at different formation process. Microb. Ecol. 2017, 73, 162–176. [Google Scholar] [CrossRef]
- Egan, A.N.; Schlueter, J.; Spooner, D.M. Applications of next-generation sequencing in plant biology. Am. J. Bot. 2012, 99, 175–185. [Google Scholar] [CrossRef] [PubMed]
- Maropola, M.K.A.; Ramond, J.B.; Trindade, M. Impact of metagenomic DNA extraction procedures on the identiable endophytic bacterial diversity in Sorghum bicolour (L. Moench). J. Microbiol. Met. 2015, 112, 104–117. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Escobedo, R.; Briones-Roblero, C.I.; Pineda-Mendoza, R.M.; Rivera-Orduña, F.N.; Zúñiga, G. Bacteriome from Pinus arizonica and P. durangensis: Diversity, comparison of assemblages, and overlapping degree with the gut bacterial community of a bark beetle that kills pines. Front. Microbiol. 2018, 9, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Fadrosh, D.W.; Ma, B.; Gajer, P.; Sengamalay, N.; Ott, S.; Brotman, R.M.; Ravel, J. An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome 2014, 2, 6. [Google Scholar] [CrossRef] [PubMed]
- Grada, A.; Weinbrecht, K. Next-Generation Sequencing: Methodology and Application. J. Investig. Dermatol. 2013, 133, e11. [Google Scholar] [CrossRef] [PubMed]
- Muzzey, D.; Evans, E.A.; Lieber, C. Understanding the basics of NGS: From mechanism to variant calling. Curr. Genet. Med. Rep. 2015, 3, 158–165. [Google Scholar] [CrossRef] [PubMed]
- Rhoads, A.; Au, K.F. PacBio sequencing and its applications. Genom. Proteom. Bioinform. 2015, 13, 278–289. [Google Scholar] [CrossRef]
- Brown, S.D.; Nagaraju, S.; Utturkar, S.; De Tissera, S.; Segovia, S.; Mitchell, W.; Land, M.L.; Dassanayake, A.; Köpke, M. Comparison of singe-molecule sequencing and hybrid approaches for finishing the genome of Clostridium autoethanogenum and analysis of CRISPR systems in industrial relevant Clostridia. Biotechnol. Biofuels 2014, 7, 40. [Google Scholar] [CrossRef]
- Tibpromma, S.; Hyde, K.D.; Bhat, J.D.; Mortimer, P.E.; Xu, J.; Promputtha, I.; Doilom, M.; Yang, J.; Tang, A.M.C.; Karunarathna, S.C. Identification of endophytic fungi from leaves of Pandanaceae based on their morphotypes and DNA sequence data from southern Thailand. MycoKeys 2018, 33, 25–67. [Google Scholar] [CrossRef]
- Blawid, R.; Silva, J.M.F.; Nagata, T. Discovering and sequencing new plant viral genomes by next-generation sequencing: Description of a practical pipeline. Ann. Appl. Biol. 2017, 170, 301–314. [Google Scholar] [CrossRef]
- Chaudhry, V.; Sharma, S.; Bansal, K.; Patil, P.B. Glimpse into the genomes of rice endophytic bacteria: Diversity and distribution of Firmicutes. Front. Microbiol. 2017, 7, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Annapurna, K.; Govindasamy, V.; Sharma, M.; Ghosh, A.; Chikara, S.K. Whole genome shotgun sequence of Bacillus paralicheniformis strain KMS 80, a rhizobacterial endophyte isolated from rice (Oryza sativa L.). 3 Biotech 2018, 8, 223. [Google Scholar] [CrossRef] [PubMed]
- Ren, Z.; Tang, S.; Jiang, Y.; Jiang, M.; Zheng, S.; Liu, W.; Yang, Z.; Sang, S.; Chen, Z.; Xia, T.; et al. High-throughput sequencing analysis of endophytic bacteria diversityin fruits of white and red pitayas from three different origins. Pol. J. Microbiol. 2018, 67, 27–35. [Google Scholar] [CrossRef]
- Dissanayake, A.J.; Purahong, W.; Wubet, T.; Hyde, K.D.; Zhang, W.; Xu, H.; Zhang, G.; Fu, C.; Liu, M.; Xing, Q.; et al. Direct comparison of culture-dependent and culture-independent molecular approaches reveal the diversity of fungal endophytic communities in stems of grapevine (Vitis vinifera). Fungal Divers. 2018, 90, 85–107. [Google Scholar] [CrossRef]
- Thomas, P.; Sekhar, A.C.; Shaik, S.P. High taxonomic diversity of cultivation-recalcitrant endophytic bacteria in grapevine field shoots, their in vitro introduction, and unsuspected persistence. Planta 2017, 246, 879–898. [Google Scholar] [CrossRef] [PubMed]
- Liao, H.-L.; Bonito, G.; Rojas, J.A.; Hameed, K.; Wu, S.; Schadt, C.W.; Labbe, J.L.; Tuskan, G.; Martin, F.M.; Grigoriev, I.V.; et al. Fungal endophytes of Populus trichocarpa alter host phenotype, gene expression and rhizobiome composition. Mol. Plant Microb. Interact. 2019. [Google Scholar] [CrossRef]
- Chen, Y.T.; Yuan, Q.; Shan, L.T.; Lin, M.A.; Cheng, D.Q.; Li, C.Y. Antitumor activity of bacterial exopolysaccharidesfrom the endophyte Bacillus amyloliquefaciens sp. isolated from Ophiopogon japonicas. Oncol. Lett. 2013, 5, 1787–1792. [Google Scholar] [CrossRef] [PubMed]
- Asaf, S.; Khan, A.L.; Khan, M.A.; Al-Harrasi, A.; Lee, I.J. Complete genome sequencing and analysis of endophytic Sphingomonas sp. LK11 and its potential in plant growth. 3 Biotech 2018, 8, 389. [Google Scholar] [CrossRef]
- Naylor, D.; DeGraaf, S.; Purdom, E.; Coleman-Derr, D. Drought and host selection influence bacterial community dynamics in the grass root microbiome. ISME J. 2017, 11, 2691–2704. [Google Scholar] [CrossRef]
- König, J.; Guerreiro, M.A.; Peršoh, D.; Begerow, D.; Krauss, J. Knowing your neighbourhood—the effects of Epichloë endophytes on foliar fungal assemblages in perennial ryegrass in dependence of season and land-use intensity. Peer J. 2018, 6, e4660. [Google Scholar] [CrossRef]
- Bullington, L.S.; Larkin, B.G. Using direct amplification and next-generation sequencing technology to explore foliar endophyte communities in experimentally inoculated western white pines. Fungal Ecol. 2015, 17, 170–178. [Google Scholar] [CrossRef]
- Ali, S.; Charles, T.C.; Glick, B.R. Amelioration of high salinity stress damage by plant growth-promoting bacterial endophytes that contain ACC deaminase. Plant Physiol. Biochem. 2014, 80, 160–167. [Google Scholar] [CrossRef] [PubMed]
- Pandey, R.; Chavan, P.N.; Walokar, N.M.; Sharma, N.; Tripathi, V.; Khetmalas, M.B. Pseudomonas stutzeri RP1: A versatile plant growth promoting endorhizospheric bacteria inhabiting sunflower (Helianthus annus). Res. J. Biotechnol. 2013, 8, 48–55. [Google Scholar]
- Singh, M.; Kumar, A.; Singh, R.; Pandey, K.D. Endophytic bacteria: A new source of bioactive compounds. 3 Biotech 2017, 7, 315. [Google Scholar] [CrossRef]
- Etesami, H.; Maheshwari, D.K. Use of plant growth promoting rhizobacteria (PGPRs) with multiple plant growth promoting traits in stress agriculture: Action mechanisms and future prospects. Ecotoxicol. Environ. Saf. 2018, 156, 225–246. [Google Scholar] [CrossRef]
- Zúñiga, A.; de la Fuente, F.; Lionne, F.F.C.; Bônnet, J.; de Lorenzo, V.; González, B. An engineered device for indoleacetic acid production under quorum sensing signals enables Cupriavidus pinatubonensis JMP134 to stimulate plant growth. ACS Synt. Biol. 2018, 7, 1519–1527. [Google Scholar] [CrossRef]
- Dawwam, G.E.; Elbeltagy, A.; Emara, H.M.; Abbas, I.H.; Hassan, M.M. Beneficial effect of plant growth promoting bacteriaisolated from the roots of potato plant. Ann. Agric. Sci. 2013, 58, 195–201. [Google Scholar] [CrossRef]
- Yu, J.; Yu, Z.H.; Fan, G.Q.; Wang, G.H.; Liu, X.B. Isolation and characterization of indole acetic acid producing root endophytic bacteria and their potential for promoting crop growth. J. Agric. Sci. Tech. 2016, 18, 1381–1391. [Google Scholar]
- Patil, N.B.; Gajbhiye, M.; Ahiwale, S.S.; Gunjal, A.B.; Kapadnis, B.P. Optimization of indole acetic acid (IAA) production by Acetobacter diazotrophicus L1 isolated from Sugarcane. Int. J. Environ. Sci. 2011, 2, 295–302. [Google Scholar]
- UmaMaheswari, T.; Anbukkarasi, K.; Hemalatha, T.; Chendrayan, K. Studies on phytohormone producing ability of indigenous endophytic bacteria isolated from tropical legume crops. Int. J. Curr. Microbiol. Appl. Sci. 2013, 2, 127–136. [Google Scholar]
- Franche, C.; Lindström, K.; Elmerich, C. Nitrogen-fixing bacteria associated with leguminous and non-leguminous plants. Plant Soil 2009, 321, 35–59. [Google Scholar] [CrossRef]
- Dimkpa, C.; Weinand, T.; Asch, F. Plant–rhizobacteria interactions alleviate abiotic stress conditions. Plant Cell Environ. 2009, 32, 1682–1694. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.R.S.; Rao, K.V.B. Biological nitrogen fixation: A review. Int. J. Adv. Life Sci. 2012, 1, 1–9. [Google Scholar]
- Ohyama, T.; Momose, A.; Ohtake, N.; Sueyoshi, K.; Sato, T.; Nakanishi, Y.; Asis, C.A.; Ruamsungsri, S.; Ando, S. Nitrogen fixation in sugarcane. Adv. Biol. Ecol. Nitrogen Fix. 2014, 3, 49–70. [Google Scholar]
- Aryantha, I.N.P.; Hidiyah, A.R.M. Colonization and performance of diazotroph endophytic bacteria on palm oil (Elaeis guineensis Jacq L.) leaves. In IOP Conference Series: Earth and Environmental Science; IOP Publishing Ltd.: Bristol, UK, 2018; Volume 166, Conference 1. [Google Scholar]
- Sharma, S.; Roy, S. Isolation and identification of a novel endophyte from a plant Amaranthus spinosus. Int. J. Curr. Microbiol. Appl. Sci. 2017, 4, 785–798. [Google Scholar]
- Prayitno, J.; Rolfe, B. Characterization of endophytic diazotroph bacteria isolated from rice. Hay. J. Biosci. 2010, 7, 73–78. [Google Scholar] [CrossRef]
- Saleem, M.; Arshad, M.; Hussain, S.; Bhatti, A.S. Perspective of plant growth promoting rhizobacteria (PGPR) containing ACC deaminase in stress agriculture. J. Ind. Microbiol. Biotechnol. 2007, 34, 635–648. [Google Scholar] [CrossRef]
- Long, H.H.; Schmidt, D.D.; Baldwin, I.T. Native bacterial endophytes promote host growth in a species-specific manner; phytohormone manipulations do not result in common growth responses. PLoS ONE 2008, 3, e2702. [Google Scholar] [CrossRef]
- Grobelak, A.; Kokot, P.; Świątek, J.; Jaskulak, M.; Rorat, A. Bacterial ACC deaminase activity in promoting plant growth on areas contaminated with heavy metals. J. Ecol. Engin. 2018, 19, 150–157. [Google Scholar] [CrossRef]
- Zhao, L.; Xu, Y.; Chang, J.; Li, M.; Zhang, Y.; Dang, Y.; Wang, M.; Cheng, Y.; Zhang, B. Screening, resistance and growth-promoting effect of endophytic bacteria with ACC deaminase activity isolated from soybean nodules. Wei Sheng Wu Xue Bao 2016, 56, 1009–1021. [Google Scholar]
- Ivanova, E.G.; Fedorov, D.N.; Doronina, N.V.; Trotsenko, Y.A. Production of vitamin B12 in aerobic methylotrophic bacteria. Microbiology 2006, 75, 494–496. [Google Scholar] [CrossRef]
- Mercado, B.J.; Bakker, P.A. Interaction between plants and beneficial Pseudomonas sp., exploiting bacterial traits for crop protection. Antonie Leeuwenhok. 2007, 92, 367–389. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekeran, M.; Paramasivan, M.; Chun, S.C. Bacillus subtilis CBR05 induces vitamin B6 biosynthesis in tomato through the de novo pathway in contributing disease resistance against Xanthomonas campestris pv. Vesicatoria. Sci. Rep. 2019, 9, 6495. [Google Scholar] [CrossRef]
- Black, L.J.; Lucas, R.M.; Sferriff, J.L.; Bjorn, L.O.; Bornman, J.F. In pursuit of vitamin D in plants. Nutrients 2017, 9, 136. [Google Scholar] [CrossRef] [PubMed]
- De Souza Leite, T.; Cnossen-Fassoni, A.; Pereira, O.L.; Mizubuti, E.S.; de Araujo, E.F.; de Queiroz, M.V. Novel and highly diverse fungal endophytes in soybean revealed by the consortium of two different techniques. J. Microbiol. 2013, 51, 56–69. [Google Scholar] [CrossRef]
- Flores-Felix, J.D.; Silva, R.L.; Rivera, L.P.; Marcos-Garcia, M.; Garcia-Fraile, P.; Martinez-Molina, E.; Mateos, P.F.; Velazquez, E.; Andrade, P.; Rivas, R. Plants probiotics as a tool to produce highly functional fruits: The case of Phyllobacterium and vitamin C in strawberries. PLoS ONE 2015, 10, e012228. [Google Scholar] [CrossRef]
- Vaidehi, K.; Sekar, C. Amino acid conjugated hydroxamate type of siderophore production in Methylobacterium phyllosphaerae mb-5. Cibtech J. Microbiol. 2012, 1, 24–30. [Google Scholar]
- Liaqat, F.; Eltem, R. Identification and characterization of endophytic bacteria isolated from in vitro cultures of peach and pear rootstocks. 3 Biotech 2016, 6, 120. [Google Scholar] [CrossRef]
- Goudjal, Y.; Zamoum, M.; Sabaou, N.; Mathieu, F.; Zitouni, A. Potential of endophytic Streptomyces spp. for biocontrol of Fusarium root rot disease and growth promotion of tomato seedlings. Biocon. Sci. Tech. 2016, 26, 1691–1705. [Google Scholar] [CrossRef]
- Jasim, B.; Joseph, A.A.; John, C.J.; Mathew, J.; Radhakrishnan, E.K. Isolation and characterization of plant growth promoting endophytic bacteria from the rhizome of Zingiber officinale. 3 Biotech 2014, 4, 197–204. [Google Scholar] [CrossRef]
- Zardak, S.G.; Dehnavi, M.M.; Salehi, A.; Gholamhoseini, M. Effects of using arbuscular mycorrhixal fungi to alleviate drought stress on the physiological traits and essential oil yield of fennel. Rhizosphere 2018, 6, 31–38. [Google Scholar] [CrossRef]
- Pusenkova, L.I.; Il’yasova, E.Y.; Lastochkina, O.V.; Maksimov, I.V.; Leonova, S.A. Changes in the species composition of the rhizosphere and phyllosphere of sugar beet under the impact of biological preparations based on endophytic bacteria and their metabolites. Eur. Soil Sci. 2016, 49, 1136–1144. [Google Scholar] [CrossRef]
- Gveroska, B. Fungicidal and stimulating effect of biopreparation TRIANUM P on tobacco seedlings. Tutun Tab. 2017, 67, 48–55. [Google Scholar]
- Horoszkiewicz-Janka, J.; Jajor, E. An influence of the selected bioproducts used as barley seed dressings on fungi establishment. J. Res. Appl. Agric. Eng. 2007, 52, 61–66. [Google Scholar]
- Badawi, M.H.; EL-Henawy, H.M.; Abd-Elgaffar, N.Y. Biomanagement of Fusarium solani and Rhizoctonia solani causing root rot and damping off diseases in common bean (Phaseolus vulgaris L.) via innovative rhizobacterial formulations. J. Appl. Sci. Res. 2013, 9, 321–329. [Google Scholar]
- Vessey, J.K. Plant growth promoting rhizobacteria as biofertilizers. Plant Soil 2003, 255, 571–586. [Google Scholar] [CrossRef]
- Mwajita, M.R.; Murage, H.; Tani, A.; Kahangi, E.M. Evaluation of rhizosphere, rhizoplane andphyllosphere bacteria and fungi isolated from ricein Kenya for plant growth promoters. Springer Plus 2013, 2, 1–9. [Google Scholar] [CrossRef]
- Vurukonda, S.S.K.P.; Vardharajula, S.; Shrivastava, M.; SkZ, A. Enhancement of drought stress tolerance in crops by plant growth promoting rhizobacteria. Microbiol. Res. 2016, 184, 13–24. [Google Scholar] [CrossRef]
- Li, H.; Wang, X.; Han, M.; Zhao, Z.; Wang, M.; Tang, Q.; Liu, C.; Kemp, B.; Gu, Y.; Shuang, J.; et al. Endophytic Bacillus subtilis ZZ120 and its potential application in control of replant diseases. Afric. J. Biotechnol. 2012, 11, 231–242. [Google Scholar]
- Gómez, O.C.; Luiz, J.H.H. Endophytic fungi isolated from medicinal plants: Future prospects of bioactive natural products from Tabebuia/Handroanthus endophytes. Appl. Microbiol. Biotechnol. 2018, 102, 9105. [Google Scholar] [CrossRef]
- Gai, C.S.; Dini-Andreote, F.; Andreote, F.D.; Lopes, J.R.S.; Araújo, W.L.; Miller, T.A.; Azevedo, J.L.; Lacava, P.T. Endophytic bacteria associated to sharpshooters (Hemiptera: Cicadellidae), insect vectors of Xylella fastidiosa subsp. pauca. Plant Pathol. Microbiol. 2011, 2, 1–8. [Google Scholar]
- Guerrero, A.P.M. Interrelationships Between Mutualistic Endophytic Microorganisms, the Root-knot Nematode Meloidogyne Incognita and the Sap-sucking Insect Aphis Gossypii on Tomato, Squash and Arabidopsis. Ph.D. Thesis, Universitat Bonn, Bonn, Germany, 2011; pp. 1–125. [Google Scholar]
- Muzzamal, H.; Sarwar, R.; Sajid, I.; Hasnain, S. Isolation, identification and screening of endophytic bacteria antagonistic to biofilm formers. Pak. J. Zool. 2012, 44, 249–257. [Google Scholar]
- Braga, R.M.; Padilla, G.; Araújo, W.L. The biotechnological potential of Epicoccum spp.: Diversity of secondary metabolites. Crit. Rev. Microbiol. 2018, 44, 759–778. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, C.F.; Vitorino, L.C.; Soares, M.A.; Souchie, E.L. Multifunctional potential of endophytic and rhizospheric microbial isolates associated with Butia purpurascens roots for promoting plant growth. Antoine Van Leeuwenhoek 2018, 111, 2157–2174. [Google Scholar] [CrossRef] [PubMed]
- Min, Y.J.; Park, M.S.; Fong, J.J.; Quan, Y.; Jung, S.; Lim, Y.W. Diversity and saline resistance of endophytic fungi associated with Pinus thunbergii in coastal shelterbelts of Korea. J. Microbiol. Biotechnol. 2014, 24, 324–333. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, R.J.; Henson, J.; Van Volkenburgh, E.; Hoy, M.; Wright, L.; Beckwith, F.; Kim, Y.O.; Redman, R.S. Stress tolerance in plants via habitat-adapted symbiosis. ISME J. 2008, 2, 404–416. [Google Scholar] [CrossRef] [PubMed]
- Kernaghan, G.; Mayerhofer, M.; Griffin, A. Fungal endophytes of wild and hybrid Vitis leaves and their potential for vineyard biocontrol. Can. J. Microbiol. 2017, 63, 583–595. [Google Scholar] [CrossRef]
- Lata, R.; Chowdhury, S.; Gond, S.K.; White, J.F. Induction of abiotic stress tolerance in plants by endophytic microbes. Lett. Appl. Microbiol. 2018, 66, 268–276. [Google Scholar] [CrossRef]
- Park, Y.H.; Kim, Y.; Mishra, R.C.; Bae, H. Fungal endophytes inhabiting mountain-cultivated ginseng (Panax ginseng Meyer): Diversity and biocontrol activity against ginseng pathogens. Sci. Rep. 2017, 7, 16221. [Google Scholar] [CrossRef]
- Khan, A.L.; Hamayun, M.; Ahmad, N.; Hussain, J.; Kang, S.M.; Kim, Y.H.; Adnan, M.; Tang, D.S.; Waqas, M.; Radhakrishnan, R.; et al. Salinity stress resistance offered by endophytic fungal interaction between Penicillium minioluteum LHL09 and Glycine max L. J. Microbiol. Biotechnol. 2011, 2, 893–902. [Google Scholar] [CrossRef]
- Hemida, K.A.; Reyad, A.M.M. Improvement salt tolerance of safflower plants by endophytic bacteria. J. Hortic. Plant Res. 2019, 5, 38–56. [Google Scholar] [CrossRef]
- Ahmad, N.M.; Hamayun, S.A.; Khan, A.L.; Lee, I.J.; Shin, D.H. Gibberellin-producing endophytic fungi isolated from Monochoria vaginalis. J. Microbiol. Biotechnol. 2010, 20, 1744–1749. [Google Scholar] [PubMed]
- Kane, K.H. Effects of endophyte infection on drought stress tolerance of Lolium perenne accessions from the Mediterranean region. Environ. Exp. Bot. 2011, 71, 337–344. [Google Scholar] [CrossRef]
- Hubbard, M.; Germida, J.J.; Vujanovic, V. Fungal endophytes enhance wheat heat and drought tolerance in terms of grain yield and second-generation seed viability. J. Appl. Microbiol. 2014, 116, 109–122. [Google Scholar] [CrossRef] [PubMed]
- Vigani, G.; Rolli, E.; Marasco, R.; Dell’Orto, M.; Michoud, G.; Soussi, A.; Raddadi, N.; Borin, S.; Sorlini, C.; Zocchi, G.; et al. Root bacterial endophytes confer drought resistance and enhance expression and activity of a vacuolar H+ pumping pyrophosphatase in pepper plants. Environ. Microbiol. 2018. [Google Scholar] [CrossRef]
- Vaishnav, A.; Shukla, A.K.; Sharma, A.; Kumar, R.; ·Choudhary, D.K. Endophytic bacteria in plant salt stress tolerance: Current and future prospects. J. Plant Growth Reg. 2018. [Google Scholar] [CrossRef]
- Ngumbi, E.; Kloepper, J. Bacterial-mediated drought tolerance: Current and future prospects. Appl. Soil Ecol. 2016, 105, 109–125. [Google Scholar] [CrossRef]
- Dastogeer, K.M.G.; Li, H.; Sivasithamparam, K.; Jones, M.G.K.; Wylie, S.J. Fungal endophytes and virus confer drought tolerance to Nicotiana benthamiana plants through modulating osmolytes, antioxidant enzymes and expression of host drought responsive genes. Environ. Exp. Bot. 2018, 149, 95–108. [Google Scholar] [CrossRef]
- Li, K.Q.; Xu, X.Y.; Huang, X.S. Identification of differentially expressed genes related to dehydration resistance in a highly drought-tolerant pear, Pyrus betulaefolia, as through RNA-Sequencing. PLoS ONE 2016, 11, e0149352. [Google Scholar] [CrossRef]
- Bao, A.K.; Du, B.Q.; Touil, L.; Kang, P.; Wang, Q.L.; Wang, S.M. Co-expression of tonoplast cation/H+ antiporter and H+-pyrophosphatase from xerophyte Zygophyllum xanthoxylum improves alfaalfa plant growth under salinity, drought and field conditions. Plant Biotechnol. J. 2016, 14, 964–975. [Google Scholar] [CrossRef]
- Raza, G.; Ali, K.; Asraf, M.Y.; Mansoor, S.; Javid, M.; Asad, S. Overexpression of an H+-PPase gene from Arabidopsis in sugarcane improves drought tolerance, plant growth, and photosynthetic responses. Turk. J. Biol. 2016, 40, 109–119. [Google Scholar] [CrossRef]
- Gaxiola, R.A.; Regmi, K.; Paez-Valencia, J.; Pizzio, G.; Zhang, S. Plant H +-PPases: Reversible enzymes with contrasting functions dependent on membrane environment. Mol. Plant 2016, 9, 317–319. [Google Scholar] [CrossRef] [PubMed]
- Li, H.-Y.; Wei, D.-Q.; Shen, M.; Zhou, Z.-P. Endophytes and their role in phytoremediation. Fungal Divers. 2012, 54, 11–18. [Google Scholar] [CrossRef]
- DalCorso, G.; Fasani, E.; Manara, A.; Visioli, G.; Furini, A. Heavy metal pollutions: State of the art and innovation in phytoremediation. Int. J. Mol. Sci. 2019, 20, 3412. [Google Scholar] [CrossRef]
- Nzila, A.; Razzak, S.A.; Zhu, J. Bioaugmentation: An emerging strategy of industrial wastewater treatment for reuse and dis charge. Int. J. Environ. Res. Public Health 2016, 13, 846. [Google Scholar] [CrossRef]
- Adams, G.O.; Fufeyin, P.T.; Okoro, S.E.; Ehinomen, I. Bioremediation, biostimulation and biouagumentation: A review. Int. J. Environ. Biorem. Biodegr. 2015, 3, 28–39. [Google Scholar]
- Zheng, M.; Wang, W.; Hayes, M.; Nydell, A.; Tarr, M.A.; Van Bael, S.A.; Papadopoulos, K. Degradation of Macondo 252 oil by endophytic Pseudomonas putida. J. Environ. Chem. Eng. 2018, 6, 643–648. [Google Scholar] [CrossRef]
- Pietro-Souza, W.; de Campos Pereira, F.; Mello, I.S.; Stachack, F.F.F.; Terezo, A.J.; Nunes da Cunha, C.; White, J.F.; Li, H.; Soares, M.A. Mercury resistance and bioremediation mediated by endophytic fungi. Chemosphere 2020, 240, 124874. [Google Scholar] [CrossRef]
- Wu, J.; Kamal, N.; Hao, H.; Liu, Z.; Qian, C.; Shao, Y.; Zhong, X.; Xu, B. Endophytic Bacillus megaterium BM18-2 mutated for cadmium accumulation and improving plant growth in hybrid Pennisetum. Biotechnol. Rep. 2019, e00374. [Google Scholar] [CrossRef]
- Suyamud, B.; Thiravetyan, P.; Gadd, G.F.; Panyapinyopol, B.; Inthorn, D. Bisphenol A removal from a plastic industry wastewater by Dracaena sanderiana endophytic bacteria and Bacillus cereus NI. Int. J. Phyt. 2019. [Google Scholar] [CrossRef]
- Wu, T.; Xu, J.; Liu, J.; Guo, W.-H.; Li, X.-B.; Xia, J.-B.; Xie, W.-J.; Yao, Z.-G.; Zhang, Y.-M.; Wang, R.-Q. Characterization and initial application of endophytic Bacillus safensis strain ZY16 for improving phytoremediation of oil-contaminated saline soils. Front. Microbiol. 2019, 10, 991. [Google Scholar] [CrossRef]
- Mitter, E.K.; Kataoka, R.; Renato de Freitas, J.; Germida, J.J. Potential use of endophytic root bacteria and host plants to degrade hydrocarbons. Int. J. Phyt. 2019, 21, 928–938. [Google Scholar] [CrossRef] [PubMed]
- Baoune, H.; Aparicio, J.D.; Pucci, G.; El Hadj-Khelil, A.O.; Polti, M.A. Bioremediation of petroleum-contaminated soils using Streptomyces sp. Hlh1. J. Soils Sed. 2019, 19, 2222–2230. [Google Scholar] [CrossRef]
- Iqbal, A.; Arshad, M.; Karthikeyan, R.; Gentry, T.J.; Rashid, J.; Iftikhar, A.I.; Schwab, A.P. Diesel degrading bacterial endophytes with plant growth promoting potential isolated from a petroleum storage facility. 3 Biotech 2019, 9, 35. [Google Scholar] [CrossRef] [PubMed]
- Feng, N.X.; Yu, J.; Mo, C.H.; Zhao, H.M.; Li, Y.W.; Wu, B.X.; Cai, Q.Y.; Li, H.; Zhou, D.M.; Wong, M.H. Biodegradation of di-n-butyl phthalate (DBP) by a novel endophytic Bacillus megaterium strain YJB3. Sci. Tot. Environ. 2018, 616–617, 117–127. [Google Scholar] [CrossRef]
- Huang, Y.H.; Huang, X.J.; Chen, X.H.; Cai, Q.Y.; Chen, S.; Mo, C.H.; Lü, H.; Wong, M.H. Biodegradation of di-butyl phthalate (DBP) by a novel endophytic bacterium Bacillus subtilis and its bioaugmentation for removing DBP from vegetation slurry. J. Environ. Manag. 2018, 224, 1–9. [Google Scholar] [CrossRef]
- Karnwal, A. Use of bio-chemical surfactant producing endophytic bacteria isolated from rice root for heavy metal bioremediation. Pert. J. Trop. Agric. Sci. 2018, 41, 699–714. [Google Scholar]
- Sim, C.S.F.; Chen, S.H.; Ting, A.S.Y. Endophytes: Emerging tools for the bioremediation of pollutants. In Emerging and Eco-Friendly Approaches for Waste Management; Springer: Singapore, 2018; pp. 189–217. [Google Scholar] [CrossRef]
- Fu, Y. Biotransformation of ginsenoside Rb1 to Gyp-XVII and minor ginsenoside Rg3 by endophytic bacterium Flavobacterium sp. GE 32 isolated from Panax ginseng. Lett. Appl. Microbiol. 2018, 68, 134–141. [Google Scholar]
- Sun, K.; Habteselassie, M.Y.; Liu, J.; Li, S.; Gao, Y. Subcellular distribution and biotransformation of phenanthrene in pakchoi after inoculation with endophytic Pseudomonas sp. as probed using HRMS coupled with isotope-labeling. Environ. Pollut. 2018, 237, 858–867. [Google Scholar] [CrossRef]
- Tam, N.T.; Tam, N.P.; Nguyen, V.T.; Hung, N.K.; Huy, C.N.; Ngoc, P.B.; Ha, C.H.; Tien, P.Q. Isolation and screening of endophytic bacteria from Ngoc Linh ginseng (Panax vietnamensis Ha et Grushv) for biosynthesis β-glucosidase. Tap. Chi. Sinh. Hoc. 2018, 40, 153–161. [Google Scholar] [CrossRef]
- Mohd, S.; Kushwaha, A.S.; Shukla, J.; Mandrah, K.; Shankar, J.; Armaria, N.; Sabena, P.N.; Khare, P.; Narayan, R.; Dixie, S.; et al. Fungal mediated biotransformation reduces toxicity of arsenic to soil dwelling microorganism and plant. Ecotoxicol. Environ. Saf. 2019, 176, 108–118. [Google Scholar] [CrossRef] [PubMed]
- Marconi, F.; Umpiérrez, M.L.; Gonzalez, D.; Giordano, S.R.; Rodriguez, P. Endophytic biocatalysts with enoate reductase activity isolated from Mentha pulegium. World J. Microbiol. Biotechnol. 2018, 34, 50. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Mohanta, T.K.; Asaf, S.; Rehman, N.; Al-Housni, S.; Al-Harrasi, A.; Khan, A.L.; Al-Rawahi, A. Biotransformation of benzoin by Sphingomonas sp. LK11 and ameliorative effects on growth of Cucumis sativus. Arch. Microbiol. 2019, 201, 591–601. [Google Scholar] [PubMed]
- Hu, Y.; Wang, N.; Yan, X.; Yuan, Y.; Luo, F.; Jiang, Z.; Zhou, Y. Ginsenoside Re impacts on biotransformation products of ginsenoside Rb1 by Cellulosimicrobium cellulans sp. 21 and its mechanisms. Proc. Biochem. 2019, 77, 57–62. [Google Scholar] [CrossRef]
- Ran, X.; Zhang, G.; Li, S.; Wang, J. Characterization and antitumor activity of camptothecin from endophytic fungus Fusarium solani isolated from Camptotheca acuminate. Afr. Health Sci. 2017, 17, 566–574. [Google Scholar] [CrossRef]
- Kasaei, A.; Mobini, D.M.; Mahjoubi, F.; Saffar, B. Isolation of Taxol-producing endophytic fungi from Iranian yew through novel molecular approach and their effects on human breast cancer cell line. Curr. Microbiol. 2017, 74, 702–709. [Google Scholar] [CrossRef]
- Sarang, H.; Rajani, P.; Vasanthakumari, M.M.; Kumara, P.M.; Siva, R.; Ravikanth, G.; Shaanker, U. An endophytic fungus, Gibberella moniliformis from Lawsonia inermis L. produces lawsone, an orange-red pigment. Antonie Van Leeuwenhoek 2017, 110, 853–862. [Google Scholar] [CrossRef]
- Tawfike, A.F.; Romli, M.; Clements, C.; Abbott, G.; Young, L.; Schumacher, M.; Diederich, M.; Farag, M.; Edrada-Ebel, R. Isolation of anticancer and anti-trypanosome secondary metabolites from the endophytic fungus Aspergillus flocculus via bioactivity guided isolation and MS based metabolomics. J. Chromatogr. B 2019, 1106–1107, 71–83. [Google Scholar] [CrossRef]
- Liu, H.X.; Tan, H.B.; Li, S.N.; Chen, Y.C.; Li, H.H.; Zhang, W.M. Two new metabolites from Daldinia eschscholtzii, an endophytic fungus derived from Pogostemon cablin. J. Asian Nat. Prod. Res. 2019, 21, 150–156. [Google Scholar] [CrossRef]
- Bernardi, D.I.; das Chagas, F.O.; Monteiro, A.F.; dos Santos, G.F.; de Souza Berlinck, R.G. Secondary metabolites of endophytic Actinomycetes: Isolation, synthesis, biosynthesis, and biological activities. In Progress in the Chemistry of Organic Natural Products; Kinghorn, A., Falk, H., Gibbons, S., Kobayashi, J., Asakawa, Y., Liu, J.K., Eds.; Springer Nature Switzerland: Basel, Switzerland, 2019; Volume 108, pp. 219–265. [Google Scholar]
- Wang, P.; Kong, F.; Wei, J.; Wang, Y.; Wang, W.; Hong, K.; Zhu, W. Alkaloids from the mangrove-derived actinomycete Jishengella endophytica 161111. Mar. Drug 2014, 12, 477–490. [Google Scholar] [CrossRef]
- Gonzalez-Menendez, V.; Crespo, G.; Toro, C.; Martin, J.; de Pedro, N.; Tormo, J.R.; Genilloud, O. Extending the metabolite diversity of the endophyte Dimorphosporicola tragani. Metabolites 2019, 9, 197. [Google Scholar] [CrossRef] [PubMed]
- Ramalingam, A.; Amutha, C. Antibacterial activity of bacteria associated with red seaweeds against pathogenic bacteria of poultry and cattle. Int. J. Adv. Life Sci. 2013, 6, 13–18. [Google Scholar]
- De Melo, F.M.P.; Fiore, M.F.; de Moraes, L.A.B.; Silva-Stenico, M.E.; Scramin, S.; de Araújo, T.M.; de Melo, I.S. Antifungal compound produced by the cassava endophyte Bacillus pumilus MAIIIM4A. Sci. Agricol. 2009, 66, 583–592. [Google Scholar] [CrossRef]
- Narayan, C.P.; Seung, H.J.; Jian, X.D.; Seung, H.Y. Assemblages of endophytic bacteria in chili pepper (Capsicum annuum L.) and their antifungal activity against phytopathogens in vitro. Plant Omics J. 2013, 6, 441–448. [Google Scholar]
- Christina, A.; Christapher, V.; Bhore, S.J. Endophytic bacteria as a source of novel antibiotics: An overview. Pharm. Rev. 2013, 7, 11–16. [Google Scholar]
- Shenpagam, H.N.; Devi, D.K.; Sinduja, G.; Sandhya, R. Isolation of endophytic Actinomycetesfrom medicinal plants and its mutational effect in biocontrol activity. Int. J. Pharm. Sci. Res. 2012, 3, 4338–4344. [Google Scholar]
- Gond, S.K.; Bergen, M.S.; Torres, M.S.; White, J.F. Endophytic Bacillus spp. produce antifungal lipopeptides and induce host defence gene expression in maize. Microbiol. Res. 2015, 172, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Gond, S.K.; Surendra, K.; Bergen, M.S.; Torres, M.S.; White, J.F.; Kharwar, R.N. Effect of bacterial endophyte on expression of defense genes in Indian popcorn against Fusarium moniliforme. Symbiosis 2015, 66, 133–140. [Google Scholar] [CrossRef]
- Hu, H.; Wang, C.; Li, X.; Tang, Y.; Wang, Y.; Chen, S.; Yan, S. RNA-Seq identification of candidate defense genes targeted by endophytic Bacillus cereus-mediated induced systemic resistance against Meloidogyne incognita in tomato. Pest Manag. Sci. 2018, 74, 2793–2805. [Google Scholar] [CrossRef]
- Dong, Z.Y.; Narsing Rao, M.P.; Wang, H.F.; Fang, B.Z.; Liu, Y.H.; Li, L.; Xiao, M.; Li, W.J. Transcriptomic analysis of two endophytes involved in enhancing salt stress ability of Arabidopsis thaliana. Sci. Tot. Environ. 2019, 10, 107–117. [Google Scholar] [CrossRef]
- Yuan, J.; Zhang, W.; Sun, K.; Tang, M.J.; Chen, P.X.; Li, X.; Dai, C.C. Comparative transcriptomics and proteomics of Atractylodes lancea in response to endophytic fungus Gilmaniella sp. AL12 reveals regulation in plant metabolism. Front. Microbiol. 2019, 10, 1208. [Google Scholar] [CrossRef] [PubMed]
- Mejía, L.C.; Herre, E.A.; Sparks, J.P.; Winter, K.; García, M.N.; Van Bael, S.A.; Stitt, J.; Shi, Z.; Zhang, Y.; Guiltinan, M.J.; et al. Pervasive effects of a dominant foliar endophytic fungus on host genetic and phenotypic expression in a tropical tree. Front. Microbiol. 2014, 5, 479. [Google Scholar] [PubMed]
- Sarkar, D.; Rovenich, H.; Jeena, G.; Nizam, S.; Tissier, A.; Balcke, G.U.; Mahdi, L.K.; Bonkowski, M.; Langen, G.; Zuccar, A. The inconspicuous gatekeeper: Endophytic Serendipita vermifera acts as extended plant protection barrier in the rhizosphere. New Phytol. 2019, 224, 886–901. [Google Scholar] [CrossRef]
- Malinich, E.; Wang, K.; Mukherjee, P.K.; Kolomiets, M.; Kenerley, C.H. Differential expression analysis of Trichoderma virens RNA reveals a dynamic transcriptome during colonization of Zea mays roots. BMC Genom. 2019, 20, 280. [Google Scholar] [CrossRef] [PubMed]
- Dinkins, R.D.; Nagabhyrub, P.; Youngc, C.A.; Westd, C.P.; Schardlb, C.L. Transcriptome analysis and differential expression in tall fescue harboring different endophyte strains in response to water deficit. Plant Genome 2019, 12. [Google Scholar] [CrossRef]

| Host Plant (Part) | Endophyte Strains | Function | Next Generation Sequencing Platform | Reference |
|---|---|---|---|---|
| Oryza sativa cultivar RP Bio-226 (leaves) | Sphingopyxis Pseudomonas | Secondary metabolite production | Illumina NextSeq-500 | [15] |
| Oryza sativa L. (roots) | Bacillus | Biological N fixation and PGP abilities | Illumina NextSeq-500 | [33] |
| Oryza sativa L. (roots) | Enterobacteriaceae Xanthomonadaceae Bacillaceae Heliobacteriaceae Rhizobiaceae Methylocystaceae Aeromonadaceae Lachnospiraceae Paenibacillaceae Rhodospirillaceae Burkholderiaceae | PGP abilities and role in defence mechanisms of plants | Illumina MiSeq | [13] |
| Oryza sativa L. (seeds) | Staphylococcus Bacillus | Protection against oxidative, heat and osmotic stresses. Production of auxins, gibberellins and siderophores | Illumina MiSeq | [32] |
| Aloe vera (leaves, roots, stem tissues) | Proteobacteria Actinobacteria Firmicutes Bacteriodetes | Production of bioactive compounds | Illumina MiSeq | [11] |
| White and Red Pitayas (fruits) | Cyanobacteria Proteobacteria Bacteroidetes | Improvement of fruit growth and protection | Illumina HiSeq 2500 | [34] |
| Sorghum bicolor (roots, stems) | Duganella, Aquabacterium, Bordetella, Massilia, Pantoea, Salmonella, Klebsiella, Kosakonia, Pseudomonas, Serratia, Stenotrophomonas, Agrobacterium, Ancylobacter, Brevundimonas, Pleomorphomonas, Curtobacterium, Microbacterium, Nocardia, Sediminihabitans Bacillus, Macrococcus, Staphylococcus, Exiguobacterium | PGP abilities. Production of hormones. Synthesis of fungicides and bactericides. Ability to supply nitrogen to plants. | Ion Torrent PGM | [17] |
| Sorghum bicolor L. Moench (stems, roots) | Microbacterium, Agrobacterium, Sphingobacterium, Herbaspirillum, Erwinia, Pseudomonas, Stenotrophomonas | PGP abilities | Roche 454-pyrosequencing | [23] |
| Oil palm (fruits) | Lactococcus Enterobacter, Citrobacter, Seratia, Klebsiella, Citrobacter Pseudomonas, Achromobacter | PGP abilities. Biological N fixation. Degradation of phytic acid | Roche 454-pyrosequencing | [16] |
| Pinus arizonica Pinus durangensis (roots, phloem, bark) | Thermi, Tenericutes Acetobacter, Burkholderia, Caulobacter, Pseudomonas, Ralstonia, Bradyrhizobium, Metylocapsa | Biosynthesis of secondary metabolites. Metabolism of cofactors and vitamins | Roche 454-pyrosequencing | [24] |
| Vitis vinifera cv. Summerblack (stems) | Acremonium, Alternaria, Arthrinium, Ascorhizoctonia, Aspergillus, Aureobasidium, Bipolaris, Botryosphaeria, Botrytis, Chaetomium, Cladosporium, Curvularia, Hypoxylon, Lasiodiplodia, Mycosphaerella, Nigrospora, Penicillium, Phoma, Scopulariopsis | Prevention against some fungal diseases | Illumina HiSeq2500 | [35] |
| Vitis vinifera | Coniochaeta Clonostachys Sterigmatosporidium Phaeoacremonium Fusarium Cladosporium Alternaria Acremonium Trichoderma Penicillium | Protection of plants against pathogens | Illumina MiSeq | [12] |
| Vitis vinifera L. (field shoots and callus tissues) | Actinobacteria Proteobacteria Firmicutes Bacteroidetes Deinococcus-Thermus | Carbohydrate protein and amino acid metabolism. Synthesis/conversion/utilization of nucleosides and nucleotides | Illumina HiSeq | [36] |
| Cinnamomum cassia Presl (roots) | Streptomyces | Antibiotic biosynthetic pathways. Antimicrobial and cytotoxic properties. PGP abilities. | Ion Torrent PGM | [14] |
| Populus spp. | Mortierella | PGP abilities: improved growth and drought tolerance | Illumina HiSeq platform | [37] |
| Ophiopogon japonicas | Bacillus | PGP abilities: oxidative stress, drought tolerance, heavy metal resistance | Illumina HiSeq | [38] |
| Tephrosia apollinea (leaves) | Sphingomonas | Drought tolerance | Single Molecule Real Time (SMRT) | [39] |
| Grasses (roots) | Actinobacteria | PGP abilities | Agilent 2100 | [40] |
| Preparation Name | Composition | Function | Reference |
|---|---|---|---|
| Phytosporin-M | Bacillus subtilis 26D | Displacement of pathogenic fungi and stimulation of plant growth | [74] |
| Albit | B. subtilis 2604D + B. subtilis 2605D + terpene acids, macro- and microelements | ||
| Vitaplan | B. subtilis 2604D + B. subtilis 2605D | ||
| Alirin-B | B. subtilis | Reduction of the number of micromycetes | [75] |
| Trianum P | Trichoderma harzianum T-22 | Fungicidal effect. Stimulation of tobacco seedling growth | |
| Cedomon EO | Pseudomonas chlororaphis MA 342 | Protection of barley seeds against Pyrenophora graminea, Fusarium spp., and Bipolaris sorokiniana | |
| Cropaid | Thiobacillus thiooxidans, T. thioparus, T. ferrooxidans and over 60 minerals | Stimulation of production of proteins and amino acids that increase plant resistance to cold and frost damage | [76] |
| Symbion-N | Azospirillium lipoferum | Restriction of the pathogenicity of fungi Fusarium solani and Rhizoctonia solani. Support of plant development and vigor | [77] |
| Symbion-NG | Gluconacetobacter diazotrophicus | ||
| Symbion-NR | Rhizobium phaseoli | ||
| Bio-cure B | Pseudomonas fluorencese | ||
| Bio-cure F | Trichoderma viridi | ||
| Bio-health WGP | Bacillus megatherium Trichoderma larizianum | ||
| Rhizo-N | Bacillus subtilis |
| Process | Endophytes | Function | Reference |
|---|---|---|---|
| BIOREMEDIATION | Pseudomonas putida | Degradation of alkanes | [110] |
| Aspergillus sp. A31, Curvularia geniculata P1, Lindgomycetaceae P87, Westerdykella sp. P71 | Reduction of mercury concentrations in soil by bioaccumulation | [111] | |
| Bacillus megaterium BM18-2 | Cd removal | [112] | |
| Pantoea dispersa | Bisphenol A (BPA) removal | [113] | |
| Bacillus safensis strain ZY16 | Degradation of C12–C32 n-alkanes from diesel oil | [114] | |
| Pseudomonas sp. | Hydrocarbon degradation | [115] | |
| Streptomyces sp. Hlh1 | Degradation of petroleum hydrocarbons (TPH), n-alkanes and aromatic hydrocarbons | [116] | |
| Pseudomonas sp. J10 | Diesel degradation | [117] | |
| Bacillus megaterium YJB3 | Removal of phthalic acid esters (PAE) | [118] | |
| Streptomyces | Degradation of n-alkanes (C6-C30), aromatic and polycyclic aromatic hydrocarbons | [116] | |
| Bacillus subtilis N-1 | Lowering PAE accumulation | [119] | |
| Pseudomonas fluorescence | Zn, Cr, Cd, Ni removal | [120] | |
| Lasiodiplodia sp. MXSF31, Mucor sp. | Bioaccumulation of Pb, Cd, Zn | [121] | |
| Bacillus, Klebsiella, Shewanella, Burkholderia, Pseudomonas, Sphingomonas | Removal of dyes | ||
| BIOTRANSFORMATION | Flavobacterium sp. GE 32 | Biotransformation of ginsenoside Rb1 to Gyp-XVII and ginsenoside Rg3 | [122] |
| Pseudomonas sp. Ph6-gfp | Biotransformation of phenanthrene | [123] | |
| Enterobacter sp., Serratia sp., Ochrobactrum sp., Arthrobacter sp. | Biotransformation of ginsenoside Rb1 to ginsenoside Rd and Rg3 | [124] | |
| Aspergillus flavus | Biotransformation of soluble arsenic into an immobilized arsenic particle | [125] | |
| Pseudomonas proteolytica FM18Mci1 Bacillus sp. FM18civ1 | Biotransformation of (4S)-(+)-carvon (1a) and (4R)-(-)-carvon (1b) | [126] | |
| Sphingomonas sp. LK11 | Biotransformation of benzoin to benzamide | [127] | |
| Cellulosimicrobium cellulans sp. 21 | Biotransformation of ginsenoside Rb1 into Rg3 and Re into Rg2 | [128] |
| Endophyte Strains | Host Plant | Bioactive Compound | Function | Reference |
|---|---|---|---|---|
| Fusarium solani | Camptotheca acuminata | Camptothecin | Cytotoxic activity against cancer cells | [129] |
| Cladosporium sp. | Taxus baccata | Taxol | Antimitotic effect on MCF-7 cells in a human breast cancer cell line | [130] |
| Gibberella moniliformis | Lawsonia inermis L. | Lawsone | Synthesis of natural dye with antioxidant, antiviral, antidermatophytic, tuberculostatic, and cytotoxic enzymes activity | [131] |
| Aspergillus flocculus | Markhamia platycalyx | cis-4-hydroxymellein, 5-hydroxymellein, diorcinol, botryoisocoumarin A, mellein | Bioactive anticancer and anti-trypanosome properties | [132] |
| Daldinia eschscholtzii A630 | Pogostemon cablin | Eschscholin A, 3-ene-2-methyl-2H-1-benzopyran-5-ol, 3,5-dihydroxy-2-methyl-4H-chromen-4-one | Antibacterial activities | [133] |
| Dimorphosporicola tragani CF-090383 | Arthrocnemum macrostachyum | Cerulenin | Inhibition of fatty acid synthesis | [136] |
| Jishengella endophytica 16111 | Xylocarpus granatum | Perlolyrine | Antiviral effect | [135] |
| Streptomyces sp. TP-A0569 | Allium fistulosum | Fistupyrone | Plant protection against pathogenic fungi | [134] |
| Streptomyces sp. TP-A0556 | Aucuba japonica | Coumarins TPU-0031-A and B | Antibiotic activity against Gram-positive and Gram-negative bacteria | |
| Streptomyces hygroscopicus TP-A0451 | Pteridium aquilinum | Pteridic acids A and B, Pterocidin | Plant growth-promoting properties |
| Host Plant | Endophytes | Type of Interaction | Reference |
|---|---|---|---|
| Maize Zea mays L. Indian popcorn | Bacillus spp. Bacillus amyloliquefaciens | Antifungal lipopeptides (Iturin A, Bacillomycin, Fengycin) induce the up-regulation of pathogenesis-related genes of host plants Gene expression induced by jasmonic acid | [142] [143] |
| Tomato Solanum lycopersicon Hezuo903 | Bacillus cereus BCM2 | Induction of phenylalanine ammonia-lyase genes (defense system) | [144] |
| Arabidopsis thaliana | Arthrobacter endophyticus SYSU 33332, Nocardiopsis alba SYSU 333140 | Induction of up-regulated water and potassium ion uptake, chlorophyll a reductase, peptidemethionine (R)-S-oxide reductase genes during salt stress | [145] |
| Medicinal herb Atractylodes lancea | Gilmaniella sp. AL12 | Induction of down-regulated plant immunity genes and up-regulated primary metabolism (carbon fixation, carbohydrate metabolism, and energy metabolism) genes | [146] |
| Cacao Theobroma cacao | Colletotrichum tropicale strain 5101 CBS 124949 | Up- and down-regulation of scores of host genes involved in defense against biotic stresses (i.e., pathogens and herbivores, ethylene signaling, and defense response pathways) as well as signaling proteins (i.e., receptor kinases) | [147] |
| Barley Hordeum vulgare L. cv Golden Promise, 43 hereafter Hv | Serendipita vermifera | Induction of expression encoding hydrolytic enzyme genes, reduction of pathogen infection | [148] |
| Maize Zea mays L. B73 | Trichoderma virens | Regulation and modulation of phytohormones and cell wall degrading encoding genes | [149] |
| Tall fescue Festuca arundinacea vr. Schreb | Endophytic nontoxic strain NTE 19 | Induction of stress pathways in response to water deficit | [150] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuźniar, A.; Włodarczyk, K.; Wolińska, A. Agricultural and Other Biotechnological Applications Resulting from Trophic Plant-Endophyte Interactions. Agronomy 2019, 9, 779. https://doi.org/10.3390/agronomy9120779
Kuźniar A, Włodarczyk K, Wolińska A. Agricultural and Other Biotechnological Applications Resulting from Trophic Plant-Endophyte Interactions. Agronomy. 2019; 9(12):779. https://doi.org/10.3390/agronomy9120779
Chicago/Turabian StyleKuźniar, Agnieszka, Kinga Włodarczyk, and Agnieszka Wolińska. 2019. "Agricultural and Other Biotechnological Applications Resulting from Trophic Plant-Endophyte Interactions" Agronomy 9, no. 12: 779. https://doi.org/10.3390/agronomy9120779
APA StyleKuźniar, A., Włodarczyk, K., & Wolińska, A. (2019). Agricultural and Other Biotechnological Applications Resulting from Trophic Plant-Endophyte Interactions. Agronomy, 9(12), 779. https://doi.org/10.3390/agronomy9120779







