Identification and Expression Analysis of the NAC Gene Family in Coffea canephora
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material Collection and Cold Treatment
2.2. RNA Extraction from Coffee Beans, Library Construction, Sequencing and Assembly
2.3. Data Collection and Identification of NAC Genes
2.4. Phylogenetic Analysis
2.5. Chromosome Location, Gene Structure, and Conserved Motifs in the CocNAC Family
2.6. qRT-PCR
3. Results
3.1. Identification of Putative NAC Genes in Coffea canephora
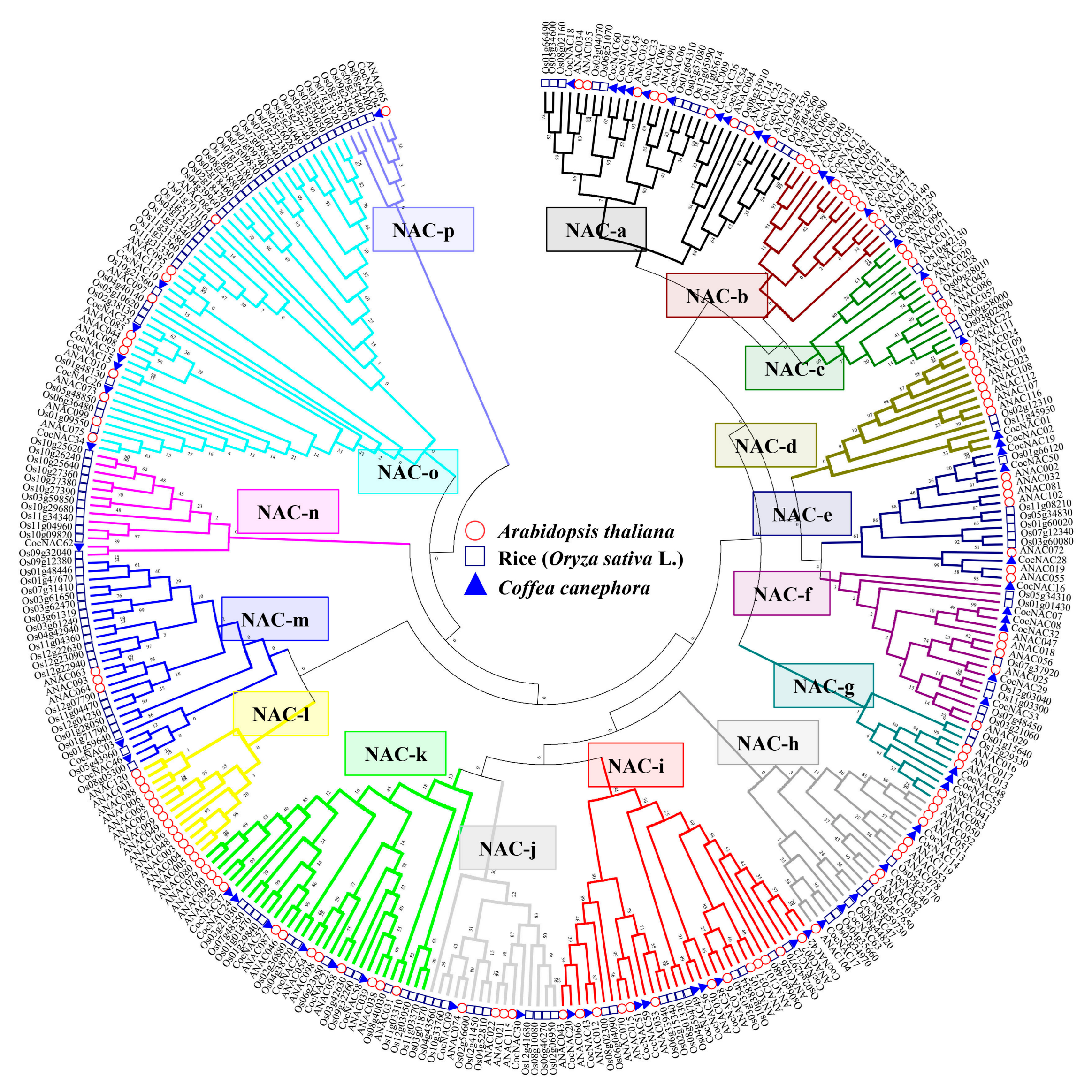
3.2. Phylogenetic Analysis of the CocNACs
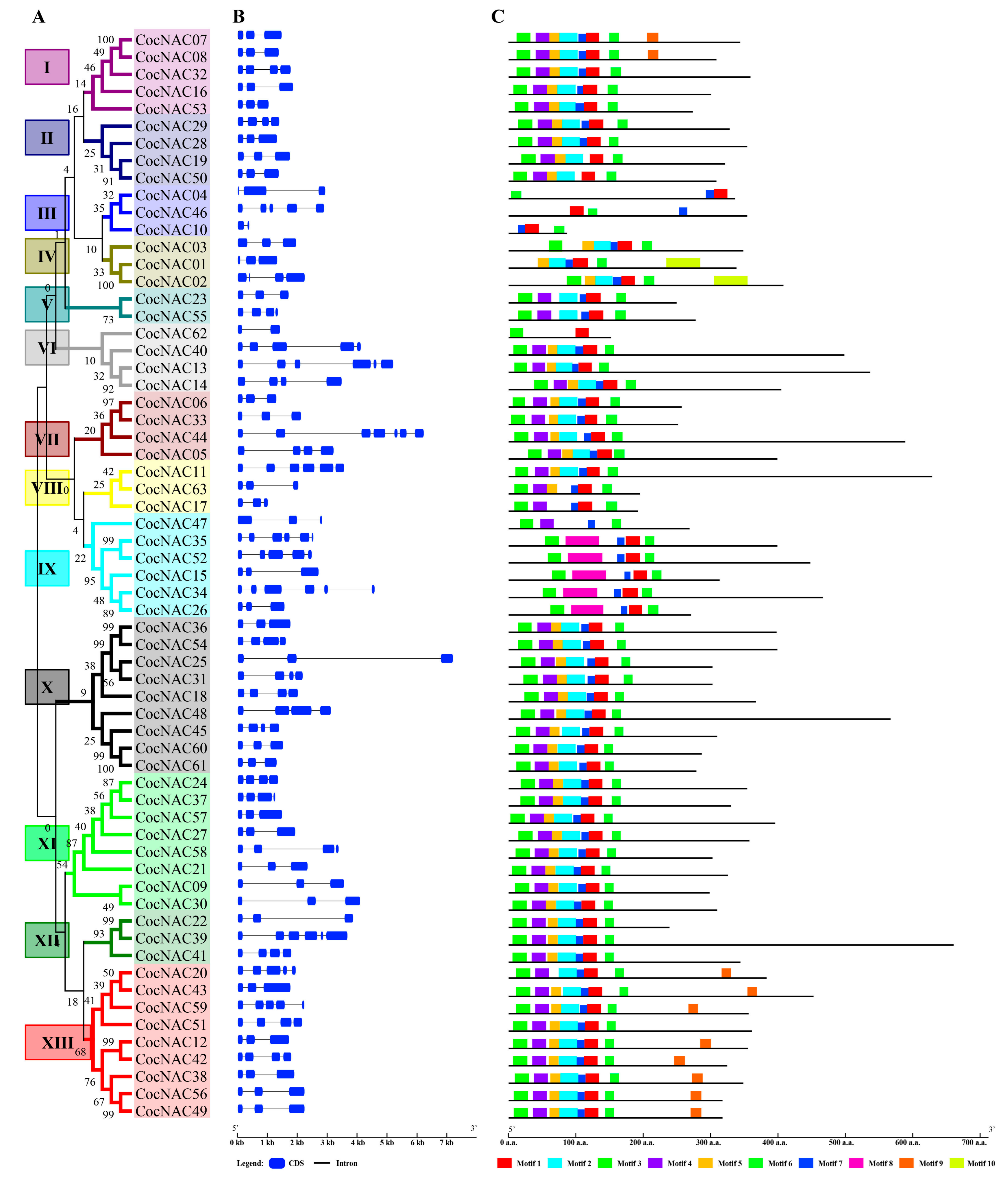
3.3. Gene Structure and Protein Motif Analysis of C. canephorae NAC Gene Family
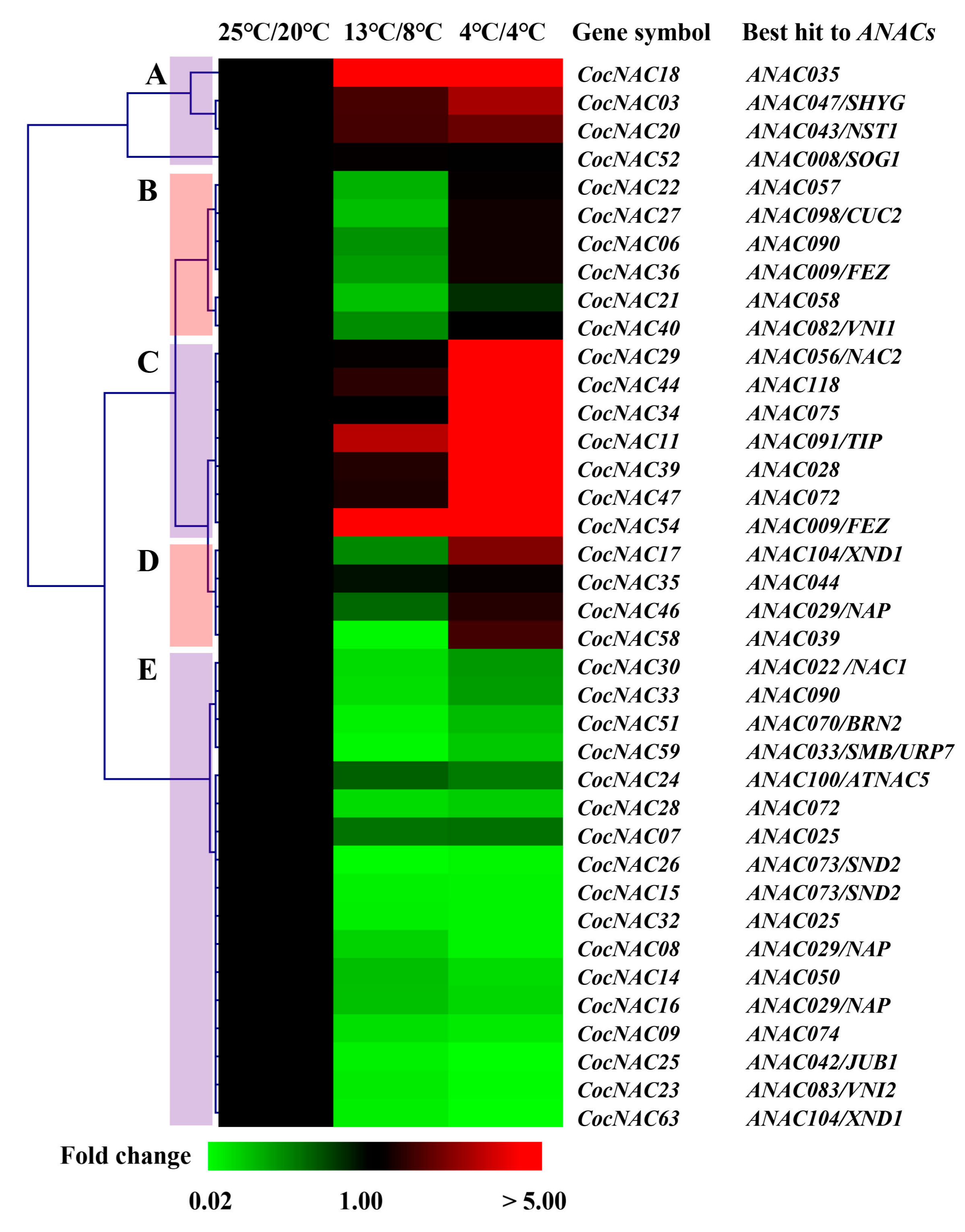
3.4. qPCR Analysis of CocNAC Genes under Low Temperature Treatment
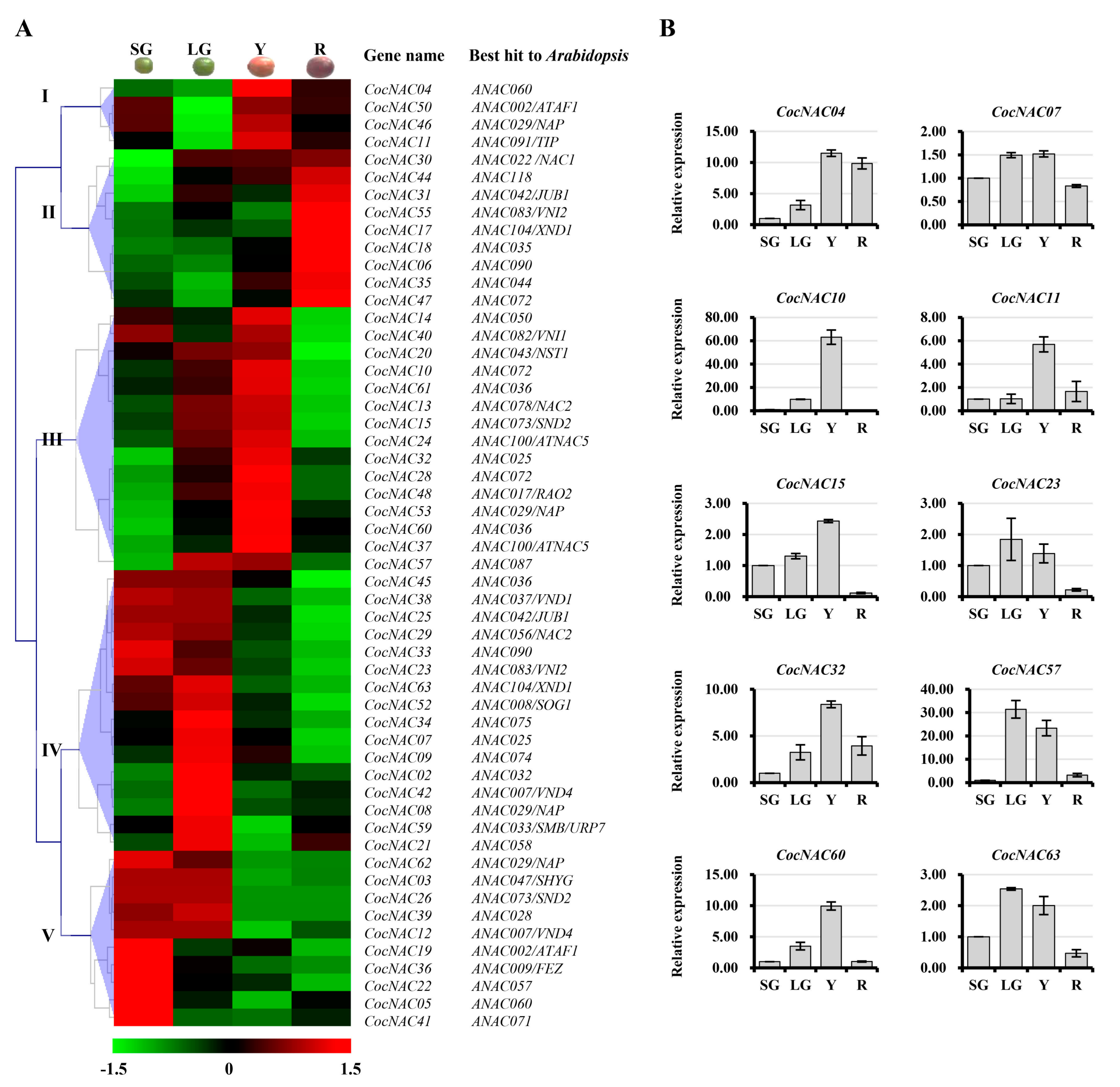
3.5. CocNAC Genes Involved in Coffee Bean Development
4. Discussion
4.1. CocNAC Gene Identification and Evolutionary Analysis in C. canephora
4.2. Identification of CocNACs Responding to Cold Stress in C. canephora
4.3. CocNAC Genes May Play an Important Role in Coffee Bean Development
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Jensen, M.K.; Hagedorn, P.H.; de Torres-Zabala, M.; Grant, M.R.; Rung, J.H.; Collinge, D.B.; Lyngkjaer, M.F. Transcriptional regulation by an NAC (NAM-ATAF1,2-CUC2) transcription factor attenuates ABA signalling for efficient basal defence towards Blumeria graminis f. sp. hordei in Arabidopsis. Plant J. 2008, 56, 867–880. [Google Scholar] [CrossRef] [PubMed]
- Sablowski, R.W.; Meyerowitz, E.M. A homolog of NO APICAL MERISTEM is an immediate target of the floral homeotic genes APETALA3/PISTILLATA. Cell 1998, 92, 93–103. [Google Scholar] [CrossRef]
- Souer, E.; van Houwelingen, A.; Kloos, D.; Mol, J.; Koes, R. The no apical meristem gene of Petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell 1996, 85, 159–170. [Google Scholar] [CrossRef]
- Ooka, H.; Satoh, K.; Doi, K.; Nagata, T.; Otomo, Y.; Murakami, K.; Matsubara, K.; Osato, N.; Kawai, J.; Carninci, P.; et al. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 2003, 10, 239–247. [Google Scholar] [CrossRef]
- Duval, M.; Hsieh, T.F.; Kim, S.Y.; Thomas, T.L. Molecular characterization of AtNAM: A member of the Arabidopsis NAC domain superfamily. Plant Mol. Biol. 2002, 50, 237–248. [Google Scholar] [CrossRef]
- Nuruzzaman, M.; Manimekalai, R.; Sharoni, A.M.; Satoh, K.; Kondoh, H.; Ooka, H.; Kikuchi, S. Genome-wide analysis of NAC transcription factor family in rice. Gene 2010, 465, 30–44. [Google Scholar] [CrossRef]
- Hussain, R.M.; Ali, M.; Feng, X.; Li, X. The essence of NAC gene family to the cultivation of drought-resistant soybean (Glycine max L. Merr.) cultivars. BMC Plant Biol. 2017, 17, 55. [Google Scholar] [CrossRef]
- Gong, X.; Zhao, L.; Song, X.; Lin, Z.; Gu, B.; Yan, J.; Zhang, S.; Tao, S.; Huang, X. Genome-wide analyses and expression patterns under abiotic stress of NAC transcription factors in white pear (Pyrus bretschneideri). BMC Plant Biol. 2019, 19, 161. [Google Scholar] [CrossRef]
- Liu, M.; Ma, Z.; Sun, W.; Huang, L.; Wu, Q.; Tang, Z.; Bu, T.; Li, C.; Chen, H. Genome-wide analysis of the NAC transcription factor family in Tartary buckwheat (Fagopyrum tataricum). BMC Genom. 2019, 20, 113. [Google Scholar] [CrossRef]
- Hu, R.; Qi, G.; Kong, Y.; Kong, D.; Gao, Q.; Zhou, G. Comprehensive Analysis of NAC Domain Transcription Factor Gene Family in Populus trichocarpa. BMC Plant Biol. 2010, 10, 145. [Google Scholar] [CrossRef]
- Wang, N.; Zheng, Y.; Xin, H.; Fang, L.; Li, S. Comprehensive analysis of NAC domain transcription factor gene family in Vitis vinifera. Plant Cell Rep. 2013, 32, 61–75. [Google Scholar] [CrossRef] [PubMed]
- Zhuo, X.; Zheng, T.; Zhang, Z.; Zhang, Y.; Jiang, L.; Ahmad, S.; Sun, L.; Wang, J.; Cheng, T.; Zhang, Q. Genome-Wide Analysis of the NAC Transcription Factor Gene Family Reveals Differential Expression Patterns and Cold-Stress Responses in the Woody Plant Prunus mume. Genes 2018, 9, 494. [Google Scholar] [CrossRef] [PubMed]
- Lv, X.; Lan, S.; Guy, K.M.; Yang, J.; Zhang, M.; Hu, Z. Global Expressions Landscape of NAC Transcription Factor Family and Their Responses to Abiotic Stresses in Citrullus lanatus. Sci. Rep. 2016, 6, 30574. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Wu, Q.; Teng, L.; Tang, F.; Pi, Z.; Shen, S. Transcriptional regulation of the paper mulberry under cold stress as revealed by a comprehensive analysis of transcription factors. BMC Plant Biol. 2015, 15, 108. [Google Scholar] [CrossRef] [PubMed]
- Calzadilla, P.I.; Maiale, S.J.; Ruiz, O.A.; Escaray, F.J. Transcriptome Response Mediated by Cold Stress in Lotus japonicus. Front. Plant Sci. 2016, 7, 374. [Google Scholar] [CrossRef]
- Hu, W.; Wei, Y.; Xia, Z.; Yan, Y.; Hou, X.; Zou, M.; Lu, C.; Wang, W.; Peng, M. Genome-Wide Identification and Expression Analysis of the NAC Transcription Factor Family in Cassava. PLoS ONE 2015, 10, e0136993. [Google Scholar] [CrossRef]
- Huang, L.; Hong, Y.; Zhang, H.; Li, D.; Song, F. Rice NAC transcription factor ONAC095 plays opposite roles in drought and cold stress tolerance. BMC Plant Biol. 2016, 16, 203. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, Y.; Shi, C.; Huang, Z.; Zhang, Y.; Li, S.; Li, Y.; Ye, J.; Yu, C.; Li, Z. SOAPnuke: A MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. GigaScience 2018, 7, 1–6. [Google Scholar] [CrossRef]
- Zheng, X.; Chen, B.; Lu, G.; Han, B. Overexpression of a NAC transcription factor enhances rice drought and salt tolerance. Biochem. Biophys. Res. Commun. 2009, 379, 985–989. [Google Scholar] [CrossRef]
- Sperotto, R.A.; Ricachenevsky, F.K.; Duarte, G.L.; Boff, T.; Lopes, K.L.; Sperb, E.R.; Grusak, M.A.; Fett, J.P. Identification of up-regulated genes in flag leaves during rice grain filling and characterization of OsNAC5, a new ABA-dependent transcription factor. Planta 2009, 230, 985–1002. [Google Scholar] [CrossRef]
- Sindhu, A.; Chintamanani, S.; Brandt, A.S.; Zanis, M.J.; Scofield, S.R.; Johal, G.S. A guardian of grasses: Specific origin and conservation of a unique disease-resistance gene in the grass lineage. Proc. Natl. Acad. Sci. USA 2008, 105, 1762–1767. [Google Scholar] [CrossRef]
- Hu, H.; You, J.; Fang, Y.; Zhu, X.; Qi, Z.; Xiong, L. Characterization of transcription factor gene SNAC2 conferring cold and salt tolerance in rice. Plant Mol. Biol. 2008, 67, 169–181. [Google Scholar] [CrossRef] [PubMed]
- Jensen, M.K.; Kjaersgaard, T.; Nielsen, M.M.; Galberg, P.; Petersen, K.; O’Shea, C.; Skriver, K. The Arabidopsis thaliana NAC transcription factor family: Structure-function relationships and determinants of ANAC019 stress signalling. Biochem. J. 2010, 426, 183–196. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.L.; Xu, K.D.; Zhao, L.J.; Pan, Y.Z.; Jiang, B.B.; Zhang, H.Q.; Liu, G.L. Overexpression of a novel chrysanthemum NAC transcription factor gene enhances salt tolerance in tobacco. Biotechnol. Lett. 2011, 33, 2073–2082. [Google Scholar] [CrossRef]
- Hao, Y.J.; Wei, W.; Song, Q.X.; Chen, H.W.; Zhang, Y.Q.; Wang, F.; Zou, H.F.; Lei, G.; Tian, A.G.; Zhang, W.K.; et al. Soybean NAC transcription factors promote abiotic stress tolerance and lateral root formation in transgenic plants. Plant J. 2011, 68, 302–313. [Google Scholar] [CrossRef]
- Lu, M.; Ying, S.; Zhang, D.F.; Shi, Y.S.; Song, Y.C.; Wang, T.Y.; Li, Y. A maize stress-responsive NAC transcription factor, ZmSNAC1, confers enhanced tolerance to dehydration in transgenic Arabidopsis. Plant Cell Rep. 2012, 31, 1701–1711. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Zhang, H.; Qian, X.; Li, A.; Zhao, G.; Jing, R. TaNAC2, a NAC-type wheat transcription factor conferring enhanced multiple abiotic stress tolerances in Arabidopsis. J. Exp. Bot. 2012, 63, 2933–2946. [Google Scholar] [CrossRef]
- Shan, W.; Kuang, J.; Lu, W.; Chen, J. Banana fruit NAC transcription factor MaNAC1 is a direct target of MaICE1 and involved in cold stress through interacting with MaCBF1. Plant Cell Environ. 2014, 37, 2116–2127. [Google Scholar] [CrossRef]
- Li, X.L.; Yang, X.; Hu, Y.X.; Yu, X.D.; Li, Q.L. A novel NAC transcription factor from Suaeda liaotungensis K. enhanced transgenic Arabidopsis drought, salt, and cold stress tolerance. Plant Cell Rep. 2014, 33, 767–778. [Google Scholar] [CrossRef]
- Yang, X.; Wang, X.; Ji, L.; Yi, Z.; Fu, C.; Ran, J.; Hu, R.; Zhou, G. Overexpression of a Miscanthus lutarioriparius NAC gene MlNAC5 confers enhanced drought and cold tolerance in Arabidopsis. Plant Cell Rep. 2015, 34, 943–958. [Google Scholar] [CrossRef]
- Jin, C.; Li, K.Q.; Xu, X.Y.; Zhang, H.P.; Chen, H.X.; Chen, Y.H.; Hao, J.; Wang, Y.; Huang, X.S.; Zhang, S.L. A Novel NAC Transcription Factor, PbeNAC1, of Pyrus betulifolia Confers Cold and Drought Tolerance via Interacting with PbeDREBs and Activating the Expression of Stress-Responsive Genes. Front. Plant Sci. 2017, 8, 1049. [Google Scholar] [CrossRef] [PubMed]
- Sant’Ana, G.C.; Pereira, L.F.P.; Pot, D.; Ivamoto, S.T.; Domingues, D.S.; Ferreira, R.V.; Pagiatto, N.F.; da Silva, B.S.R.; Nogueira, L.M.; Kitzberger, C.S.G.; et al. Genome-wide association study reveals candidate genes influencing lipids and diterpenes contents in Coffea arabica L. Sci. Rep. 2018, 8, 465. [Google Scholar] [CrossRef] [PubMed]
- Cheng, B.; Furtado, A.; Smyth, H.E.; Henry, R.J. Influence of genotype and environment on coffee quality. Trends Food Sci. Technol. 2016, 57, 20–30. [Google Scholar] [CrossRef]
- Joët, T.; Laffargue, A.; Salmona, J.; Doulbeau, S.; Descroix, F.; Bertrand, B.; de Kochko, A.; Dussert, S. Metabolic pathways in tropical dicotyledonous albuminous seeds: Coffea arabica as a case study. New Phytol. 2009, 182, 146–162. [Google Scholar] [CrossRef] [PubMed]
- Tran, H.T.M.; Lee, L.S.; Furtado, A.; Smyth, H.E.; Henry, R.J. Advances in genomics for the improvement of quality in Coffee. J. Sci. Food Agric. 2016, 96, 3300–3312. [Google Scholar] [CrossRef]
- Kunieda, T.; Mitsuda, N.; Ohme-Takagi, M.; Takeda, S.; Aida, M.; Tasaka, M.; Kondo, M.; Nishimura, M.; Hara-Nishimura, I. NAC family proteins NARS1/NAC2 and NARS2/NAM in the outer integument regulate embryogenesis in Arabidopsis. Plant Cell 2008, 20, 2631–2642. [Google Scholar] [CrossRef]
- Meng, Q.; Zhang, C.; Gai, J.; Yu, D. Molecular cloning, sequence characterization and tissue-specific expression of six NAC-like genes in soybean (Glycine max (L.) Merr.). J. Plant Physiol. 2007, 164, 1002–1012. [Google Scholar] [CrossRef]
- Zhang, Z.; Dong, J.; Ji, C.; Wu, Y.; Messing, J. NAC-type transcription factors regulate accumulation of starch and protein in maize seeds. Proc. Natl. Acad. Sci. USA 2019, 116, 11223–11228. [Google Scholar] [CrossRef]
- Moyano, E.; Martínez-Rivas, F.J.; Blanco-Portales, R.; Molina-Hidalgo, F.J.; Ric-Varas, P.; Matas-Arroyo, A.J.; Caballero, J.L.; Muñoz-Blanco, J.; Rodríguez-Franco, A. Genome-wide analysis of the NAC transcription factor family and their expression during the development and ripening of the Fragaria × ananassa fruits. PLoS ONE 2018, 13, e0196953. [Google Scholar] [CrossRef]
- Murozuka, E.; Massange-Sanchez, J.A.; Nielsen, K.; Gregersen, P.L.; Braumann, I. Genome wide characterization of barley NAC transcription factors enables the identification of grain-specific transcription factors exclusive for the Poaceae family of monocotyledonous plants. PLoS ONE 2018, 13, e0209769. [Google Scholar] [CrossRef]
- Borrill, P.; Harrington, S.A.; Uauy, C. Genome-Wide Sequence and Expression Analysis of the NAC Transcription Factor Family in Polyploid Wheat. G3 Genes Genomes Genet. 2017, 7, 3019–3029. [Google Scholar] [CrossRef] [PubMed]
- Mathew, I.E.; Das, S.; Mahto, A.; Agarwal, P. Three Rice NAC Transcription Factors Heteromerize and Are Associated with Seed Size. Front. Plant Sci. 2016, 7, 1638. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, R.; Werr, W. Pattern Formation in the Monocot Embryo as Revealed by NAMand CUC3 Orthologues from Zea mays L. Plant Mol. Biol. 2005, 58, 669–685. [Google Scholar] [CrossRef] [PubMed]
- Ramalho, J.C.; DaMatta, F.M.; Rodrigues, A.P.; Scotti-Campos, P.; Pais, I.; Batista-Santos, P.; Partelli, F.L.; Ribeiro, A.; Lidon, F.C.; Leitão, A.E. Cold impact and acclimation response of Coffea spp. plants. Theor. Exp. Plant Physiol. 2014, 26, 5–18. [Google Scholar] [CrossRef]
- Batista-Santos, P.; Lidon, F.C.; Fortunato, A.; Leitao, A.E.; Lopes, E.; Partelli, F.; Ribeiro, A.I.; Ramalho, J.C. The impact of cold on photosynthesis in genotypes of Coffea spp.--photosystem sensitivity, photoprotective mechanisms and gene expression. J. Plant Physiol. 2011, 168, 792–806. [Google Scholar] [CrossRef] [PubMed]
- Privat, I.; Foucrier, S.; Prins, A.; Epalle, T.; Eychenne, M.; Kandalaft, L.; Caillet, V.; Lin, C.; Tanksley, S.; Foyer, C.; et al. Differential regulation of grain sucrose accumulation and metabolism in Coffea arabica (Arabica) and Coffea canephora (Robusta) revealed through gene expression and enzyme activity analysis. New Phytol. 2008, 178, 781–797. [Google Scholar] [CrossRef]
- Santos, T.B.; de Lima, R.B.; Nagashima, G.T.; Petkowicz, C.L.; Carpentieri-Pipolo, V.; Pereira, L.F.; Domingues, D.S.; Vieira, L.G. Galactinol synthase transcriptional profile in two genotypes of Coffea canephora with contrasting tolerance to drought. Genet Mol. Biol. 2015, 38, 182–190. [Google Scholar] [CrossRef]
- Castro Caicedo, B.L.; Cortina Guerrero, H.A.; Roux, J.; Wingfield, M.J. New coffee (Coffea arabica) genotypes derived from Coffea canephora exhibiting high levels of resistance to leaf rust and Ceratocystis canker. Trop. Plant Pathol. 2013, 38, 485–494. [Google Scholar] [CrossRef]
- Damatta, F.M.; Ramalho, J.C. Impacts of drought and temperature stress on coffee physiology and production: A review. Braz. J. Plant Physiol. 2006, 18, 55–81. [Google Scholar] [CrossRef]
- Figueiredo, S.A.; Lashermes, P.; Aragão, F.J. Molecular characterization and functional analysis of the β-galactosidase gene during Coffea arabica (L.) fruit development. J. Exp. Bot. 2011, 62, 2691–2703. [Google Scholar] [CrossRef]
- Salmona, J.; Dussert, S.; Descroix, F.; de Kochko, A.; Bertrand, B.; Joet, T. Deciphering transcriptional networks that govern Coffea arabica seed development using combined cDNA array and real-time RT-PCR approaches. Plant Mol. Biol. 2008, 66, 105–124. [Google Scholar] [CrossRef] [PubMed]
- Simkin, A.J.; Qian, T.; Caillet, V.; Michoux, F.; Ben Amor, M.; Lin, C.; Tanksley, S.; McCarthy, J. Oleosin gene family of Coffea canephora: Quantitative expression analysis of five oleosin genes in developing and germinating coffee grain. J. Plant Physiol. 2006, 163, 691–708. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Denoeud, F.; Carretero-Paulet, L.; Dereeper, A.; Droc, G.; Guyot, R.; Pietrella, M.; Zheng, C.; Alberti, A.; Anthony, F.; Aprea, G.; et al. The coffee genome provides insight into the convergent evolution of caffeine biosynthesis. Science 2014, 345, 1181–1184. [Google Scholar] [CrossRef]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Dong, X.; Yang, Y.; Zhang, Z.; Xiao, Z.; Bai, X.; Gao, J.; Hur, Y.; Hao, S.; He, F. Genome-Wide Identification of WRKY Genes and Their Response to Cold Stress in Coffea canephora. Forests 2019, 10, 335. [Google Scholar] [CrossRef]
- Chen, M.; Tan, Q.; Sun, M.; Li, D.; Fu, X.; Chen, X.; Xiao, W.; Li, L.; Gao, D. Genome-wide identification of WRKY family genes in peach and analysis of WRKY expression during bud dormancy. Mol. Genet. Genom. 2016, 291, 1319–1332. [Google Scholar] [CrossRef] [PubMed]
- Saeed, A.I.; Sharov, V.; White, J.; Li, J.; Liang, W.; Bhagabati, N.; Braisted, J.; Klapa, M.; Currier, T.; Thiagarajan, M.; et al. TM4: A free, open-source system for microarray data management and analysis. Biotechniques 2003, 34, 374–378. [Google Scholar] [CrossRef] [PubMed]
- Dalman, K.; Wind, J.J.; Nemesio-Gorriz, M.; Hammerbacher, A.; Lunden, K.; Ezcurra, I.; Elfstrand, M. Overexpression of PaNAC03, a stress induced NAC gene family transcription factor in Norway spruce leads to reduced flavonol biosynthesis and aberrant embryo development. BMC Plant Biol. 2017, 17, 6. [Google Scholar] [CrossRef]
- Wang, Z.; Rashotte, A.M.; Moss, A.G.; Dane, F. Two NAC transcription factors from Citrullus colocynthis, CcNAC1, CcNAC2 implicated in multiple stress responses. Acta Physiol. Plant. 2014, 36, 621–634. [Google Scholar] [CrossRef]
- Wang, Z.; Rashotte, A.M.; Dane, F. Citrullus colocynthis NAC transcription factors CcNAC1 and CcNAC2 are involved in light and auxin signaling. Plant Cell Rep. 2014, 33, 1673–1686. [Google Scholar] [CrossRef]
- Blanc, G.; Hokamp, K.; Wolfe, K.H. A Recent Polyploidy Superimposed on Older Large-Scale Duplications in the Arabidopsis Genome. Genome Res. 2003, 13, 137–144. [Google Scholar] [CrossRef]
- Diao, W.; Snyder, J.C.; Wang, S.; Liu, J.; Pan, B.; Guo, G.; Ge, W.; Dawood, M. Genome-Wide Analyses of the NAC Transcription Factor Gene Family in Pepper (Capsicum annuum L.): Chromosome Location, Phylogeny, Structure, Expression Patterns, Cis-Elements in the Promoter, and Interaction Network. Int. J. Mol. Sci. 2018, 19, 1028. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Li, X.; Chao, J.; Zhang, Z.; Wang, W.; Guo, Y. NAC Family Transcription Factors in Tobacco and Their Potential Role in Regulating Leaf Senescence. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Shang, X.; Cao, Y.; Ma, L. Alternative Splicing in Plant Genes: A Means of Regulating the Environmental Fitness of Plants. Int. J. Mol. Sci. 2017, 18, 432. [Google Scholar] [CrossRef] [PubMed]
- Laloum, T.; Martín, G.; Duque, P. Alternative Splicing Control of Abiotic Stress Responses. Trends Plant Sci. 2018, 23, 140–150. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.J.; Rio, D.C. Mechanisms and Regulation of Alternative Pre-mRNA Splicing. Annu. Rev. Biochem. 2015, 84, 291–323. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Duan, X.; Jiang, H.; Sun, Y.; Tang, Y.; Yuan, Z.; Guo, J.; Liang, W.; Chen, L.; Yin, J.; et al. Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis. Plant Physiol. 2006, 141, 1167–1184. [Google Scholar] [CrossRef]
- Kazemi-Shahandashti, S.S.; Maali-Amiri, R. Global insights of protein responses to cold stress in plants: Signaling, defence, and degradation. J. Plant Physiol. 2018, 226, 123–135. [Google Scholar] [CrossRef]
- Chinnusamy, V.; Zhu, J.; Zhu, J.K. Cold stress regulation of gene expression in plants. Trends Plant Sci. 2007, 12, 444–451. [Google Scholar] [CrossRef]
- Liu, Y.; Dang, P.; Liu, L.; He, C. Cold acclimation by the CBF-COR pathway in a changing climate: Lessons from Arabidopsis thaliana. Plant Cell Rep. 2019, 38, 511–519. [Google Scholar] [CrossRef]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci. 2005, 10, 88–94. [Google Scholar] [CrossRef]
- Ramalho, J.C.; Rodrigues, A.P.; Lidon, F.C.; Marques, L.; Leitao, A.E.; Fortunato, A.S.; Pais, I.P.; Silva, M.J.; Scotticampos, P.; Lopes, A.M. Stress cross-response of the antioxidative system promoted by superimposed drought and cold conditions in Coffea spp. PLoS ONE 2018, 13, e0198694. [Google Scholar] [CrossRef] [PubMed]
- An, J.P.; Li, R.; Qu, F.J.; You, C.X.; Wang, X.F.; Hao, Y.J. An apple NAC transcription factor negatively regulates cold tolerance via CBF-dependent pathway. J. Plant Physiol. 2018, 221, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Yoo, S.Y.; Kim, Y.; Kim, S.Y.; Lee, J.S.; Ahn, J.H. Control of flowering time and cold response by a NAC-domain protein in Arabidopsis. PLoS ONE 2007, 2, e642. [Google Scholar] [CrossRef] [PubMed]
- Fujita, M.; Fujita, Y.; Maruyama, K.; Seki, M.; Hiratsu, K.; Ohmetakagi, M.; Tran, L.P.; Yamaguchishinozaki, K.; Shinozaki, K. A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J. 2004, 39, 863–876. [Google Scholar] [CrossRef]
- Yang, S.B.; Seo, P.J.; Yoon, H.; Park, C. The Arabidopsis NAC Transcription Factor VNI2 Integrates Abscisic Acid Signals into Leaf Senescence via the COR/RD Genes. Plant Cell 2011, 23, 2155–2168. [Google Scholar] [CrossRef]
- Le, B.H.; Cheng, C.; Bui, A.Q.; Wagmaister, J.A.; Henry, K.F.; Pelletier, J.; Kwong, L.; Belmonte, M.; Kirkbride, R.; Horvath, S.; et al. Global analysis of gene activity during Arabidopsis seed development and identification of seed-specific transcription factors. Proc. Natl. Acad. Sci. USA 2010, 107, 8063–8070. [Google Scholar] [CrossRef]


| Gene Symbol | Gene ID | Peptide Length (a.a.) | Chr. No. | Introns | PI | Mw (KDa) | Best hit to ANACs (Blastp) | ||
|---|---|---|---|---|---|---|---|---|---|
| Gene ID | Gene Name | E-Value | |||||||
| CocNAC01 | Cc01_g04540 | 340 | Chr1 | 2 | 8.63 | 38.95 | AT1G52890.1 | ANAC019 | 5.3 × 10−44 |
| CocNAC02 | Cc01_g04560 | 410 | Chr1 | 3 | 5.04 | 47.10 | AT1G77450.1 | ANAC032 | 9.6 × 10−46 |
| CocNAC03 | Cc01_g04590 | 350 | Chr1 | 2 | 5.84 | 39.74 | AT3G04070.2 | ANAC047 | 5.2 × 10−39 |
| CocNAC04 | Cc01_g08720 | 338 | Chr1 | 2 | 8.91 | 35.87 | AT3G44290.2 | ANAC060 | 3.0 × 10−12 |
| CocNAC05 | Cc01_g08730 | 401 | Chr1 | 3 | 5.66 | 44.83 | AT3G44290.1 | ANAC060 | 6.3 × 10−86 |
| CocNAC06 | Cc01_g08900 | 258 | Chr1 | 2 | 6.99 | 29.72 | AT5G22380.1 | ANAC090 | 4.5 × 10−86 |
| CocNAC07 | Cc01_g11320 | 346 | Chr1 | 2 | 8.57 | 38.95 | AT1G61110.1 | ANAC025 | 2.6 × 10−74 |
| CocNAC08 | Cc01_g11350 | 310 | Chr1 | 2 | 6.67 | 34.92 | AT1G69490.1 | ANAC029 | 2.4 × 10−70 |
| CocNAC09 | Cc01_g18220 | 300 | Chr1 | 2 | 5.85 | 33.36 | AT4G28530.1 | ANAC074 | 4.0 × 10−89 |
| CocNAC10 | Cc01_g19370 | 87 | Chr1 | 1 | 8.71 | 9.91 | AT4G27410.3 | ANAC072 | 5.7 × 10−08 |
| CocNAC11 | Cc02_g00700 | 632 | Chr2 | 5 | 5.24 | 69.87 | AT5G24590.2 | ANAC091 | 3.9 × 10−86 |
| CocNAC12 | Cc02_g04160 | 357 | Chr2 | 2 | 6.02 | 41.16 | AT1G12260.1 | ANAC007 | 3.5 × 10−149 |
| CocNAC13 | Cc02_g10630 | 583 | Chr2 | 5 | 4.64 | 64.88 | AT5G04410.1 | ANAC078 | 7.2 × 10−146 |
| CocNAC14 | Cc02_g10640 | 407 | Chr2 | 3 | 5.35 | 45.14 | AT3G10480.2 | ANAC050 | 5.5 × 10−117 |
| CocNAC15 | Cc02_g13910 | 315 | Chr2 | 2 | 8.18 | 35.09 | AT4G28500.1 | ANAC073 | 5.7 × 10−121 |
| CocNAC16 | Cc02_g14010 | 302 | Chr2 | 2 | 8.96 | 34.10 | AT1G69490.1 | ANAC029 | 6.6 × 10−74 |
| CocNAC17 | Cc02_g21890 | 193 | Chr2 | 2 | 5.02 | 21.94 | AT5G64530.1 | ANAC104 | 1.9 × 10−86 |
| CocNAC18 | Cc02_g23810 | 369 | Chr2 | 3 | 8.26 | 41.94 | AT2G02450.2 | ANAC035 | 1.7 × 10−121 |
| CocNAC19 | Cc02_g33930 | 323 | Chr2 | 2 | 5.95 | 37.36 | AT1G01720.1 | ANAC002 | 6.3 × 10−123 |
| CocNAC20 | Cc02_g39920 | 385 | Chr2 | 4 | 8.29 | 43.69 | AT2G46770.1 | ANAC043 | 6.0 × 10−115 |
| CocNAC21 | Cc04_g07090 | 327 | Chr4 | 2 | 6.98 | 36.58 | AT3G18400.1 | ANAC058 | 1.5 × 10−111 |
| CocNAC22 | Cc04_g08930 | 240 | Chr4 | 2 | 4.89 | 27.62 | AT3G17730.1 | ANAC057 | 1.4 × 10−130 |
| CocNAC23 | Cc04_g09840 | 251 | Chr4 | 2 | 8.85 | 28.23 | AT5G13180.1 | ANAC083 | 5.5 × 10−80 |
| CocNAC24 | Cc04_g16290 | 356 | Chr4 | 3 | 8.18 | 39.65 | AT5G61430.1 | ANAC100 | 1.5 × 10−133 |
| CocNAC25 | Cc05_g01340 | 304 | Chr5 | 2 | 8.97 | 34.27 | AT2G43000.1 | ANAC042 | 1.1 × 10−88 |
| CocNAC26 | Cc05_g07770 | 272 | Chr5 | 2 | 8.27 | 30.58 | AT4G28500.1 | ANAC073 | 6.2 × 10−117 |
| CocNAC27 | Cc05_g11720 | 359 | Chr5 | 2 | 8.03 | 39.35 | AT5G53950.1 | ANAC098 | 1.2 × 10−112 |
| CocNAC28 | Cc05_g12500 | 356 | Chr5 | 2 | 8.61 | 39.61 | AT4G27410.2 | ANAC072 | 9.2 × 10−132 |
| CocNAC29 | Cc05_g12510 | 330 | Chr5 | 3 | 8.98 | 36.51 | AT3G15510.1 | ANAC056 | 3.8 × 10−120 |
| CocNAC30 | Cc05_g15960 | 311 | Chr5 | 2 | 8.33 | 35.23 | AT1G56010.2 | ANAC022 | 1.5 × 10−108 |
| CocNAC31 | Cc05_g16190 | 304 | Chr5 | 3 | 8.44 | 34.48 | AT2G43000.1 | ANAC042 | 5.0 × 10−96 |
| CocNAC32 | Cc06_g03790 | 361 | Chr6 | 3 | 8.81 | 40.62 | AT1G61110.1 | ANAC025 | 1.3 × 10−77 |
| CocNAC33 | Cc06_g06140 | 253 | Chr6 | 2 | 5.73 | 28.74 | AT5G22380.1 | ANAC090 | 4.5 × 10−81 |
| CocNAC34 | Cc06_g10730 | 469 | Chr6 | 5 | 6.32 | 52.21 | AT4G29230.2 | ANAC075 | 0.0 |
| CocNAC35 | Cc07_g01550 | 401 | Chr7 | 5 | 5.47 | 44.36 | AT3G01600.1 | ANAC044 | 1.9 × 10−98 |
| CocNAC36 | Cc07_g03320 | 400 | Chr7 | 2 | 6.20 | 44.57 | AT1G26870.1 | ANAC009 | 1.5 × 10−99 |
| CocNAC37 | Cc07_g03570 | 332 | Chr7 | 3 | 5.67 | 37.39 | AT5G61430.1 | ANAC100 | 7.9 × 10−114 |
| CocNAC38 | Cc07_g10520 | 350 | Chr7 | 2 | 5.28 | 39.80 | AT2G18060.3 | ANAC037 | 8.0 × 10−151 |
| CocNAC39 | Cc07_g12760 | 664 | Chr7 | 5 | 5.54 | 75.49 | AT1G65910.1 | ANAC028 | 1.6 × 10−155 |
| CocNAC40 | Cc07_g18360 | 501 | Chr7 | 4 | 4.76 | 54.76 | AT5G09330.4 | ANAC082 | 7.1 × 10−89 |
| CocNAC41 | Cc08_g02440 | 346 | Chr8 | 3 | 5.53 | 39.04 | AT4G17980.1 | ANAC071 | 2.5 × 10−99 |
| CocNAC42 | Cc08_g12970 | 326 | Chr8 | 3 | 6.41 | 37.67 | AT1G12260.2 | ANAC007 | 2.5 × 10−139 |
| CocNAC43 | Cc08_g16900 | 455 | Chr8 | 2 | 5.78 | 49.95 | AT2G46770.1 | ANAC043 | 4.2 × 10−114 |
| CocNAC44 | Cc08_g17070 | 592 | Chr8 | 6 | 5.43 | 65.88 | AT4G35580.1 | ANAC118 | 1.0 × 10−122 |
| CocNAC45 | Cc09_g10560 | 311 | Chr9 | 3 | 8.12 | 35.18 | AT2G17040.1 | ANAC036 | 3.2 × 10−89 |
| CocNAC46 | Cc10_g03330 | 356 | Chr10 | 4 | 6.62 | 38.73 | AT1G69490.1 | ANAC029 | 8.0 × 10−13 |
| CocNAC47 | Cc10_g04680 | 270 | Chr10 | 2 | 7.03 | 30.40 | AT4G27410.2 | ANAC072 | 2.1 × 10−22 |
| CocNAC48 | Cc10_g06320 | 570 | Chr10 | 3 | 4.69 | 63.48 | AT1G34190.1 | ANAC017 | 1.3 × 10−141 |
| CocNAC49 | Cc10_g11470 | 319 | Chr10 | 2 | 6.32 | 36.73 | AT1G71930.1 | ANAC030 | 3.9 × 10−115 |
| CocNAC50 | Cc10_g12200 | 310 | Chr10 | 2 | 6.16 | 35.20 | AT1G01720.1 | ANAC002 | 9.5 × 10−141 |
| CocNAC51 | Cc10_g16190 | 363 | Chr10 | 3 | 7.00 | 40.85 | AT4G10350.1 | ANAC070 | 4.4 × 10−140 |
| CocNAC52 | Cc11_g10610 | 450 | Chr11 | 4 | 4.95 | 50.75 | AT1G25580.1 | ANAC008 | 0.0 |
| CocNAC53 | Cc11_g12740 | 275 | Chr11 | 2 | 7.63 | 31.52 | AT1G69490.1 | ANAC029 | 9.6 × 10−112 |
| CocNAC54 | Cc11_g13040 | 401 | Chr11 | 3 | 6.45 | 45.19 | AT1G26870.1 | ANAC009 | 7.4 × 10−110 |
| CocNAC55 | Cc11_g17410 | 279 | Chr11 | 3 | 9.37 | 30.75 | AT5G13180.1 | ANAC083 | 4.4 × 10−84 |
| CocNAC56 | Cc00_g05780 | 319 | Chr un | 2 | 5.88 | 36.63 | AT1G71930.1 | ANAC030 | 2.9 × 10−115 |
| CocNAC57 | Cc00_g08030 | 398 | Chr un | 2 | 6.03 | 44.68 | AT5G18270.1 | ANAC087 | 9.4 × 10−109 |
| CocNAC58 | Cc00_g08040 | 304 | Chr un | 2 | 9.00 | 34.03 | AT2G24430.2 | ANAC039 | 5.1 × 10−118 |
| CocNAC59 | Cc00_g09070 | 358 | Chr un | 4 | 6.48 | 40.65 | AT1G79580.5 | ANAC033 | 2.0 × 10−112 |
| CocNAC60 | Cc00_g14270 | 288 | Chr un | 2 | 8.49 | 33.07 | AT2G17040.1 | ANAC036 | 1.8 × 10−92 |
| CocNAC61 | Cc00_g14290 | 280 | Chr un | 2 | 7.68 | 31.80 | AT2G17040.1 | ANAC036 | 2.7 × 10−85 |
| CocNAC62 | Cc00_g26010 | 153 | Chr un | 1 | 9.79 | 17.94 | AT1G69490.1 | ANAC029 | 3.0 × 10−29 |
| CocNAC63 | Cc00_g30840 | 196 | Chr un | 2 | 4.91 | 22.64 | AT5G64530.1 | ANAC104 | 7.2 × 10−47 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dong, X.; Jiang, Y.; Yang, Y.; Xiao, Z.; Bai, X.; Gao, J.; Tan, S.; Hur, Y.; Hao, S.; He, F. Identification and Expression Analysis of the NAC Gene Family in Coffea canephora. Agronomy 2019, 9, 670. https://doi.org/10.3390/agronomy9110670
Dong X, Jiang Y, Yang Y, Xiao Z, Bai X, Gao J, Tan S, Hur Y, Hao S, He F. Identification and Expression Analysis of the NAC Gene Family in Coffea canephora. Agronomy. 2019; 9(11):670. https://doi.org/10.3390/agronomy9110670
Chicago/Turabian StyleDong, Xiangshu, Yuan Jiang, Yanan Yang, Ziwei Xiao, Xuehui Bai, Jing Gao, Shirui Tan, Yoonkang Hur, Shumei Hao, and Feifei He. 2019. "Identification and Expression Analysis of the NAC Gene Family in Coffea canephora" Agronomy 9, no. 11: 670. https://doi.org/10.3390/agronomy9110670
APA StyleDong, X., Jiang, Y., Yang, Y., Xiao, Z., Bai, X., Gao, J., Tan, S., Hur, Y., Hao, S., & He, F. (2019). Identification and Expression Analysis of the NAC Gene Family in Coffea canephora. Agronomy, 9(11), 670. https://doi.org/10.3390/agronomy9110670





