Quantitative Trait Loci (QTL) for Forage Traits in Intermediate Wheatgrass When Grown as Spaced-Plants versus Monoculture and Polyculture Swards
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Phenotypic Evaluations
2.3. Statistical and Genetic Analysis
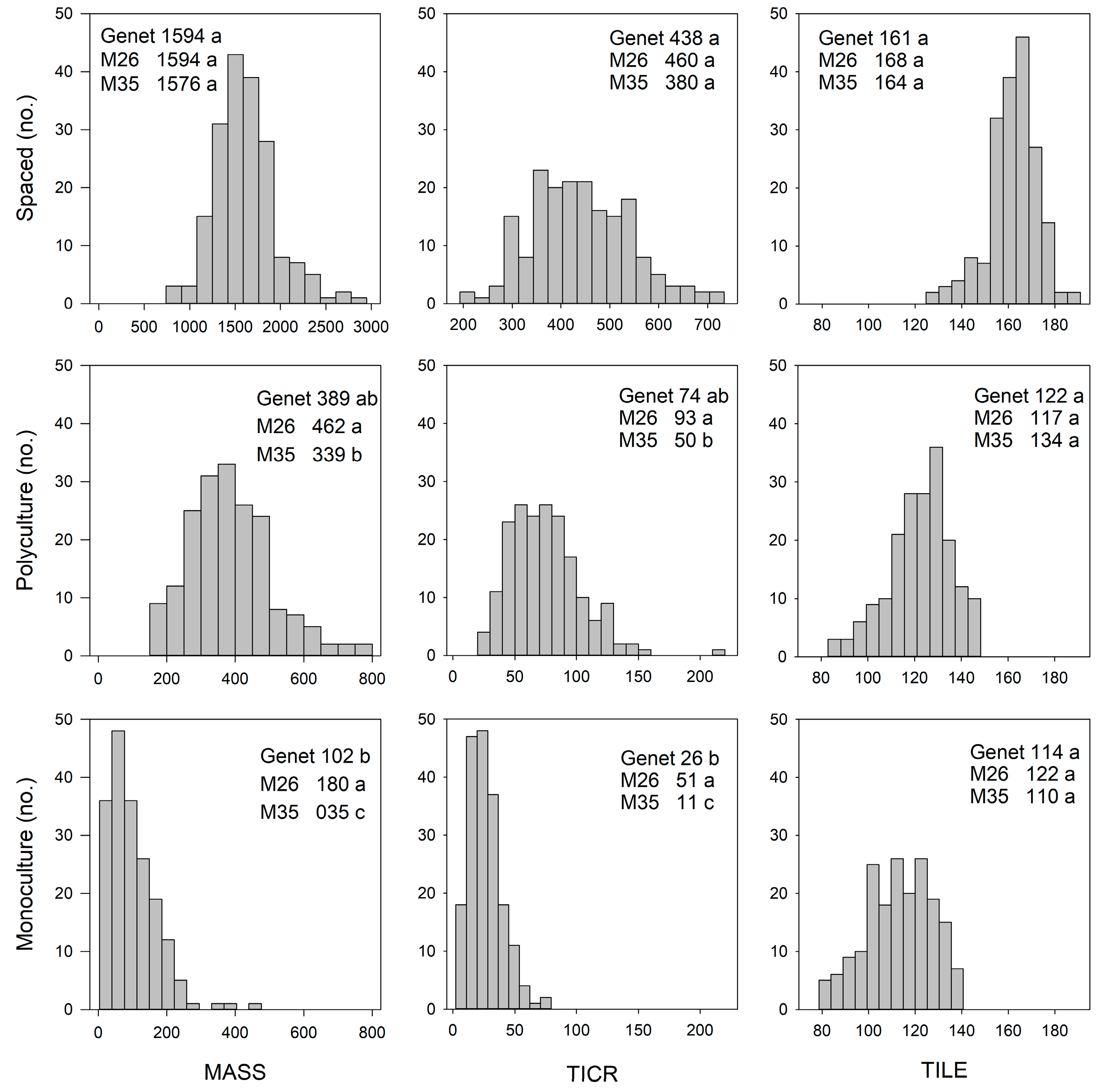
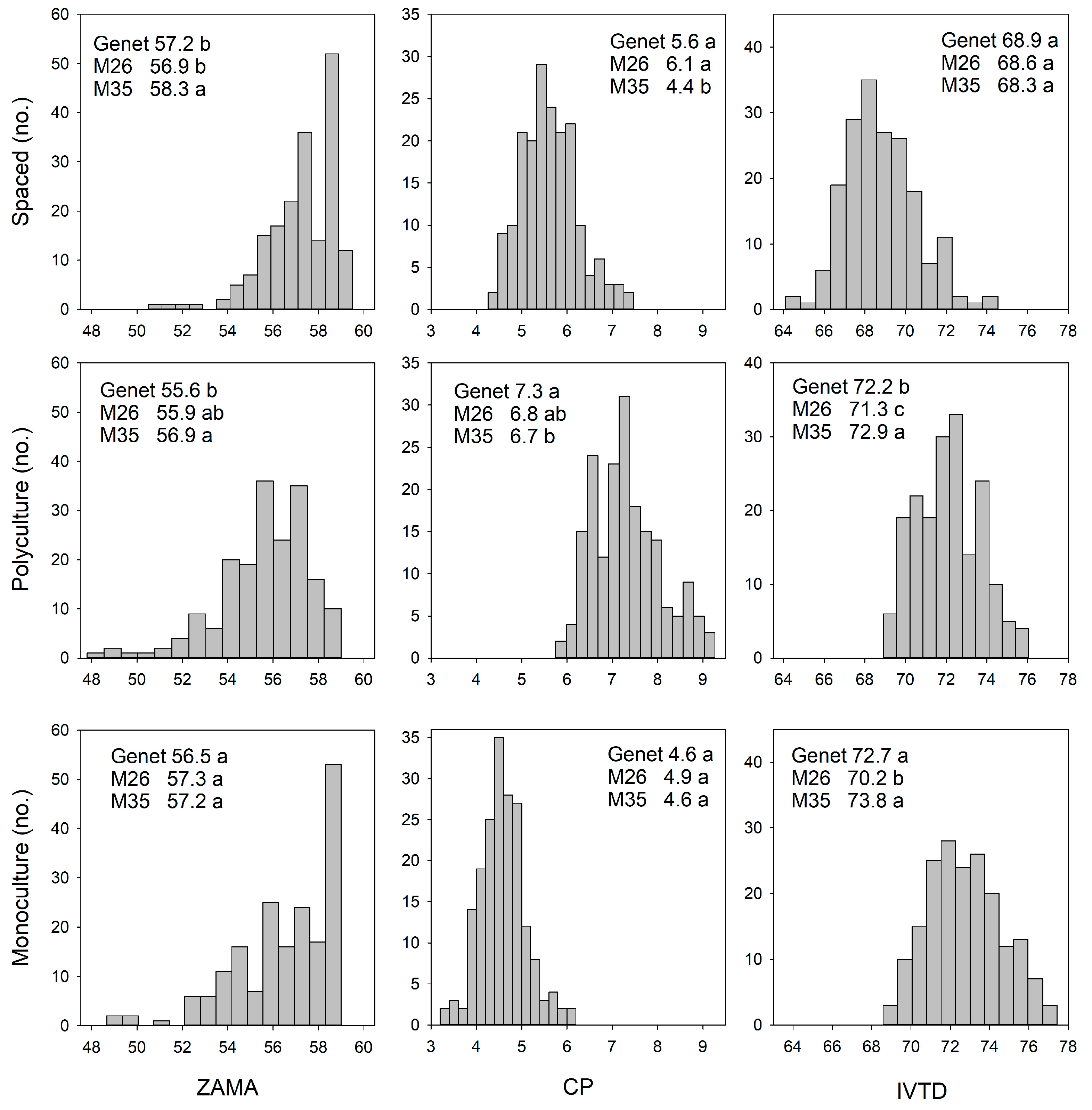
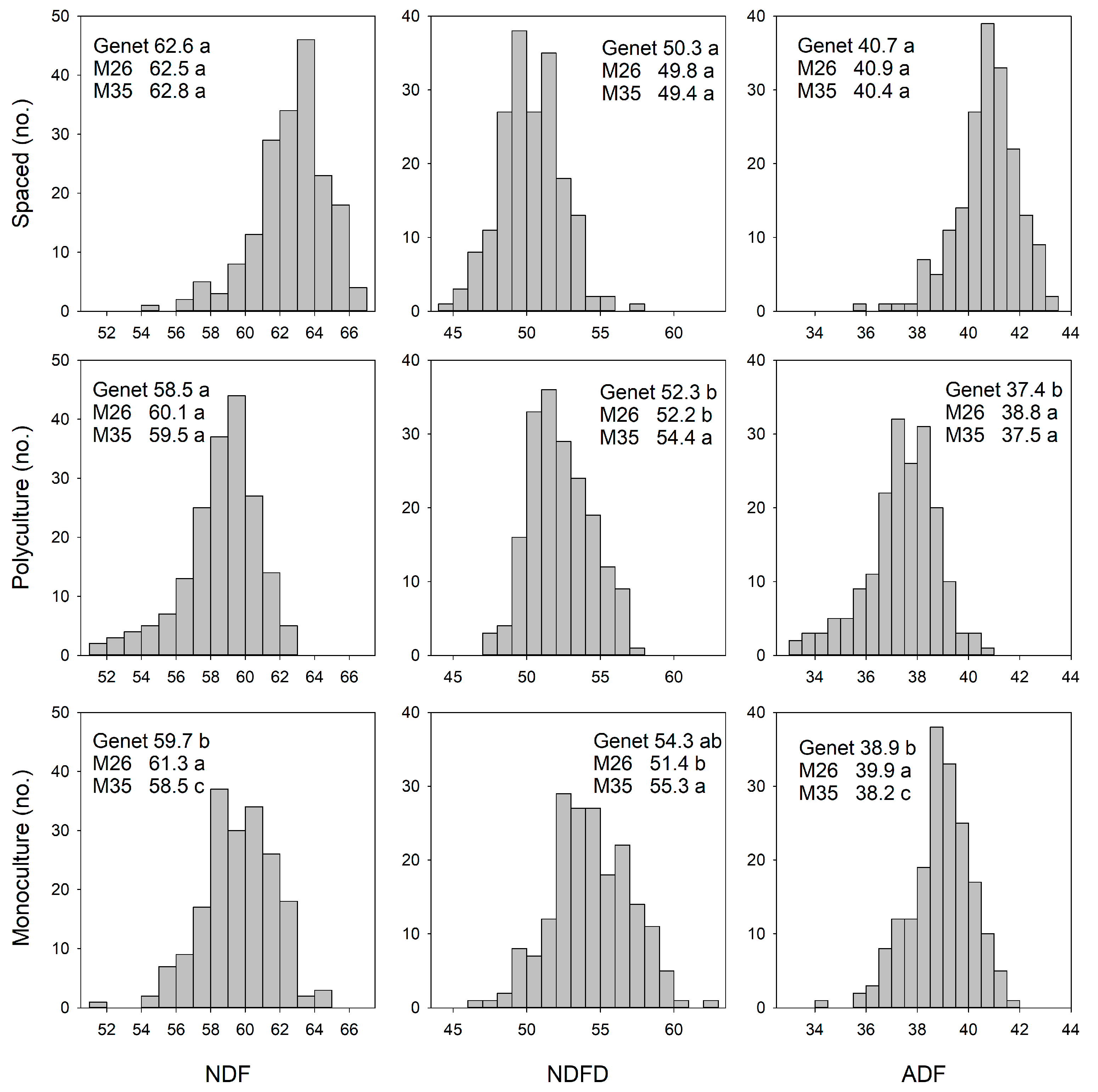
3. Results
3.1. Phenotypic and Genetic Variation: Relationship Among Environments
3.1.1. Biomass and Morphological Traits
3.1.2. Forage Nutritive Value
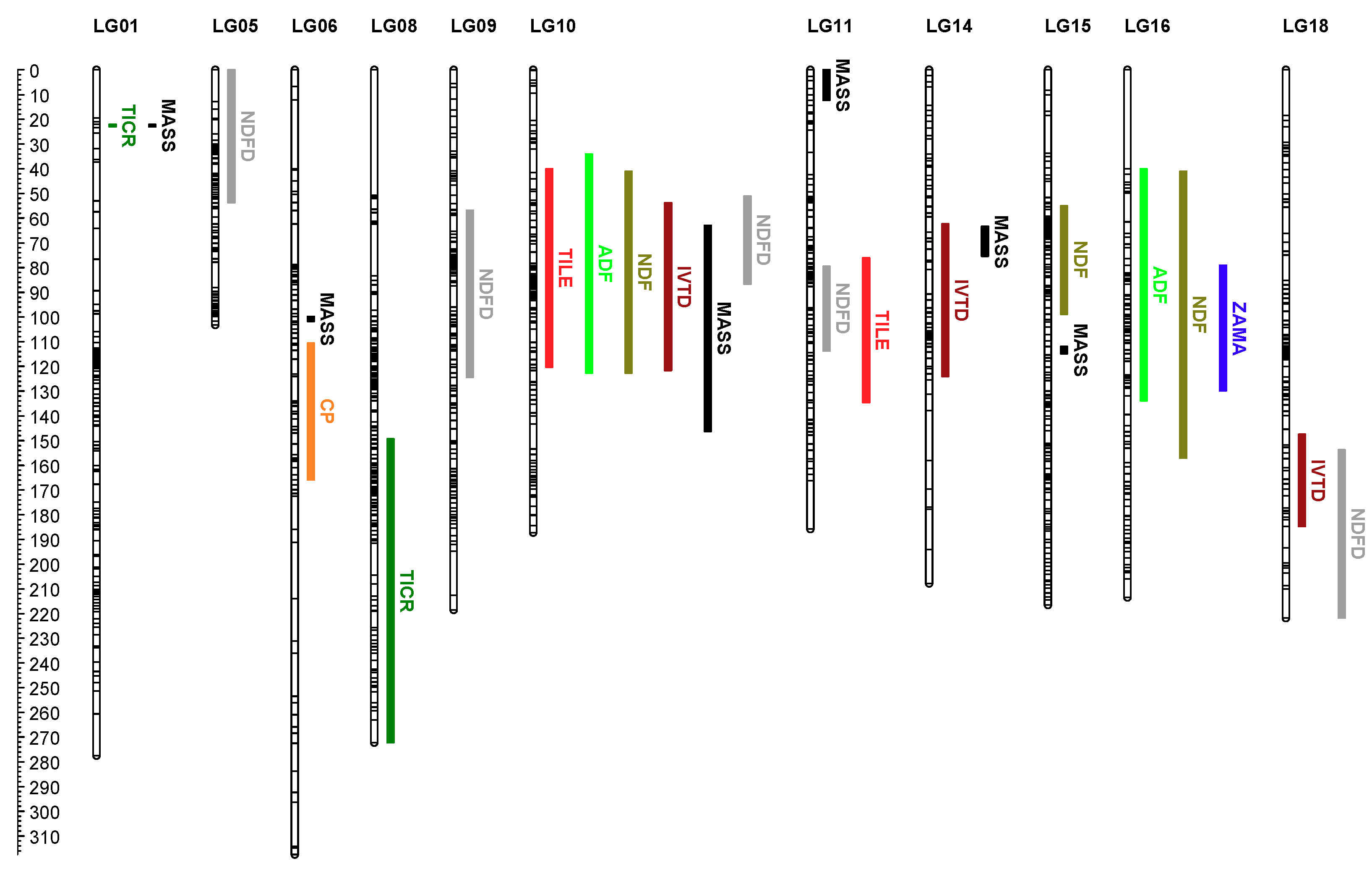
3.2. QTL Analysis
3.2.1. Biomass and Morphological Trait QTL
3.2.2. Forage Nutritive Value QTL
3.3. QTL × Environment Interactions
4. Discussion
4.1. Genetic Control in Spaced-Plants and Swards
4.1.1. Spaced-Plant vs. Sward: Biomass and Morphological Traits
4.1.2. Spaced-Plant vs. Sward: Forage Nutritive Value
4.2. Genetic Control in Grass Monoculture and Grass−Legume Polyculture
4.2.1. Monoculture vs. Polyculture: Biomass and Morphological Traits
4.2.2. Monoculture vs. Polyculture: Forage Nutritive Value
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| Trait | Spaced-Plants | Polyculture | Monoculture | |||
|---|---|---|---|---|---|---|
| 2016 | 2017 | 2016 | 2017 | 2016 | 2017 | |
| MASS | 1275 ± 455 | 1934 ± 592 | 123 ± 94 | 658 ± 304 | 70 ± 79 | 137 ± 152 |
| TILE | 164 ± 14 | 160 ± 17 | 111 ± 18 | 133 ± 19 | 116 ± 19 | 112 ± 20 |
| TICR | 435 ± 106 | 558 ± 217 | 30 ± 29 | 119 ± 68 | 24 ± 19 | 30 ± 26 |
| ZAMA | 58 ± 1 | 56 ± 2 | 55 ± 2 | 56 ± 3 | 57 ± 3 | 56 ± 3 |
| CP | 65 ± 13 | 47 ± 8 | 84 ± 12 | 63 ± 10 | 50 ± 11 | 41 ± 7 |
| NDF | 599 ± 33 | 654 ± 25 | 545 ± 33 | 626 ± 26 | 599 ± 27 | 595 ± 33 |
| ADF | 394 ± 19 | 421 ± 15 | 349 ± 22 | 399 ± 17 | 386 ± 16 | 393 ± 19 |
| ADL | 88 ± 7 | 95 ± 6 | 85 ± 9 | 95 ± 7 | 79 ± 11 | 93 ± 10 |
| IVTD | 694 ± 28 | 683 ± 22 | 730 ± 20 | 713 ± 23 | 726 ± 24 | 728 ± 31 |
| NDFD | 490 ± 33 | 515 ± 27 | 503 ± 37 | 541 ± 27 | 542 ± 38 | 543 ± 40 |
| ME | 2.08 ± 0.07 | 2.04 ± 0.07 | 2.21 ± 0.06 | 2.11 ± 0.06 | 2.17 ± 0.08 | 2.14 ± 0.07 |
| LG | MASS | TILE | TICR | ZAMA | CP | NDF | ADF | ADL | IVTD | NDFD |
|---|---|---|---|---|---|---|---|---|---|---|
| GW † | 4.9 | 4.9 | 5 | 5.2 | 5 | 5.2 | 5.1 | 4.8 | 4.9 | 4.8 |
| 1 | 3.3 | 3.4 | 3.4 | 3.6 | 3.5 | 3.5 | 3.5 | 3.3 | 3.3 | 3.4 |
| 2 | 3.3 | 3.2 | 3.4 | 3.3 | 3.4 | 3.2 | 3.3 | 3.2 | 3.2 | 3.4 |
| 3 | 3.2 | 3.4 | 3.3 | 3.5 | 3.4 | 3.4 | 3.4 | 3.3 | 3.3 | 3.2 |
| 4 | 3.5 | 3.5 | 3.5 | 3.7 | 3.5 | 3.5 | 3.5 | 3.4 | 3.5 | 3.5 |
| 5 | 3.1 | 3.3 | 3.3 | 3.4 | 3.2 | 3.4 | 3.4 | 3.3 | 3.3 | 3.3 |
| 6 | 3.4 | 3.4 | 3.5 | 3.6 | 3.5 | 3.6 | 3.7 | 3.4 | 3.3 | 3.4 |
| 7 | 3.3 | 3.2 | 3.3 | 3.3 | 3.3 | 3.2 | 3.4 | 3.4 | 3.4 | 3.3 |
| 8 | 3.6 | 3.4 | 3.5 | 3.8 | 3.7 | 3.7 | 3.6 | 3.7 | 3.6 | 3.6 |
| 9 | 3.3 | 3.3 | 3.5 | 3.7 | 3.5 | 3.7 | 3.6 | 3.4 | 3.6 | 3.4 |
| 10 | 3.3 | 3.3 | 3.2 | 3.3 | 3.3 | 3.3 | 3.3 | 3.4 | 3.5 | 3.4 |
| 11 | 3.3 | 3.3 | 3.3 | 3.3 | 3.2 | 3.3 | 3.4 | 3.3 | 3.3 | 3.4 |
| 12 | 3.3 | 3.1 | 3.1 | 3.3 | 3.2 | 3.3 | 3.3 | 3.2 | 3.2 | 3.2 |
| 13 | 3.3 | 3.3 | 3.4 | 3.9 | 3.5 | 3.6 | 3.8 | 3.3 | 3.5 | 3.3 |
| 14 | 3.2 | 3.2 | 3.2 | 3.4 | 3.2 | 3.3 | 3.3 | 3.3 | 3.3 | 3.4 |
| 15 | 3.4 | 3.4 | 3.5 | 3.7 | 3.5 | 3.4 | 3.4 | 3.4 | 3.3 | 3.5 |
| 16 | 3.2 | 3.1 | 3 | 3.4 | 3.3 | 3.2 | 3.4 | 3.2 | 3.2 | 3.2 |
| 17 | 3.5 | 3.4 | 3.3 | 3.7 | 3.4 | 3.5 | 3.3 | 3.4 | 3.3 | 3.4 |
| 18 | 3.3 | 3.3 | 3.4 | 3.4 | 3.3 | 3.2 | 3.3 | 3.4 | 3.4 | 3.3 |
| 19 | 3.3 | 3.3 | 3.4 | 3.6 | 3.4 | 3.3 | 3.3 | 3.3 | 3.4 | 3.3 |
| 20 | 3.5 | 3.5 | 3.4 | 3.8 | 3.6 | 3.8 | 3.6 | 3.4 | 3.4 | 3.5 |
| 21 | 3.4 | 3.2 | 3.2 | 3.5 | 3.4 | 3.5 | 3.3 | 3.3 | 3.4 | 3.4 |
| Trait | Linkage Group | Spaced−Plants | Polyculture | Monoculture | Across Environments | Q × E |
|---|---|---|---|---|---|---|
| MASS | 1 | (0.01, −0.30) | (0.00, 0.02) | (0.01, −0.33) ** | (0.01, −0.30) | NS |
| 6 | (−0.03, 0.00) | (−0.01, 0.01) | (−0.13, 0.12) ** | (−0.08, 0.09) | NS | |
| 10 | (−0.08, 0.09) ** | (−0.02, 0.02) * | (0.01, 0.01) * | (−0.04, 0.04) ** | 0.0001 | |
| 11 | (−0.01, −0.02) | (0.00, −0.02) * | (0.00, −0.01) ** | (0.00, −0.01) | NS | |
| 14 | (−0.01, −0.08) * | (0.01, −0.02) | (0.00, −0.01) ** | (0.00, −0.03) * | 0.0004 | |
| 15 | (−0.11, 0.02) | (−0.01, −0.01) | (−0.13, 0.11) ** | (−0.08, 0.04) | 0.0007 | |
| TILE | 10 | (−3.13, 2.15) ** | (−3.92, 2.72) ** | (−3.69, 2.8) * | (−3.24, 2.34) ** | NS |
| 11 | (2.32, 0.54) * | (2.84, −1.97) * | (2.78, −2.74) * | (2.61, −2.28) ** | NS | |
| TICR | 1 | (31.42, −56.82) | (0.58, −32.17) | (1.09, −79.75) ** | (12.03, −199.51) | NS |
| 8 | (34.5, 29.51) ** | (2.66, 11.88) | (0.40, 5.66) | (10.43, 6.09) * | NS | |
| ZAMA | 16 | (−0.40, 0.38) ** | (2.73, −5.06) | (−0.29, −1.19) | (−0.43, 0.29) * | NS |
| CP | 6 | (0.13, −0.12) * | (0.18, 0.15) ** | (0.08, 0.08) * | (0.13, 0.07) ** | NS |
| NDF | 10 | (−0.42, 0.53) ** | (−0.36, 0.64) ** | (−0.30, 0.73) ** | (−0.35, 0.57) ** | NS |
| 15 | (−0.08, 0.43) | (−0.09, 0.24) | (−0.04, 0.81) ** | (−0.05, 0.51) | NS | |
| 16 | (0.44, −0.62) * | (0.18, −0.48) | (0.44, −0.73) ** | (0.38, −0.61) ** | NS | |
| ADF | 10 | (−0.21, 0.27) ** | (−0.24, 0.44) ** | (−0.14, 0.43) ** | (−0.05, 0.45) ** | NS |
| 16 | (0.24, −0.32) * | (0.10, −0.3) | (0.26, −0.43) ** | (0.23, −0.36) ** | NS | |
| 10 | (0.30, −0.51) ** | (0.39, −0.41) ** | (0.59, −0.76) ** | (0.33, −0.48) ** | NS | |
| 14 | (−0.24, 0.46) * | (−0.14, 0.4) * | (−0.08, 0.56) ** | (−0.23, 0.48) ** | NS | |
| 18 | (0.18, 0.53) * | (0.36, 0.49) ** | (0.24, 0.56) | (0.25, 0.48) ** | NS | |
| NDFD | 5 | (0.19, 0.19) ** | (0.14, 0.48) * | (0.08, 0.33) | (0.21, 0.4) * | NS |
| 9 | (−0.17, −0.78) ** | (0.27, −0.85) ** | (0.16, −0.64) | (0.22, −0.74) ** | NS | |
| 10 | (0.25, −0.35) | (0.29, −0.09) | (0.47, −0.49) ** | (0.32, −0.27) | 0.0003 | |
| 11 | (−0.40, −0.34) ** | (−0.47, −0.39) * | (−0.41, 0.03) | (−0.40, −0.25) * | NS | |
| 18 | (0.37, 0.51) * | (0.50, 0.49) ** | (0.41, 0.49) * | (0.40, 0.51) ** | NS |
References
- Casler, M.D.; Carlson, I.T.; Berg, C.C.; Sleper, D.A.; Barker, R.E. Convergent-divergent selection for seed production and forage traits in orchardgrass: I. direct selection responses. Crop Sci. 1997, 37, 1047–1053. [Google Scholar] [CrossRef]
- Waldron, B.L.; Robins, J.G.; Peel, M.D.; Jensen, K.B. Predicted efficiency of spaced-plant selection to indirectly improve tall fescue sward yield and quality. Crop Sci. 2008, 48, 443–449. [Google Scholar] [CrossRef]
- Hill, J. The three C’s—Competition, coexistence and coevolution—And their impact on the breeding of forage crop mixtures. Theor. Appl. Genet. 1990, 79, 168–176. [Google Scholar] [CrossRef] [PubMed]
- Waldron, B.L.; Peel, M.D.; Larson, S.R.; Mott, I.W.; Creech, J.E. Tall fescue forage mass in a grass-legume mixture: Predicted efficiency of indirect selection. Euphytica 2017, 213, 67. [Google Scholar] [CrossRef]
- Annicchiarico, P. Breeding white clover for increased ability to compete with associated grasses. J. Agric. Sci. 2003, 140, 255–266. [Google Scholar] [CrossRef]
- De Leon, N.; Jannink, J.-L.; Edwards, J.W.; Kaeppler, S.M. Introduction to a Special Issue on Genotype by Environment Interaction. Crop Sci. 2016, 56, 2081–2089. [Google Scholar] [CrossRef]
- Sukumaran, S.; Crossa, J.; Jarquin, D.; Lopes, M.; Reynolds, M.P. Genomic prediction with pedigree and genotype × environment interaction in spring wheat grown in South and West Asia, North Africa, and Mexico. G3 Genes Genomes Genet. 2017, 7, 481–495. [Google Scholar] [CrossRef]
- Lacaze, X.; Roumet, P. Environment characterisation for the interpretation of environmental effect and genotype × environment interaction. Theor. Appl. Genet. 2004, 109, 1632–1640. [Google Scholar] [CrossRef] [PubMed]
- Vargas, M.; van Eeuwijk, F.A.; Crossa, J.; Ribaut, J.-M. Mapping QTLs and QTL × environment interaction for CIMMYT maize drought stress program using factorial regression and partial least squares methods. Theor. Appl. Genet. 2006, 112, 1009–1023. [Google Scholar] [CrossRef]
- van Eeuwijk, F.A.; Malosetti, M.; Yin, X.; Struik, P.C.; Stam, P. Statistical models for genotype by environment data: From conventional ANOVA models to eco-physiological QTL models. Aust. J. Agric. Res. 2005, 56, 883–894. [Google Scholar] [CrossRef]
- Jensen, K.B.; Yan, X.; Larson, S.R.; Wang, R.R.C.; Robins, J.G. Agronomic and genetic diversity in intermediate wheatgrass (Thinopyrum intermedium). Plant Breed. 2016, 135, 751–758. [Google Scholar] [CrossRef]
- Kantarski, T.; Larson, S.; Zhang, X.; DeHaan, L.; Borevitz, J.; Anderson, J.; Poland, J. Development of the first consensus genetic map of intermediate wheatgrass (Thinopyrum intermedium) using genotyping-by-sequencing. Theor. Appl. Genet. 2017, 130, 137–150. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.R.C.; Larson, S.R.; Jensen, K.B.; Bushman, B.S.; Dehaan, L.R.; Wang, S.; Yan, X. Genome Evolution of Intermediate Wheatgrass as Revealed by EST-SSR Markers Developed from Its Three Progenitor Diploid Species. Genome 2015, 58, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Larson, S.; Pearson, C.; Jensen, K.; Jones, T.; Mott, I.; Robbins, M.; Staub, J.; Waldron, B. Development and testing of cool−season grass species, varieties and hybrids for biomass feedstock production in western North America. Agronomy 2017, 7, 3. [Google Scholar] [CrossRef]
- Robins, J.G. Cool-season grasses produce more total biomass across the growing season than do warm-season grasses when managed with an applied irrigation gradient. Biomass Bioenergy 2010, 34, 500–505. [Google Scholar] [CrossRef]
- Harmoney, K.R. Cool-season grass biomass in the southern mixed-grass prairie region of the USA. BioEnergy Res. 2015, 8, 203–210. [Google Scholar] [CrossRef]
- Wang, G.J.; Nyren, P.; Xue, Q.W.; Aberle, E.; Eriksmoen, E.; Tjelde, T.; Liebig, M.; Nichols, K.; Nyren, A. Establishment and yield of perennial grass monocultures and binary mixtures for bioenergy in North Dakota. Agron. J. 2014, 106, 1605–1613. [Google Scholar] [CrossRef]
- Monono, E.M.; Nyren, P.E.; Berti, M.T.; Pryor, S.W. Variability in biomass yield, chemical composition, and ethanol potential of individual and mixed herbaceous biomass species grown in North Dakota. Ind. Crops Prod. 2013, 41, 331–339. [Google Scholar] [CrossRef]
- Lee, D.; Owens, V.N.; Boe, A.; Koo, B.C. Biomass and seed yields of big bluestem, switchgrass, and intermediate wheatgrass in response to manure and harvest timing at two topographic positions. GCB Bioenergy 2009, 1, 171–179. [Google Scholar] [CrossRef]
- DeHaan, L.R.; Van Tassel, D.L.; Anderson, J.A.; Asselin, S.R.; Barnes, R.; Baute, G.J.; Cattani, D.J.; Culman, S.W.; Dorn, K.M.; Hulke, B.S.; et al. A pipeline strategy for grain crop domestication. Crop Sci. 2016, 56, 917–930. [Google Scholar] [CrossRef]
- Cattani Doug, D.J. Selection of a perennial grain for seed productivity across years: Intermediate wheatgrass as a test species. Can. J. Plant Sci. 2017, 97, 516–524. [Google Scholar] [CrossRef]
- Jungers, J.M.; DeHaan, L.R.; Betts, K.J.; Sheaffer, C.C.; Wyse, D.L. Intermediate wheatgrass grain and forage yield responses to nitrogen fertilization. Agron. J. 2017, 109, 462–472. [Google Scholar] [CrossRef]
- Cox, T.S.; Glover, J.D.; van Tassel, D.L.; Cox, C.M.; DeHaan, L.R. Prospects for developing perennial grain crops. Bioscience 2006, 56, 649–659. [Google Scholar] [CrossRef]
- Zhang, X.; Larson, S.R.; Gao, L.; Teh, S.L.; DeHaan, L.R.; Fraser, M.; Sallam, A.; Kantarski, T.; Frels, K.; Poland, J.; et al. Uncovering the Genetic Architecture of Seed Weight and Size in Intermediate Wheatgrass through Linkage and Association Mapping. Plant Genome 2017. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.F.; Sallam, A.; Gao, L.L.; Kantarski, T.; Poland, J.; DeHaan, L.R.; Wyse, D.L.; Anderson, J.A. Establishment and optimization of genomic selection to accelerate the domestication and improvement of intermediate wheatgrass. Plant Genome 2016. [Google Scholar] [CrossRef] [PubMed]
- Larson, S.; DeHaan, L.; Poland, J.; Zhang, X.; Dorn, K.; Kantarski, T.; Anderson, J.; Schmutz, J.; Grimwood, J.; Jenkins, J.; et al. Genome mapping of quantitative trait loci (QTL) controlling domestication traits of intermediate wheatgrass (Thinopyrum intermedium). Theor. Appl. Genet. 2019, 132, 2325–2351. [Google Scholar] [CrossRef]
- Van Dijk, G.E.; Winkelhorst, G.D. Testing perennial ryegrass (Lolium perenne L.) as spaced plants in swards. Euphytica 1978, 27, 855–860. [Google Scholar] [CrossRef]
- Asay, K.H.; Jensen, K.B.; Horton, H.W.; Johnson, D.A.; Chatterton, N.J. Registration of ‘Roadcrest’ crested wheatgrass. Crop Sci. 1999, 39, 1535. [Google Scholar] [CrossRef]
- Peel, M.D.; Asay, K.H.; Waldron, B.L.; Jensen, K.B.; Robins, J.G.; Mott, I.W. ‘Don’, a diploid falcata alfalfa for western U.S. rangelands. J. Plant Regist. 2009, 3, 115–118. [Google Scholar] [CrossRef]
- Zadokst, J.C.; Chang, T.; Konzak, C.F. A decimal code for the growth stages of cereals. Weed Res. 1974, 14, 415–421. [Google Scholar] [CrossRef]
- Anonymous. Neutral Detergent Fiber in Feeds: Filter Bag Technique; Ankom Technology Corporation: Macedon, NY, USA, 2005. [Google Scholar]
- Anonymous. Vitro True Digestibility Using the DAISYII Incubator; Ankom Technology Corporation: Macedon, NY, USA, 2005. [Google Scholar]
- Anonymous. Acid Detergent Fiber in Feeds: Filter Bag Technique; Ankom Technology Corporation: Macedon, NY, USA, 2005. [Google Scholar]
- Anonymous. Method for Determining Acid Detergent Lignin in the DaisyII incubator; Ankom Technology Corporation: Macedon, NY, USA, 2005. [Google Scholar]
- Goering, H.K.; Van Soest, P.J. Forage fiber analysis (apparatus, reagents, procedures, and some applications). In ARS Agriculture Handbook; USDA, Ed.; US Govt. Printing Office: Washington, DC, USA, 1970; Volume 379. [Google Scholar]
- National Research Council. Nutrient Requirements of Beef Cattle: Seventh Revised Edition: Update 2000; The National Academies Press: Washington, DC, USA, 2000; p. 248. [Google Scholar]
- Saha, U.K.; Sonon, L.S.; Hancock, D.W.; Hill, N.S.; Stewart, L.; Heusner, G.L.; Kissel, D.E. Common terms used in animal feeding and nutrition. In University of Georgia Coop. Ext. Bulletin; University of Georgia: Athens, GA, USA, 2010; p. 19. [Google Scholar]
- Holland, J.B.; Nyquist, W.E.; Cervantes-Martínez, C.T. Estimating and interpreting heritability for plant breeding: An update. Plant Breed. Rev. 2010, 22, 9–112. [Google Scholar] [CrossRef]
- Holland, J.B. Estimating genotypic correlations and their standard errors using multivariate restricted maximum likelihood estimation with SAS proc MIXED. Crop Sci. 2006, 46, 642–654. [Google Scholar] [CrossRef]
- Van Ooijen, J.W. MapQLT 6, Software for the Mapping of Quantitative Trait Loci in Experimental Populations of Diploid Species; Kyazma, B.V., Ed.; Kyazma: Wageningen, The Netherlands, 2009. [Google Scholar]
- Van Ooijen, J.W. JoinMap 4, Software for the Calculation of Genetic Linkage Maps in Experimental Populations; Kyazma, B.V., Ed.; Kyazma: Wageningen, The Netherlands, 2006. [Google Scholar]
- Voorrips, R.E. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef] [PubMed]
- Hayward, M.D.; Vivero, J.L. Selection for yield in Lolium perenne. II. Performance of spaced plant selections under competitive conditions. Euphytica 1984, 33, 787–800. [Google Scholar] [CrossRef]
- Larson, S.R.; Jensen, K.B.; Robins, J.G.; Waldron, B.L. Genes and Quantitative Trait Loci Controlling Biomass Yield and Forage Quality Traits in Perennial Wildrye. Crop Sci. 2014, 54, 111–126. [Google Scholar] [CrossRef]
- Carlsson, G.; Huss-Danell, K. Nitrogen fixation in perennial forage legumes in the field. Plant Soil 2003, 253, 353–372. [Google Scholar] [CrossRef]
- Heichel, G.H.; Henjum, K.I. Dinitrogen fixation, nitrogen transfer, and productivity of forage legume−grass communities. Crop Sci. 1991, 31, 202–208. [Google Scholar] [CrossRef]
- Zuppinger-Dingley, D.; Flynn, D.F.B.; Brandl, H.; Schmid, B. Selection in monoculture vs. mixture alters plant metabolic fingerprints. J. Plant Ecol. 2015, 8, 549–557. [Google Scholar] [CrossRef]
- Zemenchik, R.A.; Albrecht, K.A.; Shaver, R.D. Improved Nutritive Value of Kura Clover—And Birdsfoot Trefoil–Grass Mixtures Compared with Grass Monocultures Contrib. of the Wisconsin Agric. Exp. Stn. Research was partially funded by Hatch Project no. 5168 and 3270. Agron. J. 2002, 94, 1131–1138. [Google Scholar] [CrossRef]
- Waldron, B.L.; Bingham, T.J.; Creech, J.E.; Peel, M.D.; Miller, R.L.; Jensen, K.B.; ZoBell, D.R.; Eun, J.S.; Heaton, K.; Snyder, D.L. Binary mixtures of alfalfa and birdsfoot trefoil with tall fescue: Herbage traits associated with the improved growth performance of beef steers. Grassl. Sci. 2019. [Google Scholar] [CrossRef]

| Trait Description | Trait Abbreviation | Units |
|---|---|---|
| Biomass | MASS | g plot−1 |
| Tiller length | TILE | cm |
| Tillers crown−1 | TICR | no. |
| Zadok’s maturity | ZAMA | 0−99 |
| Crude protein | CP | g kg−1 DM |
| Neutral detergent fiber | NDF | g kg−1 DM |
| Acid detergent fiber | ADF | g kg−1 DM |
| Acid detergent lignin | ADL | g kg−1 DM |
| In vitro true digestibility | IVTD | g kg−1 DM |
| Neutral detergent fiber digestibility | NDFD | g kg−1 NDF |
| Metabolizable energy | ME | Mcal kg−1 DM |
| Trait | MASS | TILE | TICR | ZAMA | CP | NDF | ADF | ADL | IVTD | NDFD | ME |
|---|---|---|---|---|---|---|---|---|---|---|---|
| MASS | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | |
| TILE | 0.73 | *** | *** | *** | *** | *** | *** | *** | *** | *** | |
| TICR | 0.91 | 0.74 | *** | *** | *** | *** | *** | *** | *** | *** | |
| ZAMA | 0.16 | 0.39 | 0.18 | *** | *** | *** | *** | *** | *** | *** | |
| CP | −0.18 | −0.21 | −0.19 | −0.26 | *** | *** | — | *** | *** | *** | |
| NDF | 0.54 | 0.51 | 0.61 | 0.14 | −0.58 | *** | *** | *** | *** | *** | |
| ADF | 0.56 | 0.55 | 0.61 | 0.19 | −0.65 | 0.94 | *** | *** | *** | *** | |
| ADL | 0.26 | 0.17 | 0.33 | −0.07 | −0.01 | 0.25 | 0.32 | *** | * | *** | |
| IVTD | −0.60 | −0.66 | −0.68 | −0.28 | 0.28 | −0.64 | −0.62 | −0.15 | *** | *** | |
| NDFD | −0.26 | −0.39 | −0.29 | −0.24 | −0.18 | 0.10 | 0.08 | 0.04 | 0.70 | *** | |
| ME | −0.59 | −0.59 | −0.66 | −0.18 | 0.33 | −0.68 | −0.74 | −0.37 | 0.87 | 0.49 |
| Spaced-Plants | Poly-Culture | Mono-Culture | Spaced-Plants vs. Polyculture | Spaced-Plants vs. Monoculture | Monoculture vs. Polyculture | ||||
|---|---|---|---|---|---|---|---|---|---|
| Trait | H | H | H | rP | rG | rP | rG | rP | rG |
| MASS | 0.73 ± 0.03 | 0.52 ± 0.07 | 0.61 ± 0.05 | 0.15 ± 0.03 | 0.37 ± 0.14 | 0.15 ± 0.03 | 0.30 ± 0.12 | 0.21 ± 0.03 | 0.48 ± 0.15 |
| TILE | 0.85 ± 0.02 | 0.84 ± 0.02 | 0.79 ± 0.02 | 0.42 ± 0.03 | 0.91 ± 0.04 | 0.32 ± 0.04 | 0.78 ± 0.06 | 0.34 ± 0.03 | 0.85 ± 0.05 |
| TICR | 0.53 ± 0.07 | 0.50 ± 0.08 | 0.64 ± 0.04 | 0.12 ± 0.03 | 0.59 ± 0.14 | 0.10 ± 0.03 | 0.47 ± 0.13 | 0.25 ± 0.03 | 0.62 ± 0.12 |
| ZAMA | 0.84 ± 0.02 | 0.87 ± 0.02 | 0.70 ± 0.03 | 0.58 ± 0.03 | 0.98 ± 0.02 | 0.50 ± 0.03 | 0.97 ± 0.03 | 0.41 ± 0.03 | 0.98 ± 0.04 |
| CP | 0.66 ± 0.04 | 0.73 ± 0.04 | 0.59 ± 0.07 | 0.23 ± 0.03 | 0.80 ± 0.08 | 0.11 ± 0.03 | 0.53 ± 0.12 | 0.16 ± 0.03 | 0.57 ± 0.11 |
| NDF | 0.82 ± 0.02 | 0.83 ± 0.03 | 0.69 ± 0.05 | 0.39 ± 0.03 | 0.93 ± 0.04 | 0.31 ± 0.04 | 0.92 ± 0.06 | 0.32 ± 0.03 | 0.80 ± 0.07 |
| ADF | 0.81 ± 0.02 | 0.81 ± 0.02 | 0.74 ± 0.04 | 0.36 ± 0.36 | 0.89 ± 0.05 | 0.32 ± 0.03 | 0.89 ± 0.06 | 0.30 ± 0.03 | 0.81 ± 0.07 |
| ADL | 0.62 ± 0.04 | 0.67 ± 0.04 | 0.46 ± 0.09 | 0.19 ± 0.03 | 0.94 ± 0.09 | 0.18 ± 0.03 | 1.00 ± 0.14 | 0.09 ± 0.03 | 0.66 ± 0.14 |
| IVTD | 0.81 ± 0.02 | 0.78 ± 0.03 | 0.72 ± 0.04 | 0.42 ± 0.03 | 1.01 ± 0.04 | 0.24 ± 0.04 | 0.87 ± 0.06 | 0.36 ± 0.03 | 0.90 ± 0.05 |
| NDFD | 0.79 ± 0.02 | 0.71 ± 0.04 | 0.75 ± 0.03 | 0.29 ± 0.03 | 0.94 ± 0.06 | 0.24 ± 0.04 | 0.82 ± 0.06 | 0.22 ± 0.03 | 0.65 ± 0.08 |
| ME | 0.82 ± 0.03 | 0.77 ± 0.03 | 0.78 ± 0.03 | 0.44 ± 0.03 | 0.97 ± 0.04 | 0.29 ± 0.04 | 0.84 ± 0.05 | 0.38 ± 0.03 | 0.94 ± 0.05 |
| Spaced-Plants | Polyculture | Monoculture | Across-Environments | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Trait | LG | LOD | cM | %R2 | LOD | cM | %R2 | LOD | cM | %R2 | LOD | cM | %R2 | Marker Interval |
| MASS | 1 | — | — | 5.8 ** | 21.99 | 13.0 | — | 21.99–22.99 | ||||||
| 6 | — | — | 5.1 ** | 99.75 | 11.6 | — | 99.75–101.75 | |||||||
| 10 | 5.4 ** | 86.04 | 11.4 | 3.8 * | 86.04 | 3.5 * | 129.41 | 6.5 ** | 86.04 | 14.4 | 62.85–146.27 | |||
| 11 | — | 4.0 * | 7.56 | 5.4 ** | 7.56 | 12.3 | — | 0.00–12.32 | ||||||
| 14 | 3.3 * | 72.46 | — | 5.1 ** | 72.46 | 11.7 | 4.3 * | 72.46 | 63.35–75.46 | |||||
| 15 | — | — | 5.9 ** | 111.89 | 13.4 | — | 111.89–114.89 | |||||||
| TILE | 10 | 5.9 ** | 86.04 | 13.6 | 5.5 ** | 69.07 | 12.8 | 4.6 * | 73.7 | 5.7 ** | 86.04 | 13.2 | 40.14–120.25 | |
| 11 | 4.4 * | 133.65 | 4.5 * | 107.80 | 3.5 * | 82.99 | 5.2 ** | 82.99 | 12.1 | 76.11–134.64 | ||||
| TICR | 1 | — | — | 5.1 ** | 21.99 | 11..8 | — | 21.97–22.99 | ||||||
| 8 | 5.2 ** | 234.70 | 11.8 | — | — | 4.7 * | 234.70 | 149.27–272.21 | ||||||
| ZAMA | 16 | 6.0 ** | 106.27 | — | — | 4.2 * | 107.53 | 79.07–129.89 | ||||||
| CP | 6 | 3.8 * | 157.71 | 6.5 ** | 133.95 | 15.0 | 4.1 * | 133.95 | 5.7 ** | 133.95 | 13.2 | 110.46–165.88 | ||
| NDF | 10 | 7.8 ** | 114.23 | 17.6 | 6.9 ** | 114.23 | 15.8 | 5.8 ** | 60.85 | 13.4 | 8.1 ** | 114.23 | 18.1 | 41.14–122.72 |
| 15 | — | — | 5.7 ** | 69.65 | 13.3 | — | 54.96–98.79 | |||||||
| 16 | 4.9 * | 79.07 | — | 7.8 ** | 107.58 | 17.6 | 5.4 ** | 79.07 | 12.6 | 41.09–157.11 | ||||
| ADF | 10 | 5.8 ** | 114.23 | 13.4 | 6.1 ** | 78.31 | 14.2 | 5.5 ** | 60.85 | 12.7 | 6.9 ** | 48.31 | 15.7 | 34.14–122.72 |
| 16 | 4.8 * | 70.47 | — | 7.6 ** | 109.4 | 17.3 | 5.1 | 79.07 | 11.9 | 40.09–133.9 | ||||
| IVTD | 10 | 5.4 ** | 83.29 | 12.6 | 6.1 ** | 82.73 | 14.1 | 8.7 ** | 78.31 | 19.5 | 7.3 ** | 78.31 | 16.6 | 53.83–121.72 |
| 14 | 3.8 * | 108.63 | 3.5 * | 90.65 | 5.2 ** | 68.06 | 12.2 | 5.0 ** | 108.63 | 11.6 | 62.35–123.98 | |||
| 18 | 4.0 * | 172.33 | 6.1 ** | 172.33 | 14.1 | — | 5.0 ** | 172.23 | 11.6 | 147.49–184.83 | ||||
| NDFD | 5 | 4.9 ** | 19.73 | 11.6 | 3.8 * | 0.0 | — | 4.1 * | 0.0 | 0.00–53.77 | ||||
| 9 | 6.6 ** | 67.87 | 15.2 | 8.3 ** | 79.28 | 18.7 | — | 7.3 ** | 79.28 | 16.6 | 56.76–124.52 | |||
| 10 | — | — | 5.3 ** | 66.52 | 12.4 | — | 51.08–86.72 | |||||||
| 11 | 5.6 ** | 106.04 | 13.0 | 4.6 * | 83.06 | — | 4.7 * | 106.04 | 79.49–113.73 | |||||
| 18 | 3.7 * | 172.33 | 4.9 ** | 177.28 | 11.4 | 3.8 * | 209.97 | 5.3 ** | 177.28 | 12.4 | 153.73–221.76 | |||
| Trait/LG † | |||||||
|---|---|---|---|---|---|---|---|
| Marker Description | MASS/10 | MASS/10 | MASS/10 | MASS/14 | MASS/14 | MASS/15 | NDFD/10 |
| Marker | TP513463 | TP301824 | TP799882 | TP524699 | TP529335 | TP880985 | TP301824 |
| Position (cM)/%R2 | 87.08/ 18.7 | 73.70/ 13.1 | 86.04/ 23.3 | 65.59/ 11.4 | 72.46/ 11.5 | 61.89/ 12.4 | 73.70/ 21.2 |
| Allele | hh, hk, kk | hh, hk, kk | hh, hk, kk | nn, np | nn, np | hh, hk, kk | hh, hk, kk |
| Env. effect | |||||||
| Spaced | 1598.5 A | 1597.6 A | 1599.8 A | 1609.8 A | 1610.5 A | 1581.9 A | 50.4 C |
| Polyculture | 383.8 B | 379.1 B | 381.8 B | 392.4 B | 390.9 B | 386.2 B | 52.2 B |
| Monoculture | 99.2 C | 97.6 C | 99.4 C | 105.0 C | 104.7 C | 97.9 C | 54.6 A |
| SEM | 15.2 | 17.4 | 17.6 | 16.5 | 17.1 | 18.8 | 0.17 |
| p-value | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 |
| Allele effect | |||||||
| ll, nn, or hh | 752.7 A | 627 C | 754.2 A | 735.2 A | 735.7 A | 640.2 B | 53.3 A |
| lm, np, or hk | 707.1 B | 706.5 B | 706.2 B | 669.6 B | 668.5 B | 709.9 A | 51.9 B |
| kk | 621.8 C | 740.8 A | 620.7 C | N/A | N/A | 715.9 A | 52.0 B |
| SEM | 15.1 | 17.4 | 17.5 | 15.4 | 15.8 | 18.6 | 0.17 |
| p-value | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | 0.0006 | <0.0001 |
| Env. × Allele effect | |||||||
| Spaced × ll, nn, or hh | 1739.2 A | 1481.3 C | 1744.6 A | 1674.7 A | 1686.9 A | 1461.9 B | 51.2 F |
| Spaced × lm, np, or hk | 1604.6 B | 1610.2 B | 1597.0 B | 1544.9 B | 1534.1 B | 1617.8 A | 49.9 G |
| Spaced × kk | 1451.6 C | 1701.3 A | 1458.0 C | N/A | N/A | 1666.1 A | 49.9 G |
| Poly × ll, nn, or hh | 407.5 D | 330.1 E | 408.1 D | 414.4 C | 407.4 C | 368.7 C | 52.6 D |
| Poly × lm, np, or hk | 406.7 D | 399.2 D | 409.3 D | 370.4 D | 374.5 C | 399.8 C | 52.1 E |
| Poly × kk | 337.4 E | 408 D | 328.1 E | N/A | N/A | 390.1 C | 51.9 E |
| Mono × ll, nn, or hh | 111.3 F | 69.7 F | 109.9 F | 116.4 E | 112.7 D | 90.1 D | 55.9 A |
| Mono × lm, np, or hk | 110 F | 110.1 F | 112.4 F | 93.6 E | 96.8 D | 112.2 D | 53.7 C |
| Mono × kk | 76.3 F | 113.1 F | 75.9 F | N/A | N/A | 91.4 D | 54.2 B |
| SEM | 18.8 | 23.6 | 24.1 | 19.4 | 20.6 | 26.4 | 0.22 |
| p-value | <0.0001 | 0.0008 | <0.0001 | 0.0004 | <0.0001 | 0.0007 | 0.0003 |
| Bonferroni p-value | 0.0031 | 0.0031 | 0.0031 | 0.0031 | 0.0031 | 0.0031 | 0.0026 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mortenson, J.S.; Waldron, B.L.; Larson, S.R.; Jensen, K.B.; DeHaan, L.R.; Peel, M.D.; Johnson, P.G.; Creech, J.E. Quantitative Trait Loci (QTL) for Forage Traits in Intermediate Wheatgrass When Grown as Spaced-Plants versus Monoculture and Polyculture Swards. Agronomy 2019, 9, 580. https://doi.org/10.3390/agronomy9100580
Mortenson JS, Waldron BL, Larson SR, Jensen KB, DeHaan LR, Peel MD, Johnson PG, Creech JE. Quantitative Trait Loci (QTL) for Forage Traits in Intermediate Wheatgrass When Grown as Spaced-Plants versus Monoculture and Polyculture Swards. Agronomy. 2019; 9(10):580. https://doi.org/10.3390/agronomy9100580
Chicago/Turabian StyleMortenson, John S., Blair L. Waldron, Steve R. Larson, Kevin B. Jensen, Lee R. DeHaan, Michael D. Peel, Paul G. Johnson, and J. Earl Creech. 2019. "Quantitative Trait Loci (QTL) for Forage Traits in Intermediate Wheatgrass When Grown as Spaced-Plants versus Monoculture and Polyculture Swards" Agronomy 9, no. 10: 580. https://doi.org/10.3390/agronomy9100580
APA StyleMortenson, J. S., Waldron, B. L., Larson, S. R., Jensen, K. B., DeHaan, L. R., Peel, M. D., Johnson, P. G., & Creech, J. E. (2019). Quantitative Trait Loci (QTL) for Forage Traits in Intermediate Wheatgrass When Grown as Spaced-Plants versus Monoculture and Polyculture Swards. Agronomy, 9(10), 580. https://doi.org/10.3390/agronomy9100580







