Barnyardgrass Root Recognition Behaviour for Rice Allelopathy
Abstract
1. Introduction
2. Results
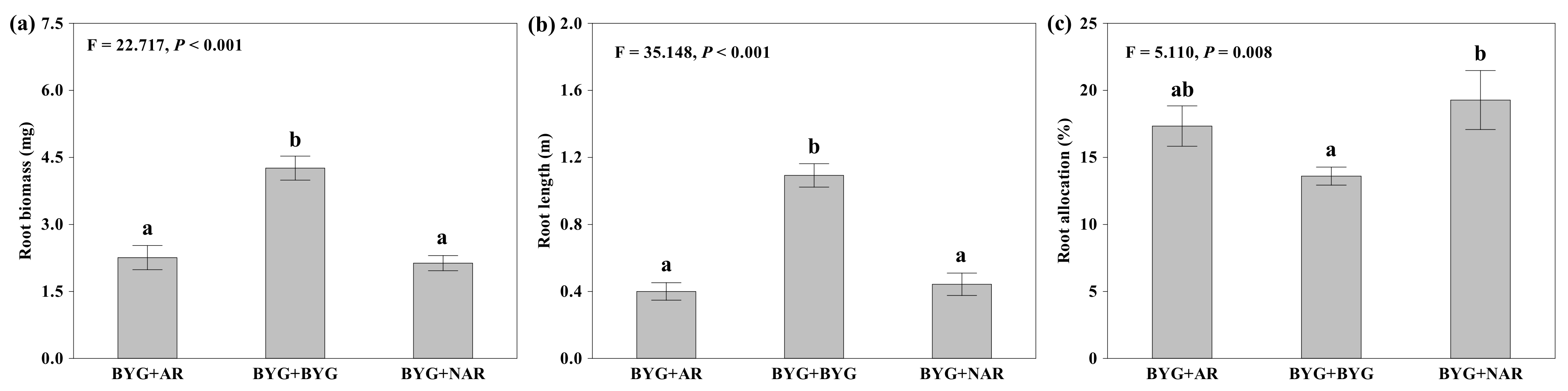
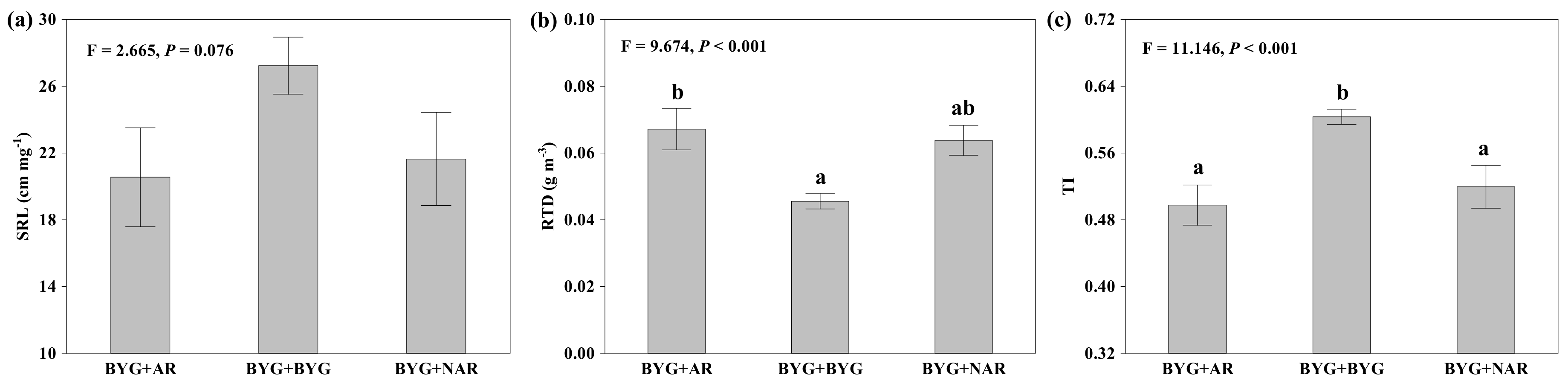
2.1. Root Proliferation and Morphological Traits
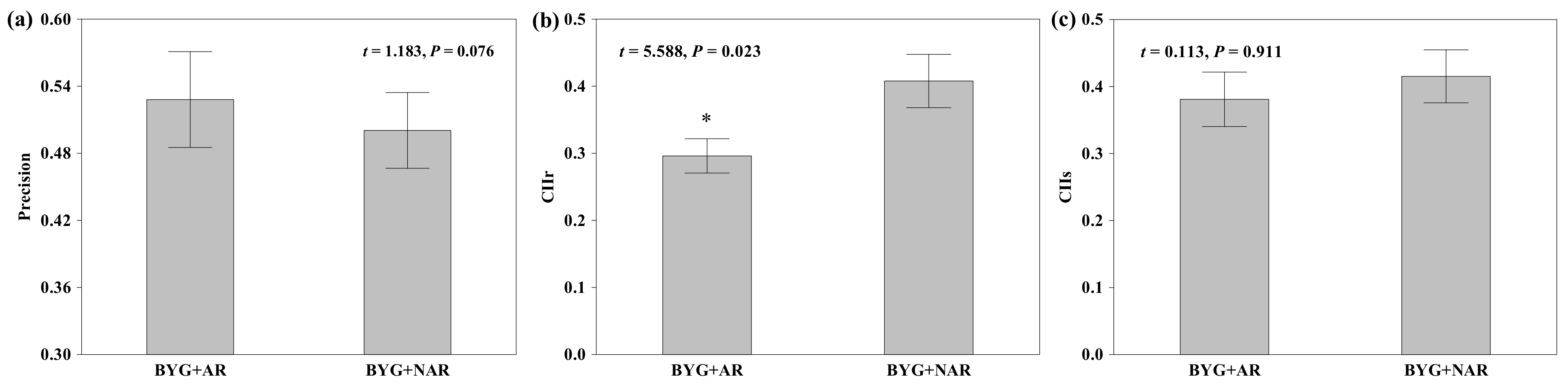
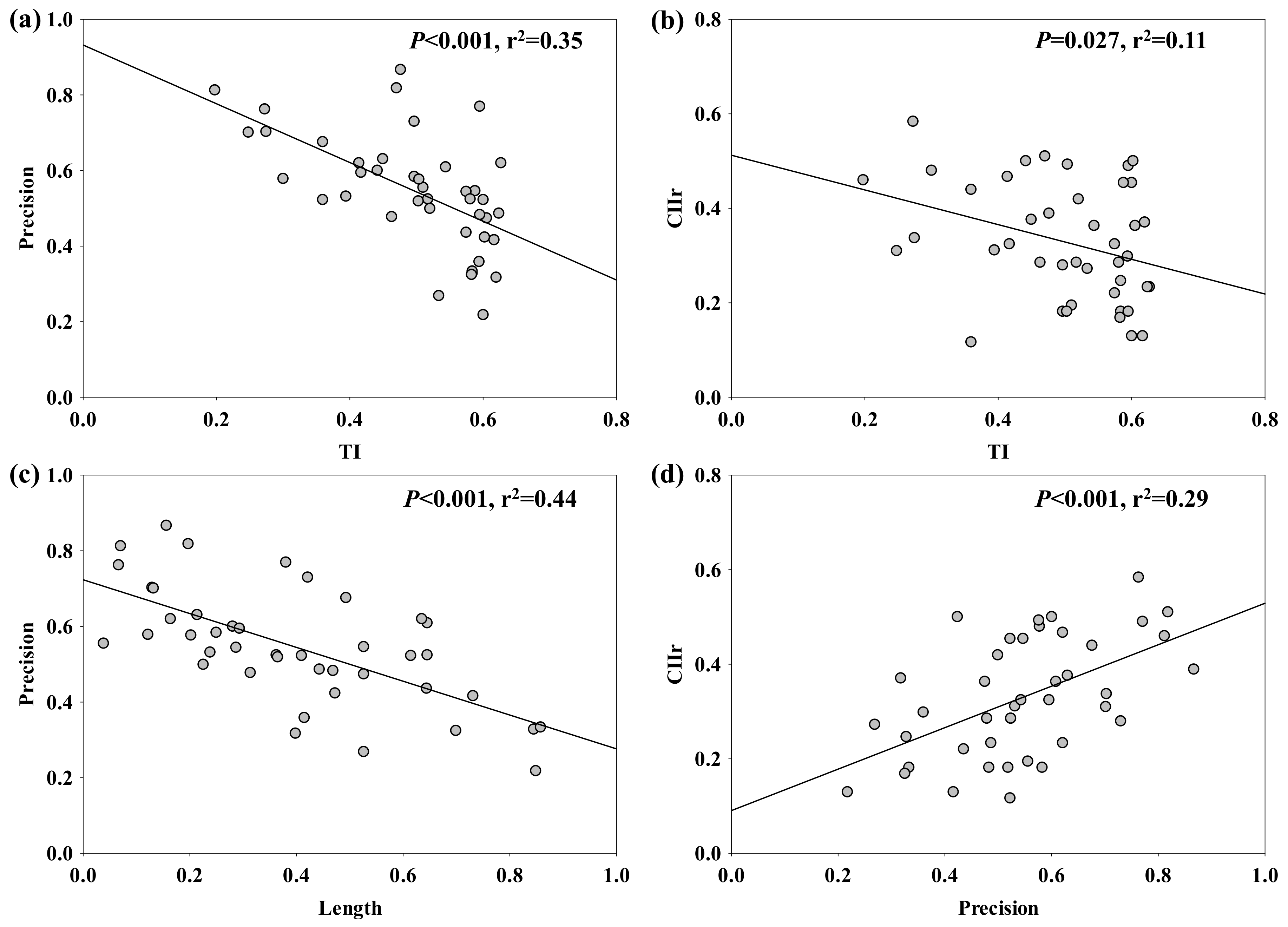
2.2. Root Behaviour of Nutrient Foraging and Competition
3. Discussion
3.1. Root Proliferation Response to Neighbouring Roots of Allelopathic Rice
3.2. Impact of Root Recognition on Nutrient Foraging and Competition
4. Materials and Methods
4.1. Plant and Soil Sample
4.2. Plant Growth, Sampling and Traits Calculating
4.3. Data Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Hutchings, M.J.; de Kroon, H. Foraging in plants: The role of morphological plasticity in resource acquisition. Adv. Ecol. Res. 1994, 25, 159–238. [Google Scholar] [CrossRef]
- Hodge, A. The plastic plant: Root responses to heterogeneous supplies of nutrients. New Phytol. 2004, 162, 9–24. [Google Scholar] [CrossRef]
- Mommer, L.; Visser, E.J.; van Ruijven, J.; de Caluwe, H.; Pierik, R.; de Kroon, H. Contrasting root behaviour in two grass species: A test of functionality in dynamic heterogeneous conditions. Plant Soil 2011, 344, 347–360. [Google Scholar] [CrossRef]
- Robinson, D. The responses of plants to non-uniform supplies of nutrients. New Phytol. 1994, 127, 635–674. [Google Scholar] [CrossRef]
- Dudley, S.A.; File, A.L. Kin recognition in an annual plant. Biol. Lett. 2007, 3, 435–438. [Google Scholar] [CrossRef] [PubMed]
- Murphy, G.P.; Dudley, S.A. Kin recognition: Competition and cooperation in Impatiens (Balsaminaceae). Am. J. Bot. 2009, 96, 1990–1996. [Google Scholar] [CrossRef] [PubMed]
- Depuydt, S. Arguments for and against self and non-self root recognition in plants. Front. Plant Sci. 2014, 5, 614. [Google Scholar] [CrossRef] [PubMed]
- Faget, M.; Nagel, K.A.; Walter, A.; Herrera, J.M.; Jahnke, S.; Schurr, U.; Temperton, V.M. Root–root interactions: Extending our perspective to be more inclusive of the range of theories in ecology and agriculture using in-vivo analyses. Ann. Bot. 2013, 112, 253–266. [Google Scholar] [CrossRef] [PubMed]
- Maina, G.G.; Brown, J.S.; Gersani, M. Intra-plant versus inter-plant root competition in beans: Avoidance, resource matching or tragedy of the commons. Plant Ecol. 2002, 160, 235–247. [Google Scholar] [CrossRef]
- Gersani, M.; Brown, J.S.; O’Brien, E.E.; Maina, G.M.; Abramsky, Z. Tragedy of the commons as a result of root competition. J. Ecol. 2001, 89, 660–669. [Google Scholar] [CrossRef]
- Falik, O.; de Kroon, H.; Novoplansky, A. Physiologically-mediated self/non self root discrimination in Trifolium repens has mixed effects on plant performance. Plant Signal. Behav. 2006, 1, 116–121. [Google Scholar] [CrossRef] [PubMed]
- Semchenko, M.; Hutchings, M.J.; John, E.A. Challenging the tragedy of the commons in root competition: Confounding effects of neighbour presence and substrate volume. J. Ecol. 2007, 95, 252–260. [Google Scholar] [CrossRef]
- Markham, J.; Halwas, S. Effect of neighbour presence and soil volume on the growth of Andropogon gerardii Vitman. Plant Ecol. Divers. 2011, 4, 265–268. [Google Scholar] [CrossRef]
- De Kroon, H. How do roots interact? Science 2007, 318, 1562–1563. [Google Scholar] [CrossRef] [PubMed]
- Semchenko, M.; John, E.A.; Hutchings, M.J. Effects of physical connection and genetic identity of neighbouring ramets on root-placement patterns in two clonal species. New Phytol. 2007, 176, 644–654. [Google Scholar] [CrossRef] [PubMed]
- Semchenko, M.; Saar, S.; Lepik, A. Plant root exudates mediate neighbor recognition and trigger complex behavioural changes. New Phytol. 2014, 204, 631–637. [Google Scholar] [CrossRef] [PubMed]
- Schenk, H.J.; Callaway, R.M.; Mahall, B.E. Spatial root segregation: Are plants territorial? Adv. Ecol. Res. 1999, 28, 145–180. [Google Scholar] [CrossRef]
- Falik, O.; Reides, P.; Gersani, M.; Novoplansky, A. Self/non-self discrimination in roots. J. Ecol. 2003, 91, 525–531. [Google Scholar] [CrossRef]
- Caffaro, M.M.; Vivanco, J.M.; Botto, J.; Rubio, G. Root architecture of Arabidopsis, is affected by competition with neighbouring plants. Plant Growth Regul. 2013, 70, 141–147. [Google Scholar] [CrossRef]
- Fang, S.; Clark, R.T.; Zheng, Y.; Iyer-Pascuzzi, A.S.; Weitz, J.S.; Kochian, L.V.; Edelsbrunner, H.; Liao, H.; Benfey, P.N. Genotypic recognition and spatial responses by rice roots. Proc. Natl. Acad. Sci. USA 2013, 110, 2670–2675. [Google Scholar] [CrossRef] [PubMed]
- Bais, H.P.; Weir, T.L.; Perry, L.G.; Gilroy, S.; Vivanco, J.M. The role of root exudates in rhizosphere interactions with plants and other organisms. Annu. Rev. Plant Biol. 2006, 57, 233–266. [Google Scholar] [CrossRef] [PubMed]
- Metlen, K.L.; Aschehoug, E.T.; Callaway, R.M. Plant behavioural ecology: Dynamic plasticity in secondary metabolites. Plan Cell Environ. 2009, 32, 641–653. [Google Scholar] [CrossRef] [PubMed]
- Kato-Noguchi, H. The chemical cross talk between rice and barnyardgrass. Plant Signal. Behav. 2011, 6, 1207–1209. [Google Scholar] [CrossRef][Green Version]
- Kong, C.H.; Liang, W.J.; Xu, X.H.; Hu, F.; Wang, P. Release and activity of allelochemicals from allelopathic rice seedlings. J. Agric. Food Chem. 2004, 52, 2861–2865. [Google Scholar] [CrossRef] [PubMed]
- Farooq, M.; Jabran, K.; Cheema, Z.A.; Wahid, A.; Siddique, K.H.M. The role of allelopathy in agricultural pest management. Pest Manag. Sci. 2011, 67, 493–506. [Google Scholar] [CrossRef] [PubMed]
- Kong, C.H.; Wang, P.; Zhao, H.; Xu, X.H.; Zhu, Y.D. Impact of allelochemical exuded from allelopathic rice on soil microbial community. Soil Biol. Biochem. 2008, 40, 1862–1869. [Google Scholar] [CrossRef]
- Kato-Noguchi, H.; Ino, T. Rice seedlings release momilactone B into the environment. Phytochemistry 2003, 63, 551–554. [Google Scholar] [CrossRef]
- Khanh, T.D.; Xuan, T.D.; Chung, I.M. Rice allelopathy and the possibility for weed management. Ann. Appl. Biol. 2007, 151, 325–339. [Google Scholar] [CrossRef]
- Xuan, T.D.; Chung, M.I.; Khanh, T.D.; Tawata, S. Identification of phytotoxic substances from early growth of barnyard grass (Echinochloa crusgalli) root exudates. J. Chem. Ecol. 2006, 32, 895–906. [Google Scholar] [CrossRef] [PubMed]
- Bajwa, A.A.; Jabran, K.; Shahid, M.; Ali, H.H.; Chauhan, B.S. Eco-biology and management of Echinochloa crus-galli. Crop Protect. 2015, 75, 151–162. [Google Scholar] [CrossRef]
- Beckie, H.J. Herbicide-resistant weeds: Management tactics and practices. Weed Technol. 2006, 20, 793–814. [Google Scholar] [CrossRef]
- Sun, B.; Wang, P.; Kong, C.H. Plant-soil feedback in the interference of allelopathic rice with barnyardgrass. Plant Soil 2014, 377, 309–321. [Google Scholar] [CrossRef]
- Yang, X.F.; Kong, C.H.; Yang, X.; Li, Y.F. Interference of allelopathic rice with penoxsulam-resistant barnyardgrass. Pest. Manag. Sci. 2017, 73, 2310–2317. [Google Scholar] [CrossRef] [PubMed]
- Biedrzycki, M.L.; Jilany, T.A.; Dudley, S.A.; Bais, H.P. Root exudates mediate kin recognition in plants. Commun. Integr. Biol. 2010, 3, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Jacob, C.E.; Tozzi, E.; Willenborg, C.J. Neighbour presence, not identity, influences root and shoot allocation in pea. PLoS ONE 2017, 12, e0173758. [Google Scholar] [CrossRef] [PubMed]
- Lamb, E.G.; Kembel, S.W.; Cahill, J.F. Shoot, but not root, competition reduces community diversity in experimental mesocosms. J. Ecol. 2009, 97, 155–163. [Google Scholar] [CrossRef]
- Kato-Noguchi, H. Barnyard grass-induced rice allelopathy and momilactone B. J. Plant Physiol. 2011, 168, 1016–1020. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.F.; Kong, C.H. Interference of allelopathic rice with paddy weeds at the root level. Plant Biol. 2017, 19, 584–591. [Google Scholar] [CrossRef] [PubMed]
- Birouste, M.; Zamora-Ledezma, E.; Bossard, C.; Pérez-Ramos, I.M.; Roumet, C. Measurement of fine root tissue density: A comparison of three methods reveals the potential of root dry matter content. Plant Soil 2014, 374, 299–314. [Google Scholar] [CrossRef]
- Westoby, M. A leaf–height–seed (LHS) plant ecology strategy scheme. Plant Soil 1998, 199, 213–227. [Google Scholar] [CrossRef]
- Craine, J.M.; Froehle, J.; Tilman, D.G.; Wedin, D.A.; Chapin, F.S., III. The relationships among root and leaf traits of 76 grassland species and relative abundance along fertility and disturbance gradients. Oikos 2001, 93, 274–285. [Google Scholar] [CrossRef]
- Pierik, R.; Visser, E.J.W.; de Kroon, H.; Voesenek, L.A.C.J. Ethylene is required in tobacco to successfully compete with proximate neighbours. Plant Cell Environ. 2003, 26, 1229–1234. [Google Scholar] [CrossRef]
- Kapulnik, Y.; Koltai, H. Strigolactone involvement in root development, response to abiotic stress, and interactions with the biotic soil environment. Plant Physiol. 2014, 166, 560–569. [Google Scholar] [CrossRef] [PubMed]
- You, L.X.; Wang, P.; Kong, C.H. The levels of jasmonic acid and salicylic acid in a rice-barnyardgrass coexistence system and their relation to rice allelochemicals. Biochem. Syst. Ecol. 2011, 39, 491–497. [Google Scholar] [CrossRef]
- Heidarzade, A.; Esmaeili, M.; Pirdashti, H. Common allelochemicals in root exudates of Barnyardgrass (Echinochloa crusgalli L.) and inhibitory potential against rice (Oryza sativa) cultivars. Int. Res. J. Appl. Basic Sci. 2012, 3, 11–17. [Google Scholar]
- Kato-Noguchi, H.; Ino, T. The chemical-mediated allelopathic interaction between rice and barnyard grass. Plant Soil 2013, 370, 267–275. [Google Scholar] [CrossRef]
- Kato-Noguchi, H.; Peters, R.J. The role of momilactones in rice allelopathy. J. Chem. Ecol. 2013, 39, 175–185. [Google Scholar] [CrossRef] [PubMed]
- Bertin, C.; Yang, X.; Weston, L.A. The role of root exudates and allelochemicals in the rhizosphere. Plant Soil 2003, 256, 67–83. [Google Scholar] [CrossRef]
- Wang, P.; Zhang, X.; Kong, C.H. The response of allelopathic rice growth and microbial feedback to barnyardgrass infestation in a paddy field experiment. Eur. J. Soil Biol. 2013, 56, 26–32. [Google Scholar] [CrossRef]
- Fitter, A.H. An architectural approach to the comparative ecology of plant-root systems. New Phytol. 1987, 106, 61–77. [Google Scholar] [CrossRef]
- Fort, F.; Cruz, P.; Jouany, C.; Field, K. Hierarchy of root functional trait values and plasticity drive early-stage competition for water and phosphorus among grasses. Funct. Ecol. 2014, 28, 1030–1040. [Google Scholar] [CrossRef]
| Root Traits | Species | Treatment | S × T | Allelopathy | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| df | F-Value | p-Value | df | F-Value | p-Value | df | F-Value | p-Value | t | df | p-Value | |
| Root biomass (mg plant−1) | 4, 8 | 4.707 | 0.029 | 2, 8 | 93.23 | <0.001 | 8, 52 | 0.826 | 0.584 | −4.967 | 40 | <0.001 |
| Root allocation (%) | 4, 8 | 1.73 | 0.235 | 2, 8 | 9.374 | 0.003 | 8, 52 | 1.849 | 0.089 | 2.061 | 40 | 0.039 |
| Root length (m) | 4, 8 | 3.956 | 0.046 | 2, 8 | 57.604 | <0.001 | 8, 52 | 1.172 | 0.334 | −0.506 | 40 | 0.615 |
| Specific root length (cm mg−1) | 4, 8 | 2.417 | 0.133 | 2, 8 | 7.291 | 0.015 | 8, 52 | 0.853 | 0.562 | −3.254 | 40 | 0.002 |
| Root tissue density (mg cm−3) | 4, 8 | 3.166 | 0.076 | 2, 8 | 17.142 | 0.001 | 8, 52 | 0.762 | 0.637 | 2.289 | 40 | 0.027 |
| Topological index | 4, 8 | 0.471 | 0.756 | 2, 8 | 11.731 | 0.004 | 8, 52 | 4.714 | <0.001 | 3.776 | 40 | 0.001 |
| Individual Root Characteristics | Abbreviation | Definition |
|---|---|---|
| Root biomass | RM | Total root dry biomass (mg plant−1) |
| Root allocation | Ra | Ratio of root mass to total mass (%) |
| Root length | Length | Total root length (m plant−1) |
| Root tips | Tips | The number of root tips |
| Specific root length | SRL | Length per unit root dry biomass (m g−1) |
| Root tissue density | RTD | Root dry biomass per unit root volume (mg cm−3) |
| Nutrient foraging behaviour characteristics | Symbol | Definition |
| Root foraging precision | Precision | Precision was measured by 1-RMt/RMc, where RMt is the root biomass of barnyardgrass in a mixed culture pot, and RMc is the corresponding value for barnyardgrass in a monoculture seedling pots(control). Greater ratios are considered to represent higher precision |
| Competition intensity index | CIIs | CIIs = (Bshoot-c − Bshoot-t)/X, where Bshoot-c is the shoot dried biomass in monoculture seedling pots (control), Bshoot-t is the mean shoot dried biomass in mixed culture seedling pots (treatment), and X is the maximum of these two values. |
| CIIr | CIIr = (Broot-c – Broot-t)/X, for evaluating root competition | |
| Topological characteristics | Symbol | Definition |
| Magnitude 1 | μ | The number of root tips (exterior links) in the system |
| External path length 1 | Pe | The sum of the number of links in all paths from each external link to the base link |
| Topological index 1 | TI | A way to quantify branching pattern using the relationship between log10 external path length and log10 magnitude, TI = log10(Pe)/log10(μ) |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, T.; Fan, B.; Wang, P. Barnyardgrass Root Recognition Behaviour for Rice Allelopathy. Agronomy 2018, 8, 39. https://doi.org/10.3390/agronomy8040039
Zhang T, Fan B, Wang P. Barnyardgrass Root Recognition Behaviour for Rice Allelopathy. Agronomy. 2018; 8(4):39. https://doi.org/10.3390/agronomy8040039
Chicago/Turabian StyleZhang, Tingshuang, Bo Fan, and Peng Wang. 2018. "Barnyardgrass Root Recognition Behaviour for Rice Allelopathy" Agronomy 8, no. 4: 39. https://doi.org/10.3390/agronomy8040039
APA StyleZhang, T., Fan, B., & Wang, P. (2018). Barnyardgrass Root Recognition Behaviour for Rice Allelopathy. Agronomy, 8(4), 39. https://doi.org/10.3390/agronomy8040039






