Genome-Wide Identification and Expression Profiling Analysis of the Galactinol Synthase Gene Family in Cassava (Manihot esculenta Crantz)
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Treatments
2.2. Identification, Isolation, and Analysis of MeGolS Family Genes
2.3. Chromosomal Localization of MeGolS Family Genes
2.4. Prediction of MeGolS Gene Structures and the Conserved Motifs
2.5. Cis-Element Analysis within the MeGolS Family Gene Promoters
2.6. The Expression Profiles of MeGolS Genes in Cassava
2.7. MeGolS Gene Expression Profiles under Drought, Cold, and Salt Stresses
2.8. Prediction of microRNAs Targeting to MeGolS Genes
3. Results
3.1. Identification and Cloning of MeGolS Family Genes in Cassava
3.2. Chromosomal Location and Distribution of MeGolS Family Genes
3.3. Analysis of Phylogenesis, Conserved Motifs, and Gene Structures of GolS Family Genes in Cassava and Arabidopsis
3.4. The Cis-Element Analysis within MeGolS Family Gene Promoters
3.5. Tissue-Specific Expression of MeGolS in Cassava
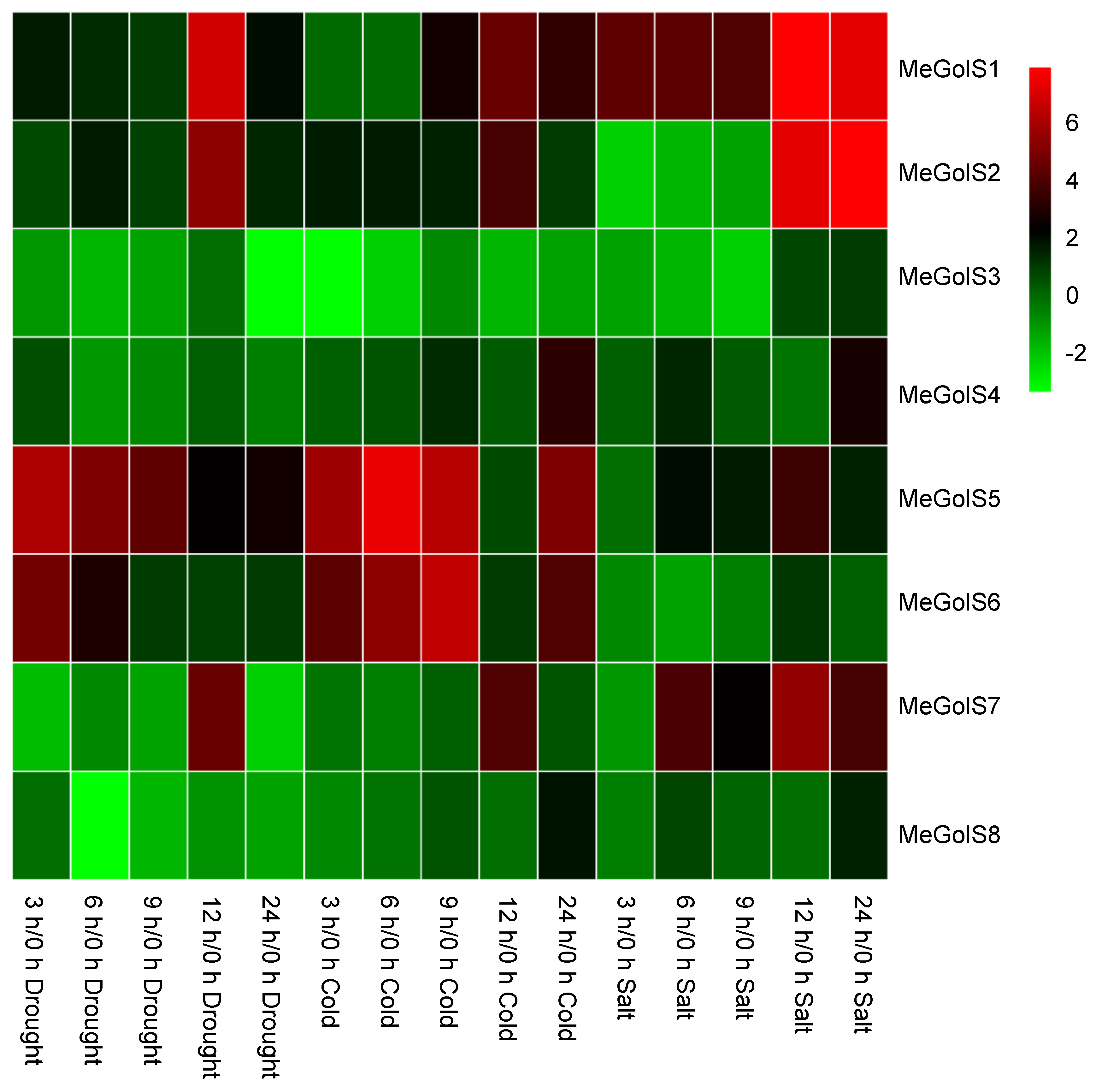
3.6. The Inducible Expressions of MeGolSs under Abiotic Stress
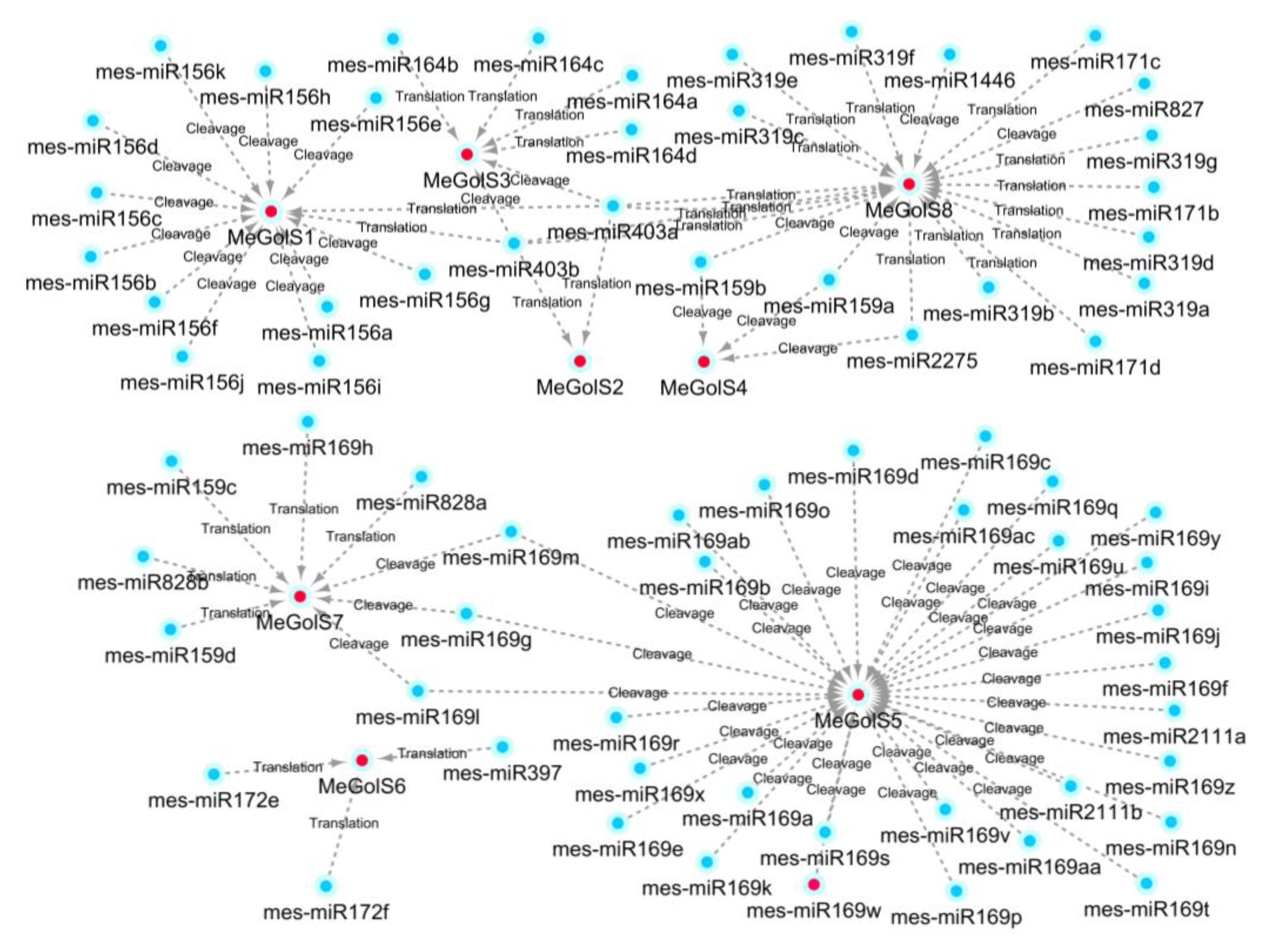
3.7. Prediction of MicroRNA Targeting MeGolS Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pluskota, W.E.; Szablińska, J.; Obendorf, R.L.; Górecki, R.J.; Lahuta, L.B. Osmotic stress induces genes, enzymes and accumulation of galactinol, raffinose and stachyose in seedlings of pea (Pisum sativum, L.). Acta Physiol. Plant. 2015, 37, 156. [Google Scholar] [CrossRef]
- Meyer, T.; Vigouroux, A.; Aumont-Niçaise, M.; Comte, G.; Vial, L.; Lavire, C.; Moréra, S. The plant defense signal galactinol is specifically used as a nutrient by the bacterial pathogen Agrobacterium fabrum. J. Biol. Chem. 2018, 293, 7930–7941. [Google Scholar] [CrossRef] [PubMed]
- Unda, F.; Canam, T.; Preston, L.; Mansfield, S.D. Isolation and characterization of galactinol synthases from hybrid poplar. J. Exp. Bot. 2012, 63, 2059–2069. [Google Scholar] [CrossRef] [PubMed]
- Sengupta, S.; Mukherjee, S.; Basak, P.; Majumder, A.L. Significance of galactinol and raffinose family oligosaccharide synthesis in plants. Front. Plant Sci. 2015, 6, 656. [Google Scholar] [CrossRef] [PubMed]
- De Souza Vidigal, D.; Willems, L.; van Arkel, J.; Dekkers, B.J.; Hilhorst, H.W.; Bentsink, L. Galactinol as marker for seed longevity. Plant Sci. 2016, 246, 112–118. [Google Scholar] [CrossRef] [PubMed]
- Pereira Lima, J.J.; Buitink, J.; Lalanne, D.; Rossi, R.F.; Pelletier, S.; da Silva, E.A.A.; Leprince, O. Molecular characterization of the acquisition of longevity during seed maturation in soybean. PLoS ONE 2017, 12, e0180282. [Google Scholar] [CrossRef] [PubMed]
- Roman, G.; Raimund, T. Raffinose family oligosaccharides act as galactose stores in seeds and are required for rapid germination ofarabidopsisin the dark. Front. Plant Sci. 2016, 7, 111. [Google Scholar]
- Kim, M.S.; Cho, S.M.; Kang, E.Y.; Im, Y.J.; Hwangbo, H.; Kim, Y.C.; Ryu, C.M.; Yang, K.Y.; Chung, G.C.; Cho, B.H. Galactinol is a signaling component of the induced systemic resistance caused by Pseudomonas chlororaphis O6 root colonization. Mol. Plant-Microbe Interact. 2008, 21, 1643–1653. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Song, X.; Bartels, D. Sugar metabolism in the desiccation tolerant grass Oropetium thomaeum in response to environmental stresses. Plant Sci. 2018, 270, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Song, C.; Chung, W.S.; Lim, C.O. Overexpression of heat shock factor gene HsfA3 increases galactinol levels and oxidative stress tolerance in Arabidopsis. Mol. Cells 2016, 39, 477–483. [Google Scholar] [CrossRef] [PubMed]
- Taji, T.; Ohsumi, C.; Iuchi, S.; Seki, M.; Kasuga, M.; Kobayashi, M.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Important roles of drought- and cold-inducible genes for galactinol synthase in stress tolerance in Arabidopsis thaliana. Plant J. 2002, 29, 417–426. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Yu, M.; Liu, M.; Zhang, R.; Sun, W.; Qian, M.; Duan, H.; Chang, W.; Ma, J.; Qu, C.; et al. Genome-wide identification, evolutionary and expression analyses of the galactinol synthase gene family in rapeseed and tobacco. Int. J. Mol. Sci. 2017, 18, 2768. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.Y.; Thacker, R.; Iii, J.W.C.; Snyder, J.C.; Meeley, R.B.; Obendorf, R.L.; Downie, B. Expression of the maize galactinol synthase gene family: (i) expression of two different genes during seed development and germination. Physiol. Plant. 2010, 121, 634–646. [Google Scholar] [CrossRef]
- Zhou, Y.; Liu, Y.; Wang, S.; Shi, C.; Zhang, R.; Rao, J.; Wang, X.; Gu, X.; Wang, Y.; Li, D.; et al. Molecular cloning and characterization of galactinol synthases in Camellia sinensis with different responses to biotic and abiotic stressors. J. Agric. Food Chem. 2017, 65, 2751. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, H.; Wang, S.; Li, H.; Xin, Q. Overexpression of a common wheat gene galactinol synthase3, enhances tolerance to zinc in Arabidopsis, and rice through the modulation of reactive oxygen species production. Plant Mol. Biol. Report. 2015, 34, 794–806. [Google Scholar] [CrossRef]
- Filiz, E.; Ozyigit, I.I.; Vatansever, R. Genome-wide identification of galactinol synthase (GolS) genes in Solanum lycopersicum and Brachypodium distachyon. Comput. Biol. Chem. 2015, 58, 149–157. [Google Scholar] [CrossRef] [PubMed]
- Salvi, P.; Kamble, N.U.; Majee, M. Stress inducible galactinol synthase of chickpea (CaGolS) implicates in heat and oxidative stress tolerance through reducing stress induced excessive reactive oxygen species accumulation. Plant Cell Physiol. 2018, 59, 155–166. [Google Scholar] [CrossRef] [PubMed]
- Nishizawa, A.; Yabuta, Y.; Yoshida, E.; Maruta, T.; Yoshimura, K.; Shigeoka, S. Arabidopsis heat shock transcription factor A2 as a key regulator in response to several types of environmental stress. Plant J. 2010, 48, 535–547. [Google Scholar] [CrossRef] [PubMed]
- Panikulangara, T.J.; Eggersschumacher, G.; Wunderlich, M.; Stransky, H.; Schöffl, F. Galactinol synthase1. A novel heat shock factor target gene responsible for heat-induced synthesis of raffinose family oligosaccharides in Arabidopsis. Plant Physiol. 2004, 136, 3148–3158. [Google Scholar] [CrossRef] [PubMed]
- Nishizawayokoi, A.; Yoshida, E.; Yabuta, Y.; Shigeoka, S. Analysis of the regulation of target genes by an Arabidopsis heat shock transcription factor, HsfA2. J. Agric. Chem. Soc. Jpn. 2009, 73, 890–895. [Google Scholar]
- Selvaraj, M.G.; Ishizaki, T.; Valencia, M.; Ogawa, S.; Dedicova, B.; Ogata, T.; Yoshiwara, K.; Maruyama, K.; Kusano, M.; Saito, K.; et al. Overexpression of an Arabidopsis thaliana galactinol synthase gene improves drought tolerance in transgenic rice and increased grain yield in the field. Plant Biotechnol. J. 2017, 15, 1465–1477. [Google Scholar] [CrossRef] [PubMed]
- Shimosaka, E.; Ozawa, K. Overexpression of cold-inducible wheat galactinol synthase confers tolerance to chilling stress in transgenic rice. Breed. Sci. 2015, 65, 363–371. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhang, L.; Chen, L.; Ma, H.; Ruan, Y.; Xu, T.; Xu, C.Q.; He, Y.; Qi, M.F. Molecular cloning and expression of an encoding galactinol synthase gene (AnGolS1) in seedling of Ammopiptanthus nanus. Sci. Rep. 2016, 6, 36113. [Google Scholar] [CrossRef] [PubMed]
- Utsumi, Y.; Tanaka, M.; Morosawa, T.; Kurotani, A.; Yoshida, T.; Mochida, K.; Matsui, A.; Umemura, Y.; Ishitani, M.; Shinozaki, K.; et al. Transcriptome analysis using a high-density oligomicroarray under drought stress in various genotypes of cassava: An important tropical crop. DNA Res. 2012, 19, 335–345. [Google Scholar] [CrossRef] [PubMed]
- El-Sharkawy, M.A. Drought-tolerant cassava for Africa, Asia, and Latin America. Bioscience 1993, 43, 441–451. [Google Scholar] [CrossRef]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018, 46, 493–496. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools in the ExPASy Server. In The Proteomics Protocols Handbook; John, M.W., Ed.; Humana Press: New York, NY, USA, 2005; pp. 571–607. [Google Scholar]
- Yu, C.S.; Cheng, C.W.; Su, W.C.; Chang, K.C.; Huang, S.W.; Hwang, J.K.; Lu, C.H. CELLO2GO: A web server for protein subcellular localization prediction with functional gene ontology annotation. PLoS ONE 2014, 9, e99368. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2014, 31, 1296. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. Meme suite: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Geislerlee, J.; Geisler, M.; Coutinho, P.M.; Segerman, B.; Nishikubo, N.; Takahashi, J.; Aspeborg, H.; Djerbi, S.; Master, E.; Andersson-Gunneras, S. Poplar carbohydrate-active enzymes. Gene identification and expression analyses. Plant Physiol. 2006, 140, 946–962. [Google Scholar] [CrossRef] [PubMed]
- You, J.; Wang, Y.; Zhang, Y.; Dossa, K.; Li, D.; Rong, Z.; Wang, L.; Zhang, X. Genome-wide identification and expression analyses of genes involved in raffinose accumulation in sesame. Sci. Rep. 2018, 8, 4331. [Google Scholar] [CrossRef] [PubMed]
- Santos, T.B.D.; de Lima, R.B.; Nagashima, G.T.; Petkowicz, C.L.; Carpentieri-pípolo, V.; Pereira, L.F.P.; Domingues, D.S.; Vieira, L.G.E. Galactinol synthase transcriptional profile in two genotypes of Coffea canephora with contrasting tolerance to drought. Genet. Mol. Biol. 2015, 38, 182–190. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Zhang, R.; Guo, S. Molecular cloning and characterization of ghgols1, a novel gene encoding galactinol synthase from cotton (Gossypium hirsutum). Plant Mol. Biol. Report. 2012, 30, 699–709. [Google Scholar] [CrossRef]
- Downie, B.; Gurusinghe, S.; Dahal, P.; Thacker, R.R.; Snyder, J.C.; Nonogaki, H. Expression of a galactinol synthase gene in tomato seeds is up-regulated before maturation desiccation and again after imbibition whenever radicle protrusion is prevented. Plant Physiol. 2003, 131, 1347–1359. [Google Scholar] [CrossRef] [PubMed]
- Lahuta, L.B.; Pluskota, W.E.; Stelmaszewska, J.; Szablińska, J. Dehydration induces expression of galactinol synthase and raffinose synthase in seedlings of pea (Pisum sativum L.). J. Plant Physiol. 2014, 171, 1306–1314. [Google Scholar] [CrossRef] [PubMed]
- Nishizawa, A.; Yabuta, Y.; Shigeoka, S. The contribution of carbohydrates including raffinose family oligosaccharides and sugar alcohols to protection of plant cells from oxidative damage. Plant Signal. Behav. 2008, 3, 1016–1018. [Google Scholar] [CrossRef]
- Lahuta, L.B.; Górecki, R.J. Raffinose in seedlings of winter vetch (Vicia villosa, Roth.) under osmotic stress and followed by recovery. Acta Physiol. Plant. 2011, 33, 725–733. [Google Scholar] [CrossRef]
- Da Maia, L.C.; Cadore, P.R.; Benitez, L.C.; Danielowski, R.; Braga, E.J.; Fagundes, P.R.; Magalhães, A.M.; de Oliveira, A.C. Transcriptome profiling of rice seedlings under cold stress. Funct. Plant Biol. 2017, 44, 419–429. [Google Scholar] [CrossRef]
- Kumar, M.; Gho, Y.S.; Jung, K.H.; Kim, S.R. Genome-wide identification and analysis of genes, conserved between japonica and indica rice cultivars, that respond to low-temperature stress at the vegetative growth stage. Front. Plant Sci. 2017, 8, 1120. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Huang, X.S. Deep sequencing-based characterization of transcriptome of pyrus ussuriensis in response to cold stress. Gene 2018, 661, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Hao, J.; Yang, J.; Dong, J.; Fei, S.Z. Characterization of BdCBF, genes and genome-wide transcriptome profiling of BdCBF3 -dependent and -independent cold stress responses in Brachypodium Distachyon. Plant Sci. 2017, 262, 52. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Zhou, Y.; Zhang, M.; Luo, X.; Xie, J. Effects of drought stress on global gene expression profile in leaf and root samples of Dongxiang wild rice (Oryza rufipogon). Biosci. Rep. 2017, 37. [Google Scholar] [CrossRef] [PubMed]
- Yadav, R.; Verma, O.P.; Padaria, J.C. Transcript profiling and gene expression analysis under drought stress in Ziziphus Nummularia, (Burm.f.) Wright & Aarn. Mol. Biol. Rep. 2018, 45, 1–12. [Google Scholar]
- Zhao, X.; Li, C.; Wan, S.; Zhang, T.; Yan, C.; Shan, S. Transcriptomic analysis and discovery of genes in the response of Arachis hypogaea to drought stress. Mol. Biol. Rep. 2018, 45, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Xu, L.; Wang, Y.; Luo, X.; Zhu, X.; Kinuthia, K.B.; Nie, S.; Feng, H.; Li, C.; Liu, L. Transcriptome-based gene expression profiling identifies differentially expressed genes critical for salt stress response in radish (Raphanus sativus L.). Plant Cell Rep. 2016, 35, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Wan, Q.; He, X.; Ning, L.; Huang, Y.; Xu, Z.; Liu, J.; Shao, H. Genome-wide characterization of the ankyrin repeats gene family under salt stress in soybean. Sci. Total Environ. 2016, 568, 899–909. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Wang, G.; Ji, J.; Shi, L.; Guan, C.; Jin, C. Comparative transcriptome analysis of transcription factors in different maize varieties under salt stress conditions. Plant Growth Regul. 2017, 81, 1–13. [Google Scholar] [CrossRef]
- Cui, J.; Ren, G.; Qiao, H.; Xiang, X.; Huang, L.; Chang, J. Comparative Transcriptome Analysis of Seedling Stage of Two Sorghum Cultivars Under Salt Stress. J. Plant Growth Regul. 2018, 6, 1–13. [Google Scholar] [CrossRef]
- Zhou, J.; Yang, Y.; Yu, J.; Wang, L.; Yu, X.; Ohtani, M.; Kusano, M.; Saito, K.; Demura, T.; Zhuge, Q. Responses of Populus trichocarpa galactinol synthase genes to abiotic stresses. J. Plant Res. 2014, 127, 347–358. [Google Scholar] [CrossRef] [PubMed]
- Mccaskill, A.; Turgeon, R. Phloem loading in Verbascum phoeniceum L. depends on the synthesis of raffinose-family oligosaccharides. Proc. Natl. Acad. Sci. USA 2007, 104, 19619–19624. [Google Scholar] [CrossRef] [PubMed]
- Sunkar, R.; Chinnusamy, V.; Zhu, J.; Zhu, J.K. Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends Plant Sci. 2007, 12, 301–309. [Google Scholar] [CrossRef] [PubMed]
- Shukla, L.I.; Chinnusamy, V.; Sunkar, R. The role of microRNAs and other endogenous small RNAs in plant stress responses. BBA–Gene Regul. Mech. 2008, 1779, 743–748. [Google Scholar] [CrossRef] [PubMed]
- Khraiwesh, B.; Zhu, J.K.; Zhu, J. Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochim. Et Biophys. Acta 2012, 1819, 137–148. [Google Scholar] [CrossRef] [PubMed]
- Sunkar, R. MicroRNAs with macro-effects on plant stress responses. Semin. Cell Dev. Biol. 2010, 21, 805–811. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, Y.; Zhou, X.H.; Liu, J. Conserved miRNAs and Their Response to Salt Stress in Wild Eggplant Solanum linnaeanum Roots. Int. J. Mol. Sci. 2014, 15, 839–849. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Xu, L.; Wang, Y.; Yu, R.; Zhu, X.; Luo, X.; Gong, Y.; Wang, R.; Limera, C.; Zhang, K.; et al. Identification of novel and salt-responsive miRNAs to explore miRNA-mediated regulatory network of salt stress response in radish (Raphanus sativus, L.). BMC Genomics 2015, 16, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.; Yu, H.; Liu, M.; Lu, Y.; Ouyang, B. Identification of salt-stress responsive microRNAs from Solanum lycopersicum, and Solanum pimpinellifolium. Plant Growth Regul. 2017, 83, 1–12. [Google Scholar] [CrossRef]
- Fu, R.; Zhang, M.; Zhao, Y.; He, X.; Ding, C.; Wang, S.; Feng, Y.; Song, X.; Li, P.; Wang, B. Identification of Salt Tolerance-related microRNAs and Their Targets in Maize (Zea mays L.) Using High-throughput Sequencing and Degradome Analysis. Front. Plant Sci. 2017, 8, 864. [Google Scholar] [CrossRef] [PubMed]
- Lv, D.K.; Bai, X.; Li, Y.; Ding, X.D.; Ge, Y.; Cai, H.; Ji, W.; Wu, N.; Zhu, Y.M. Profiling of cold-stress-responsive miRNAs in rice by microarrays. Gene 2010, 459, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhang, Y.; Ren, Y.; Xu, J.; Zhang, Z.; Wang, Y. Genome-wide identification of cold-responsive and new microRNAs in Populus tomentosa by high-throughput sequencing. Biochem. Biophys. Res. Commun. 2012, 417, 892–896. [Google Scholar] [CrossRef] [PubMed]
- Martinelli, F.; Cannarozzi, G.; Balan, B.; Siegrist, F.; Weichert, A.; Blösch, R.; Tadele, Z. Identification of miRNAs linked with the drought response of tef [Eragrostis tef, (Zucc.) Trotter]. J. Plant Physiol. 2018, 224, 163–172. [Google Scholar] [CrossRef] [PubMed]
- Kantar, M.; Lucas, S.J.; Budak, H. miRNA expression patterns of Triticum dicoccoides in response to shock drought stress. Planta 2011, 233, 471–484. [Google Scholar] [CrossRef] [PubMed]
- Xie, R.; Zhang, J.; Ma, Y.; Pan, X.; Dong, C.; Pang, S.; He, S.; Deng, L.; Yi, S.; Zheng, Y.; et al. Combined analysis of mRNA and miRNA identifies dehydration and salinity responsive key molecular players in citrus roots. Sci. Rep. 2017, 7, 42094. [Google Scholar] [CrossRef] [PubMed]
- Carolina, B.T.; Germán, P.; Sarah, A.; Fausto, R.Z.; Jorge, D.; Joe, T. Identification of cassava microRNAs under abiotic stress. Int. J. Genomics 2013, 2013, 857986. [Google Scholar]
- Khatabi, B.; Arikit, S.; Xia, R.; Winter, S.; Oumar, D.; Mongomake, K.; Meyers, B.C.; Fondong, V.N. High-resolution identification and abundance profiling of cassava (Manihot esculenta Crantz) microRNAs. BMC Genomics 2016, 17, 85. [Google Scholar] [CrossRef] [PubMed]
- Cui, N.; Sun, X.; Sun, M.; Jia, B.; Duanmu, H.; Lv, D.; Duan, X.; Zhu, Y. Overexpression of OsmiR156k, leads to reduced tolerance to cold stress in rice (Oryza sativa). Mol. Breed. 2015, 35, 214. [Google Scholar] [CrossRef]
- Arshad, M.; Feyissa, B.A.; Amyot, L.; Aung, B.; Hannoufa, A. MicroRNA156 improves drought stress tolerance in alfalfa (Medicago sativa) by silencing SPL13. Plant Sci. 2017, 258, 122–136. [Google Scholar] [CrossRef] [PubMed]
- Arshad, M.; Gruber, M.Y.; Wall, K.; Hannoufa, A. An insight into microRNA156 role in salinity stress responses of alfalfa. Front. Plant Sci. 2017, 8, 658. [Google Scholar] [CrossRef] [PubMed]
- Patade, V.Y.; Suprasanna, P. Short-term salt and peg stresses regulate expression of microRNA, miR159 in sugarcane leaves. J. Crop Sci. Biotechnol. 2010, 13, 177–182. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, N.; Mi, X.; Wu, L.; Ma, R.; Zhu, X.; Yao, L.; Jin, X.; Si, H.; Wang, D. Identification of miR159s and their target genes and expression analysis under drought stress in potato. Comput. Biol. Chem. 2014, 53, 204–213. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wang, Y.; Wang, Z.; Guo, X.; Wang, F.; Xia, X.J.; Zhou, J.; Shi, K.; Yu, J.Q.; Zhou, Y.H. Microarray and genetic analysis reveals that csa-miR159b plays a critical role in abscisic acid-mediated heat tolerance in grafted cucumber plants. Plant Cell Environ. 2016, 39, 1790–1804. [Google Scholar] [CrossRef] [PubMed]
- Luan, M.; Xu, M.; Lu, Y.; Zhang, L.; Fan, Y.; Wang, L. Expression of zma-miR169 miRNAs and their target ZmNF-YA genes in response to abiotic stress in maize leaves. Gene 2015, 555, 178–185. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhang, N.; Zhou, X.; Si, H.; Wang, D. Identification of four novel stu-miR169s and their target genes in Solanum tuberosum, and expression profiles response to drought stress. Plant Syst. Evol. 2016, 302, 55–66. [Google Scholar] [CrossRef]
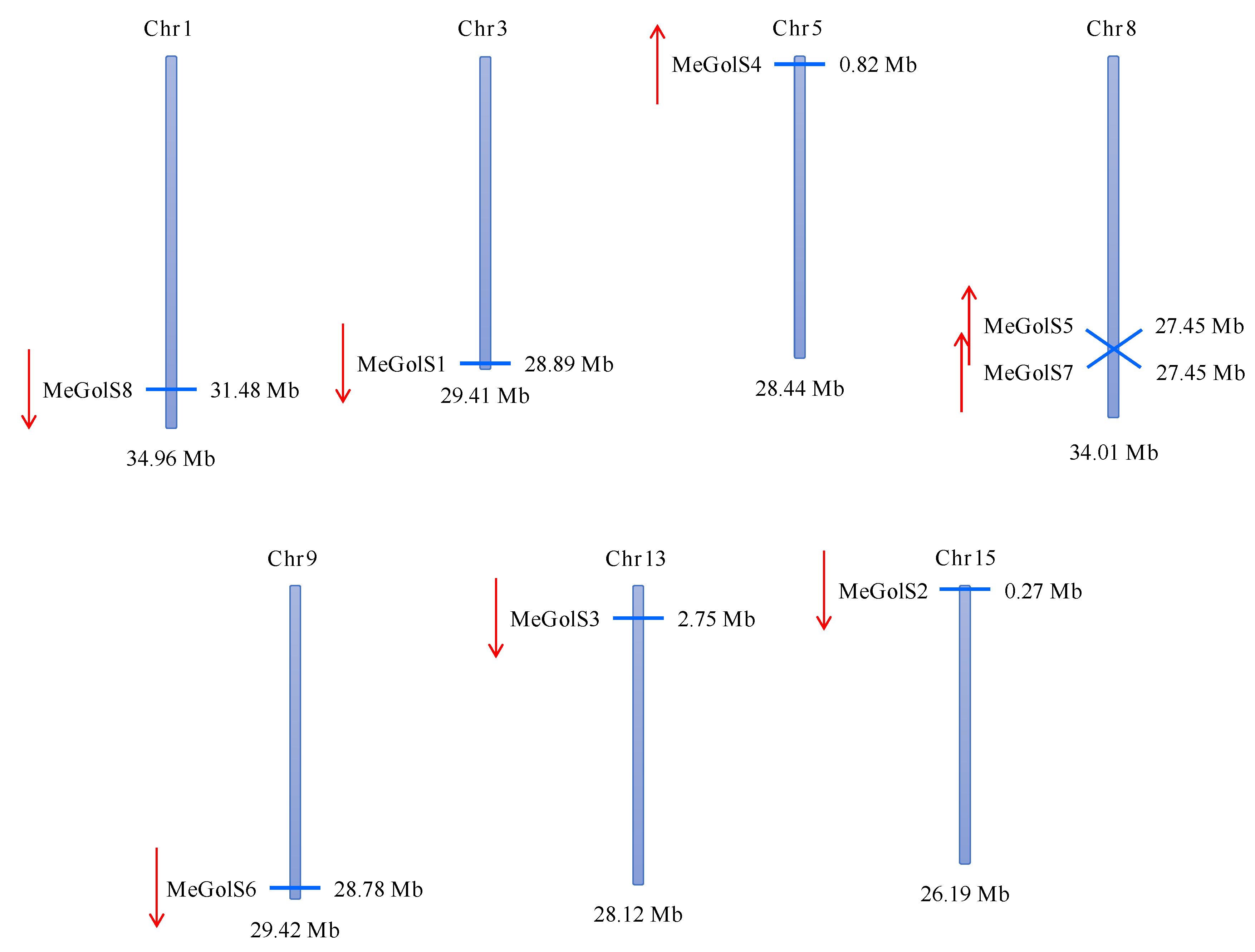
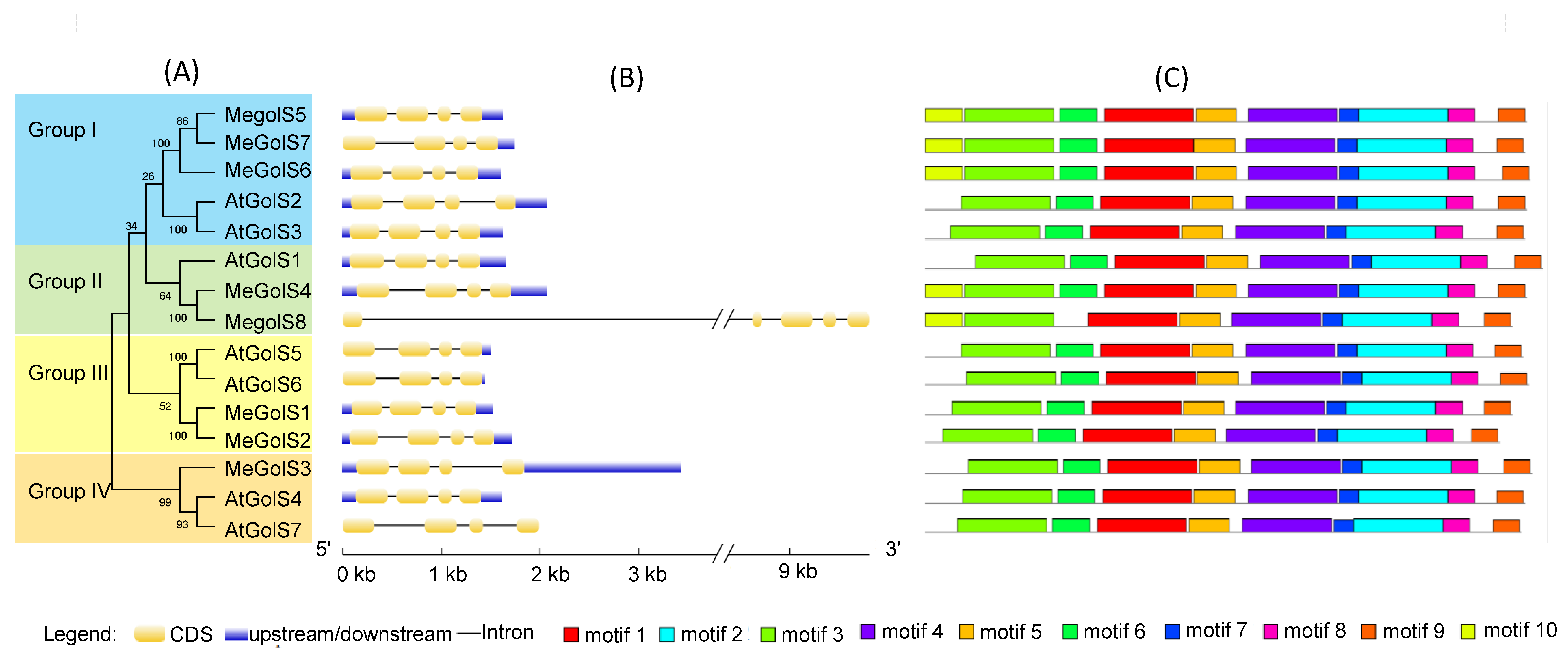
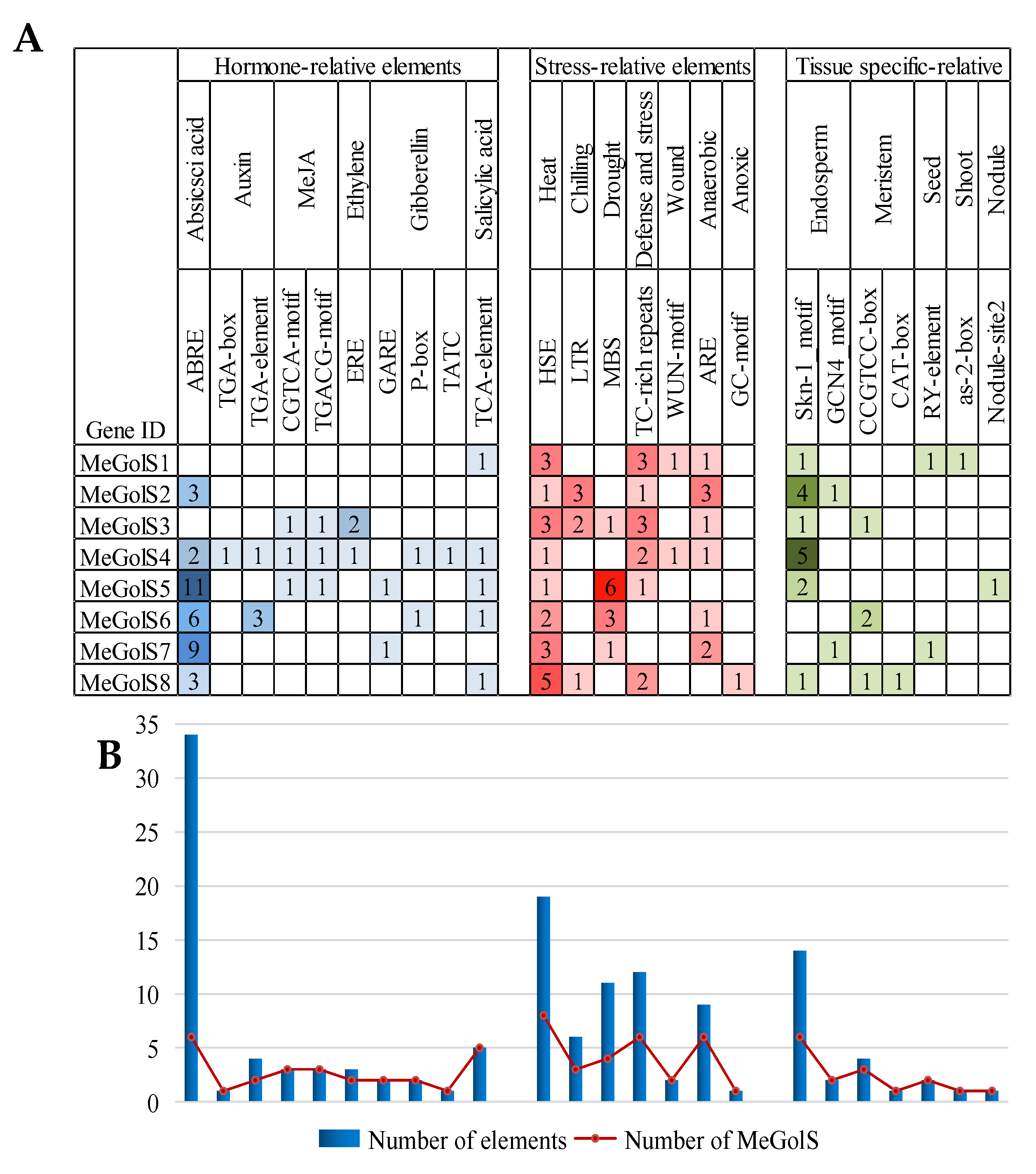
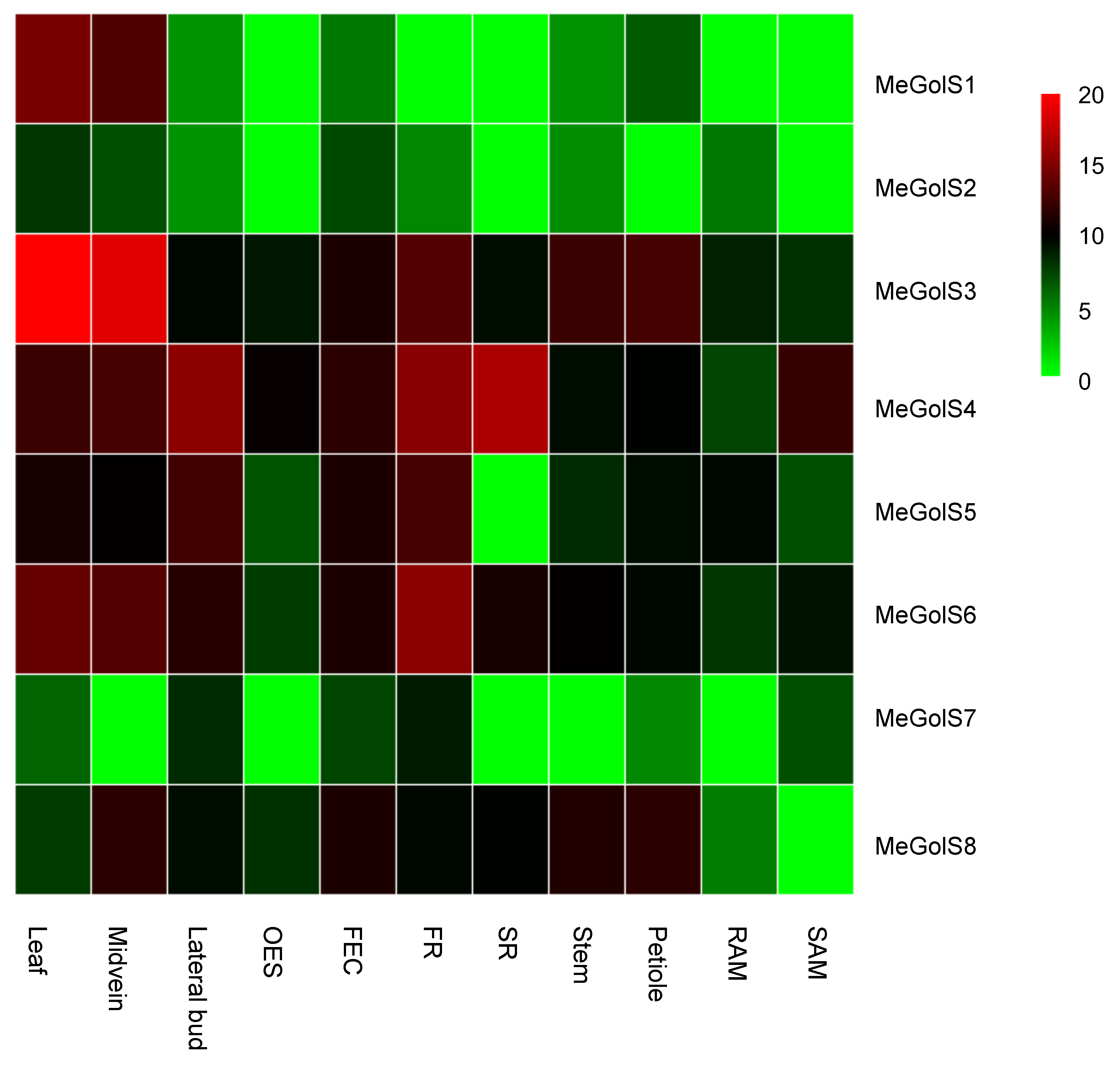
| GenBank ID | Gene Name | Locus Name | Chromosomal Location | Genomic Position (5′–3′) | gDNA (bp) | ORF (bp) | Protein Length (aa) | pI | MW (KD) | Exon | GRAVY | Subcellular Location Prediction |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| KF793045 | MeGolS1 | Manes.03G208800 | Chr03 | 28896616–28898137 | 1522 | 984 | 327 | 5.04 | 37.70 | 4 | –0.378 | Cytoplasmic |
| KF793046 | MeGolS2 | Manes.15G003200 | Chr15 | 272870–274578 | 1709 | 963 | 320 | 5.2 | 37.14 | 4 | –0.339 | Cytoplasmic |
| KJ722606 | MeGolS3 | Manes.13G029000 | Chr13 | 2755754–2759184 | 3431 | 1017 | 338 | 5.27 | 38.60 | 5 | –0.193 | Cytoplasmic, Chloroplast |
| KJ722605 | MeGolS4 | Manes.05G012000 | Chr05 | 822143–824202 | 2060 | 1008 | 335 | 5.39 | 38.30 | 4 | –0.213 | Cytoplasmic |
| KJ722604 | MeGolS5 | Manes.08G109300 | Chr08 | 27446317–27447941 | 1625 | 1008 | 335 | 5.08 | 38.10 | 4 | –0.162 | Plasma Membrane |
| KJ722603 | MeGolS6 | Manes.09G179100 | Chr09 | 28784797–28786402 | 1606 | 1014 | 337 | 5.47 | 38.19 | 4 | –0.19 | Plasma Membrane |
| KJ816317 | MeGolS7 | Manes.08G109400 | Chr08 | 27451435–27453176 | 1742 | 1005 | 334 | 5.33 | 38.10 | 4 | –0.167 | ER |
| MH535931 | MeGolS8 | Manes.01G231500 | Chr01 | 31476735–31486544 | 9810 | 984 | 327 | 5.27 | 37.35 | 5 | –0.182 | Cytoplasmic |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, R.; Yuan, S.; He, Y.; Fan, J.; Zhou, Y.; Qiu, T.; Lin, X.; Yao, Y.; Liu, J.; Fu, S.; et al. Genome-Wide Identification and Expression Profiling Analysis of the Galactinol Synthase Gene Family in Cassava (Manihot esculenta Crantz). Agronomy 2018, 8, 250. https://doi.org/10.3390/agronomy8110250
Li R, Yuan S, He Y, Fan J, Zhou Y, Qiu T, Lin X, Yao Y, Liu J, Fu S, et al. Genome-Wide Identification and Expression Profiling Analysis of the Galactinol Synthase Gene Family in Cassava (Manihot esculenta Crantz). Agronomy. 2018; 8(11):250. https://doi.org/10.3390/agronomy8110250
Chicago/Turabian StyleLi, Ruimei, Shuai Yuan, Yingdui He, Jie Fan, Yangjiao Zhou, Tingting Qiu, Xuejun Lin, Yuan Yao, Jiao Liu, Shaoping Fu, and et al. 2018. "Genome-Wide Identification and Expression Profiling Analysis of the Galactinol Synthase Gene Family in Cassava (Manihot esculenta Crantz)" Agronomy 8, no. 11: 250. https://doi.org/10.3390/agronomy8110250
APA StyleLi, R., Yuan, S., He, Y., Fan, J., Zhou, Y., Qiu, T., Lin, X., Yao, Y., Liu, J., Fu, S., Hu, X., & Guo, J. (2018). Genome-Wide Identification and Expression Profiling Analysis of the Galactinol Synthase Gene Family in Cassava (Manihot esculenta Crantz). Agronomy, 8(11), 250. https://doi.org/10.3390/agronomy8110250





