Abstract
In wheat, remobilization of nitrogen absorbed before anthesis and regulation of monocarpic senescence is a major issue in breeding for nutrient use efficiency. We identified natural variants of NAM-A1, a gene having the same role as its well-characterized homoeolog NAM-B1, a NAC transcription factor associated with senescence kinetics and nutrient remobilization to the grain. Differences in haplotype frequencies between a worldwide core collection and a panel of European elite varieties were assessed and discussed. Moreover, hypotheses for the loss of function of the most common haplotype in elite European germplasm are discussed.
1. Introduction
Breeding for enhanced nitrogen (N) use efficiency is necessary to sustainably increase worldwide cereal production. In wheat, remobilization of N absorbed before anthesis accounts for most of the N accumulated in the grain [1]. As most remobilized N is contained in the photosynthetic apparatus of the leaf, the regulation of monocarpic senescence is a major issue in breeding for N use efficiency.
In hexaploid bread wheat (Triticum aestivum L.) and tetraploid durum wheat (Triticum turgidum L. ssp durum), the No Apical Meristem (NAM) gene at the Gpc-B1 locus on chromosome arm 6BS encodes a NAC transcription factor known to accelerate senescence, to increase nutrient remobilization [2,3,4] and thus to increase grain protein concentration. Different effects of NAM-B1 were assessed depending on genotypes × environment combinations [2]. Moreover, optimal senescence kinetics can differ depending on N levels [5] leading to the hypothesis that NAM-B1 effects can also depend on the fertilization regimes.
Most bread wheats have a non-functional allele of NAM-B1 [6] and thus its physiological characterization began after a chromosome segment introgression from wild emmer wheat (Triticum turgidum L. subsp. dicoccoides) [7]. Nevertheless, hexaploid wheats have five other NAM genes, three paralogous (on chromosomes 2A, 2B and 2D) and two homoeologous (on chromosomes 6A and 6D) of which NAM-A1 (6A) has the same role as NAM-B1 [4,8]. Most studies on NAM wheat genes used mutants [4,8], near isogenic lines [9,10,11,12] or RNAi lines [3,4] and few studies have focused on the cultivated diversity [6,13]. This paper aims to characterize natural variants of NAM-A1 in hexaploid bread wheat and to hypothesize on the biological mechanisms explaining their effects.
2. Results and Discussion
2.1. SNP Detection on NAM-A1, Genotyping and Mapping
We screened the IWGSC (International Wheat Genome Sequencing Consortium) bank of genomic sequences and identified NAM-A1 in the sequence 6AS:4397602. This 29,595 bp sequence contains several transposable insertions; the coding sequence of NAM-A1 is localized between 15,502 bp and 17,060 bp and is composed of three exons for a total cDNA length of 1235 bp.
Single nucleotide polymorphism (SNP) identification was performed on 12 varieties and two high qualities SNP were detected in NAM-A1 genomic region (See Figures S1 and S2). The first SNP (SNP1) is located in NAM-A1 NAC domain (exon 2, 6AS:4397602_16233) and carries a C/T polymorphism. This SNP caused an alanine to valine substitution in the protein sequence. The second SNP (SNP2) is located at the end of the coding sequence (exon 3, 6AS:4397602_17020) corresponds to an A/deletion polymorphism which causes a reading frame shift leading to a truncated protein (see Figure S2).
Using the KASPar technology, these two SNP were genotyped on a total of 795 wheat cultivars composed of 367 accessions from a worldwide core collection [14] and 334 elite varieties with six varieties in common. Computing linkage disequilibrium between SNP located in NAM-A1 and SNP from the iSelect 90 K wheat SNP chip [15], we confirmed that our SNP tagging NAM-A1 are located on chromosome 6A and we estimated their map position at 74.24 cM (See Figure S1) in the genetic map by Wang et al. [15].
SNP frequencies are not balanced (Table 1). For SNP1, the T allele was the most frequent in the core and elite collections. For SNP2, the A allele was more frequent in the core collection and the Del allele in the elite collection. When considering haplotypes, NAM-A1c (T-A) is the most frequent haplotype in the core collection and NAM-A1d (T-Del) in the elite panel. In the core collection, accessions carrying the haplotype NAM-A1d are mainly modern Western European cultivars. In both panels, haplotype NAM-A1b is the less frequent with no accession carrying it in the elite panel and only one landrace from Georgia in the core collection. A Chi2 test shows that the observed haplotypes frequencies are not as expected from the SNP frequencies (Chi2 = 120.3, p < 0.001, both collections together, see Table S1). Although NAM-A1d is not the major haplotype in the core collection, it is over-represented in the two collections together. The NAM-A1a haplotype is also over-represented while the NAM-A1b is largely under-represented.

Table 1.
No Apical Meristem (NAM)-A1 haplotype frequencies in two collections of bread wheat genotypes. Frequency followed by the number of lines (in parenthesis).
| Genotype | Frequency | |||
|---|---|---|---|---|
| Haplotype | SNP1 | SNP2 | Core Collection | Elite |
| NAM-A1a | C | A | 0.232 (85) | 0.083 (28) |
| NAM-A1b | C | Del | 0.003 (1) | 0.000 (0) |
| NAM-A1c | T | A | 0.477 (175) | 0.189 (63) |
| NAM-A1d | T | Del | 0.215 (79) | 0.716 (239) |
| Undefined | 0.074 (27) | 0.012 (4) | ||
In the worldwide core collection, NAM-A1a is mainly found in accessions from Nepal (21 of 23 accessions), China (8 of 16 accessions) and Japan (7 of 12 accessions). Moreover, accessions carrying the haplotype NAM-A1a are mostly spring wheat. In the elite collection, NAM-A1a is over-represented in varieties with a high bread-making quality. Brevis et al. [10] showed that Gpc-B1 introgression was associated with a positive effect on several bread-making and pasta-making quality parameters. We can expect the same effect for NAM-A1. Thus, NAM-A1a may have been maintained in elite germplasm through selection for high baking quality. In addition, SNP1 is linked to the core collection genetic structure as SNP1_C is over-represented in far Eastern countries that form a cluster of diversity in the core collection [14]. Consequently, NAM-A1b under-representation could probably be explained by a Del mutation (SNP2) occurring only in the SNP1_T allelic lineage [16]. Then, over-representation of NAM-A1d in modern European elites suggests that the haplotype may have been selected. NAM-A1b could be the results of a recent recombination between NAM-A1a and NAM-A1d.
2.2. Effect of NAM-A1 Haplotypes
Focusing on 196 European elite varieties genotyped in this study with available phenotypic data [17], the highest grain protein concentration (GPC) and lowest grain yield (GY) were reached in varieties carrying the haplotype NAM-A1a (Table 2). This is caused by the well-known negative correlation between GY and GPC (i.e., [18]). The lower grain yield was linked with a reduced grain weight (TKW) not compensated by the number of grain (spike per area (SA) × kernel per spike (KS) in Table 2). Nevertheless, varieties with NAM-A1a showed also the highest grain protein deviation (GPD, [19]) and a high N harvest index associated with a low straw N content at maturity (%N_S). Varieties carrying the haplotype NAM-A1c were intermediate between those carrying NAM-A1a and NAM-A1d. This can be explained by differences in haplotype effects. However, the genetic background of varieties is also a possible explanation. In general, due to the highly unbalanced frequencies and a distribution linked to the panel’s structure as previously mentioned, we lacked power to be able to distinguish the effect of the genotypes’ genetic background and the actual effect of NAM-A1. Therefore, results presented in Table 2 should be interpreted with caution.

Table 2.
Mean agronomic values for the two NAM-A1 single nucleotide polymorphism (SNP) genotyped on 196 (16 NAM-A1a; 37 NAM-A1c; 143 NAM-A1d) European elite varieties.
| SNP1 | SNP2 | Haplotype | GY | TKW | SA | KS | GPC | GPD | NHI | %N_S |
|---|---|---|---|---|---|---|---|---|---|---|
| C | A | NAM-A1a | 6976c | 41.3b | 421a | 40.4b | 10.46a | 0.20a | 81.17ab | 0.41a |
| T | A | NAM-A1c | 7241b | 41.6b | 413a | 42.5a | 10.15b | 0.04ab | 81.47a | 0.41a |
| T | Del | NAM-A1d | 7799a | 42.7a | 411a | 43.0a | 9.79c | -0.09b | 80.98b | 0.42b |
GY, dry matter grain yield (kg/ha); TKW, thousand kernel weight (g); SA, spike per area (spike/m2); KS, kernel per spike; GPC, Grain Protein Concentration (%); GPD, Grain Protein Deviation [19]; NHI, nitrogen harvest index (%N); %N_S, straw N content at maturity (%N). Letters indicate significance group by LSD test (p < 0.05) for SNP or haplotype effects without taking into account genetic background.
Nevertheless, in agreement with the described mean values, several studies analyzing the introgression of the functional allele of Gpc-B1 in different spring hexaploid wheat [9,11,12] concluded that NAM-A1 homoeolog increased GPC and decreased TKW. However, other studies such as Waters et al. [20] showed no effect on TKW. An improved N remobilization (%N_S and NHI) was also assessed [9]. However, the effect of Gpc-B1 on grain yield across genotypes and environments was not significant [9,11,12] even if it was strongly affected by the genetic background [9]. Similarly, study of mutants concluded that functional NAM-A1 (6A) and NAM-B2 (2B) genes accelerate senescence and increase GPC with a larger phenotypic effect for NAM-A1 than NAM-B2 [4,8].
To conclude, we hypothesize that NAM-A1a could be a functional variant of the NAM-A1 gene. Accelerated senescence could have improved N remobilization and GPC but decreased TKW leading to a GY decrease as in our elite panel where varieties carrying NAM-A1a has also a lower number of grains. This is in accordance with the low frequency of NAM-A1a in elite germplasm mainly selected on GY, and its high frequency in spring Nepalese accessions cultivated within a short growing season.
2.3. 3D Conformation of NAM-A1 NAC Domain
Prediction of the NAM-A1 NAC domain 3D structure was based on the crystal structure of the rice stress responsive NAC1 (SNAC1) NAC domain [21]. Crystallographic analysis of the NAC domain of the ANAC protein [22,23] encoded by the abscisic acid-responsive NAC gene from Arabidopsis thaliana and mutants study [24] were also used.
According to their high amino acid similarity (69.7%), the topology of SNAC1 NAC domain and the predicted topology of NAM-A1 NAC domain were similar. The NAM-A1 NAC domain prediction resulted in seven twisted β-strands forming a semi-β-barrel with four α-helices (Figure 1). Although the residues of the loop region between β6–β7 in both SNAC1 and ANAC NAC domains were unobserved due to its non-participation in crystal packing [21], in NAM-A1 NAC domain an α-helix is predicted. This α4-helix is truncated in the protein encoded by the haplotypes NAM-A1c and NAM-A1d, due to SNP1 alanine to valine substitution (Figure 1). Indeed, alanine is one of the best α-helix-forming residues due to aliphatic sidechains regions. In contrast, with short sidechains that can form hydrogen bonds, valine is a poor α-helix former.
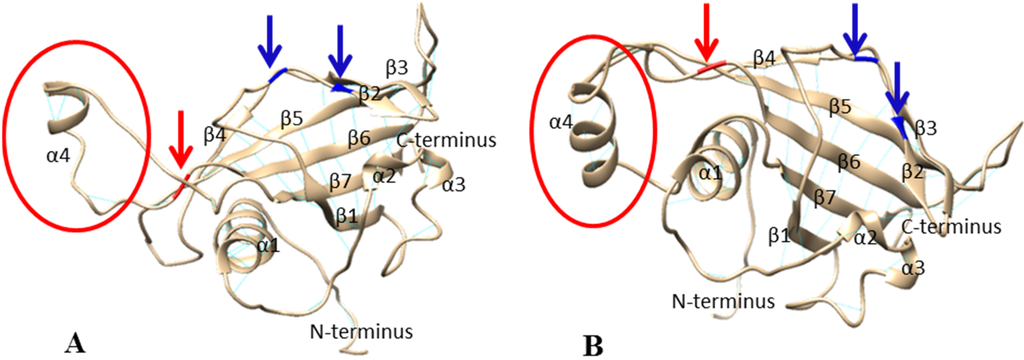
Figure 1.
Predicted 3D structure of NAM-A1 NAC domain for (A) the valine variant (SNP1_T) and (B) the alanine variant (SNP1_C). Blue arrows: Arg107 and Arg110; red arrows: variant amino acid; red circle: affected α4-helix.
Dimerization of DNA binding domains is common and can modulate the DNA-binding specificity [25]. Gel filtration studies on ANAC NAC domain [22] and SNAC1 NAC domain [21] have shown that in solution they exist as dimers that form the functional unit necessary for stable DNA binding [24]. We can reasonably presume that it is also the case for NAM-A1. The interface between the two monomers of SNAC1 consists of residues in the N-terminal loop region and two residues in the α1-helix [21]. In NAM-A1, this domain is not predicted to be affected by SNP1 variation.
Olsen et al. [24] showed that K79A/R85A/R88A and R85A/R88A were ANAC mutants that impaired DNA binding. Using these results, Chen et al. [21] hypothesized that Arg85 and Arg88 are responsible of DNA binding in SNAC1 (residues Arg107 and Arg110 in NAM-A1).
Using yeast one hybrid assay, Duval et al. [26] identified the DNA binding domain of AtNAM between Val119 and Ser183 (AtNAM numbering) and hypothesized that the region folds in a helix-turn-helix structure. In contrast, in ANAC and SNAC1, this region consists of β-sheet [21,23], but as previously mentioned the conformation of part of residues in the loop region between β6–β7 was unobserved. This missing loop region, poorly conserved between NAC domains and maybe related to their biological function [21], was predicted as being the region affected by the alanine to valine substitution discovered in NAM-A1 (SNP1).
Thus, in accordance with the lowest GPC and GPD observed (Table 2) for the NAM-A1d (SNP1_T, SNP2_del) haplotype compared to the NAM-A1a haplotype (SNP1_C, SNP2_A), we hypothesize that the valine variant of NAM-A1 NAC domain (SNP1_T) may form dimers, may bind to DNA, but its biological function may be affected. A second hypothesis could be that the more recent mutation (SNP2) leading to a slightly truncated protein may affect the transcriptional activation by the C-terminus and differences between NAM-A1a and NAM-A1c could be due to a genetic background effect. Sequence alignment of the closest NAC proteins from wheat, barley, rice and A. thaliana did not allow us to distinguish between these two hypotheses (Figure S3). Indeed, it revealed that none of these NAC proteins seems truncated in the available genotypes. The deletion (SNP2) may be unique to wheat. Moreover, wheat NAC proteins carry the alanine variant (SNP1_C) and SNP1 is not conserved across species (Figure S4). SNP1 lies outside the conserved domain, in a region that could be involved in the proteins’ biological functions as previously mentioned.
3. Experimental Section
The IWGSC (International Wheat Genome Sequencing Consortium) bank of genomic sequences was screened by Basic Local Alignment Search Tool (BLAST) using the sequence DQ869672.1 (Triticum turgidum subsp. durum NAM-A1 complete coding DNA sequence).
SNP detection was performed following sequencing of NAM-A1 in 12 varieties: Alcedo, Brigadier, Cassius, Premio, Récital, Renan, Rialto, Robigus, Sarina, Soissons, Tremie and Xi19. Genomic sequences were aligned using Chinese Spring as a reference.
The KASPar SNP Genotyping System (KBiociences, Herts, UK) was used to validate SNPs. KASPar Primers were designed with Primer picker (KBioscience) and PCR amplifications were performed on hydrocycler (LGC genomics), for 50 cycles at 57 °C and then run onto a Genotyper (Applied Biosystem).
Linkage disequilibrium between the discovered SNP on NAM-A1 and the iSelect 90 K SNP was computed using genotyping data of 281 varieties from the European elite collection. A list of all the elite varieties used in this study is provided in Table S2.
Mean agronomic values were calculated from 196 European elite varieties (16 CA; 37 TA; 143 TDel) experimented in eight combinations of year, site, and N regime [17]. Mean values were calculated using a linear model with the experiment (year_site_N) and SNP or haplotype as fixed factors.
Prediction of 3D structure was carried out using SWISS-MODEL SERVER [27] and based on the 3ulx.1.A template (X-ray, 2.60 Å) of SNAC1 [21]. Visualization was made using Chimera [28].
4. Conclusions
Grain protein concentration is maximized in varieties carrying the NAM-A1a haplotype coding for the alanine variant of NAM-A1 NAC domain and a non-truncated protein confirming the hypothesis that it may be a functional haplotype conserved in high-baking quality germplasm used in modern selection. The difference between both haplotypes coding a valine variant of NAM-A1 NAC domain (NAM-A1c and NAM-A1d) remains unclear as the effects of both mutations on protein’s function. In the context of fertilizer reduction, increasing the frequency of the NAM-A1a haplotype in elite germplasm may help to breed for an increased remobilization. However, the effect of NAM-A1 on yield seems to depend on genotypes and environments. Moreover the exact possible negative impact on grain yield is not known. It is probable that the efficient use of this allele in an elite breeding program would requires the selection of an adapted background as it was done for NAM-B1 in India [29]. Marker-assisted selection was performed for introgression into 10 wheat backgrounds. As a result, BC3 progenies were developed and evaluated enabling the identification of progenies with significantly higher GPC with no yield penalty. To conclude, further investigation at low N regime after flowering may be required to maximize the impact of remobilization on agronomic performance. This study provided the necessary tools to achieve these goals.
Acknowledgments
These analyses were a part of a PhD thesis (129/2012) supported by the ANRT (Association Nationale de la Recherche et de la Technologie). Data were obtained thanks to the support of the ANR ProtNBle project (06 GPLA016). The authors are also grateful to Hien Le for English style proofing.
Author Contributions
Fabien Cormier, MickaelThroude and Catherine Ravel: SNP detection, manuscript development. Magalie Leveugle, bioinformatics, SNP discovery, manuscript. FC: protein 3D conformation. Nadine Duranton, Florence Exbrayat: genotyping and SNP analyses. Stéphane Lafarge, Jacques Le Gouis, Sébastien Praud: supervision of experiments, manuscript development.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Sanford, D.A.; MacKown, C.T. Variation in nitrogen use efficiency among soft red winter wheat genotypes. Theor. Appl. 1986, 72, 158–163. [Google Scholar] [CrossRef]
- Uauy, C.; Brevis, J.C.; Dubcovsky, J. The high grain protein content gene Gpc-B1 accelerates senescence and has pleiotropic effects on protein content in wheat. J. Exp. Bot. 2006, 57, 2785–2794. [Google Scholar] [CrossRef] [PubMed]
- Uauy, C.; Distelfeld, A.; Fahima, T.; Blechl, A.; Dubcovsky, J. A NAC gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science 2006, 314, 1298–1301. [Google Scholar] [CrossRef] [PubMed]
- Distelfeld, A.; Pearce, S.P.; Avni, R.; Scherer, B.; Uauy, C.; Piston, F.; Slade, A.; Zhao, R.; Dubcovsky, J. Divergent functions of orthologous NAC transcription factors in wheat and rice. Plant Mol. Biol. 2012, 78, 515–524. [Google Scholar] [CrossRef] [PubMed]
- Gaju, O.; Allard, V.; Martre, P.; Snape, J.W.; Heumez, E.; Le Gouis, J.; Moreau, D.; Bogard, M.; Griffiths, S.; Orford, S.; et al. Identification of traits to improve the nitrogen-use efficiency of wheat genotypes. Field Crops Res. 2011, 123, 139–152. [Google Scholar] [CrossRef]
- Hagenblad, J.; Asplund, L.; Balfourier, F.; Ravel, C.; Leino, M. Strong presence of the high grain protein content allele of NAM-B1 in Fennoscandian wheat. Theor. Appl. Genet. 2012, 125, 1677–1686. [Google Scholar] [CrossRef] [PubMed]
- Distefeld, A.; Uauy, C.; Fahima, T.; Dubcosky, J. Physical map of the wheat high-grain protein content gene Gpc-B1 and development of a high-throughput molecular marker. New Physiol. 2006, 169, 753–763. [Google Scholar] [CrossRef]
- Avni, R.; Zhao, R.; Pearce, S.; Jun, Y.; Uauy, C.; Tabbita, F.; Fahima, T.; Slade, A.; Dubcovsky, J.; Distelfeld, A. Functional characterization of GPC-1 genes in hexaploid wheat. Planta 2014, 239, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Brevis, J.C.; Dubcovsky, J. Effects of the Chromosome Region Including the Gpc-B1 Locus on Wheat Grain and Protein Yield. Crop Sci. 2010, 50, 93–104. [Google Scholar] [CrossRef]
- Brevis, J.C.; Morris, C.F.; Manthey, F.; Dubcovsky, J. Effect of the grain protein content locus Gpc-B1 on bread and pasta quality. J. Cereal Sci. 2010, 51, 357–365. [Google Scholar] [CrossRef]
- Eagles, H.A.; McLean, R.; Eastwood, R.F.; Appelbee, M.J.; Cane, K.; Martin, P.J.; Wallwork, H. High-yielding lines of wheat carrying Gpc-B1 adapted to Mediterranean-type environments of the south and west of Australia. Crop Pasture Sci. 2014, 65, 854–861. [Google Scholar] [CrossRef]
- Tabbita, F.; Lewis, S.; Vouilloz, J.P.; Ortega, M.A.; Kade, M.; Abbate, P.E.; Barneix, A.J. Effects of the Gpc-B1 locus on high grain protein content introgressed into Argentinean wheat germplasm. Plant Breed. 2012, 132, 48–52. [Google Scholar] [CrossRef]
- Asplund, L.; Bergkvist, G.; Leino, M.W.; Westerbergh, A.; Weih, M. Swedish spring varieties with the rare high grain protein allele of NAM-B1 differ leaf senescence and grain mineral content. PLoS ONE 2013, 8, e59704. [Google Scholar] [CrossRef]
- Balfourier, F.; Roussel, V.; Strelchenko, P.; Exbrayat-Vinson, F.; Sourdille, P.; Boutet, G.; Koenig, J.; Ravel, C.; Mitrofanova, O.; Beckert, M.; et al. A worldwide bread wheat core collection arrayed in a 384-well plate. Theor. Appl. Genet. 2007, 114, 1265–1275. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Debbie, W.; Forrest, K.; Allen, A.; Chao, S.; Huang, B.E.; Maccaferri, M.; Salvi, S.; Milner, S.G.; Cattivelli, L.; et al. Characterization of polyploidy wheat genomic diversity using a high-density 90,000 single nucleotide polymorphism array. Plant Biotech. J. 2014, 12, 787–796. [Google Scholar] [CrossRef]
- Flint-Garcia, S.; Thornberry, J.M.; Buckler, E.S. Structure of linkage disequilibrium in plants. Annu. Rev. Plant Biol. 2003, 54, 357–374. [Google Scholar] [CrossRef] [PubMed]
- Cormier, F.; Faure, S.; Dubreuil, P.; Heumez, E.; Beauchêne, K.; Lafarge, S.; Praud, S.; Le Gouis, J. A multi-environmental study of recent breeding progress on nitrogen use efficiency in wheat (Triticum aestivum L.). Theor. Appl. Genet. 2013, 126, 3035–3048. [Google Scholar] [CrossRef] [PubMed]
- Feil, B. The inverse yield-protein relationships in cereals: Possibilities and limitations for genetically improving the grain protein yield. Trends Agron. 1997, 1, 103–119. [Google Scholar]
- Monaghan, J.M.; Snape, J.W.; Chojecki, A.J.S.; Kettlewell, P.S. The use of grain protein deviation for identifying wheat cultivars with high protein concentration and yield. Euphytica 2001, 122, 309–317. [Google Scholar] [CrossRef]
- Waters, B.M.; Uauy, C.; Dubcovsky, J.; Grusak, M.A. Wheat (Triticum aestivum) NAM proteins regulate the translocation of iron, zinc, and nitrogen compounds from vegetative tissues to grain. J. Exp. Bot. 2009, 60, 4263–4274. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Wang, Q.; Xiong, L.; Lou, Z. A structural view of the conserved domain of rice stress-responsive NAC1. Protein Cell 2011, 2, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Olsen, A.N.; Ernst, H.A.; Lo Leggio, L.; Johansson, E.; Larsen, S.; Skriver, K. Preliminary crystallographic analysis of the NAC domain of ANAC, a member of the plant-specific NAC transcription factor family. Acta Crystallogr. D 2004, 60, 112–115. [Google Scholar] [CrossRef] [PubMed]
- Ernst, H.A.; Olsen, A.N.; Skriver, K.; Larsen, S.; Lo Leggio, L. Structure of the conserved domain of ANAC, a member of the NAC family of transcription factors. EMBO Rep. 2004, 3, 297–303. [Google Scholar] [CrossRef]
- Olsen, A.N.; Ernst, H.A.; Leggio, L.L.; Skriver, K. NAC transcription factors: Structurally distinct, functionally diverse. Trends Plant Sci. 2005, 10, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Müller, C.W. Transcription factors: Global and detailed views. Curr. Opin. Struct. Biol. 2001, 11, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Duval, M.; Hsieh, T.F.; Kim, S.Y.; Thomas, T.L. Molecular characterization of AtNAM: A member of the Arabidopsis NAC domain superfamily. Plant Mol. Biol. 2002, 50, 237–248. [Google Scholar] [CrossRef] [PubMed]
- Arnold, K.; Bordoli, L.; Kopp, J.; Schwede, T. The SWISS-MODEL workspace: A web based environment for protein structure homology modelling. Bioinformatics 2006, 22, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Kumar, J.; Jaiswal, V.; Kumar, A.; Kumar, N.; Mir, R.R.; Kumar, S.; Dhariwal, R.; Tyagi, S.; Khandelwal, M.; Prabhu, K.V.; et al. Introgression of a major gene for high grain protein content in some Indian bread wheat cultivars. Field Crops Res. 2011, 123, 226–233. [Google Scholar] [CrossRef]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).