Physiological and Molecular Mechanisms of Ethylene in Sculpting Rice Root System Architecture
Abstract
1. Introduction
Literature Search Strategy
2. Ethylene Biosynthesis and Signaling
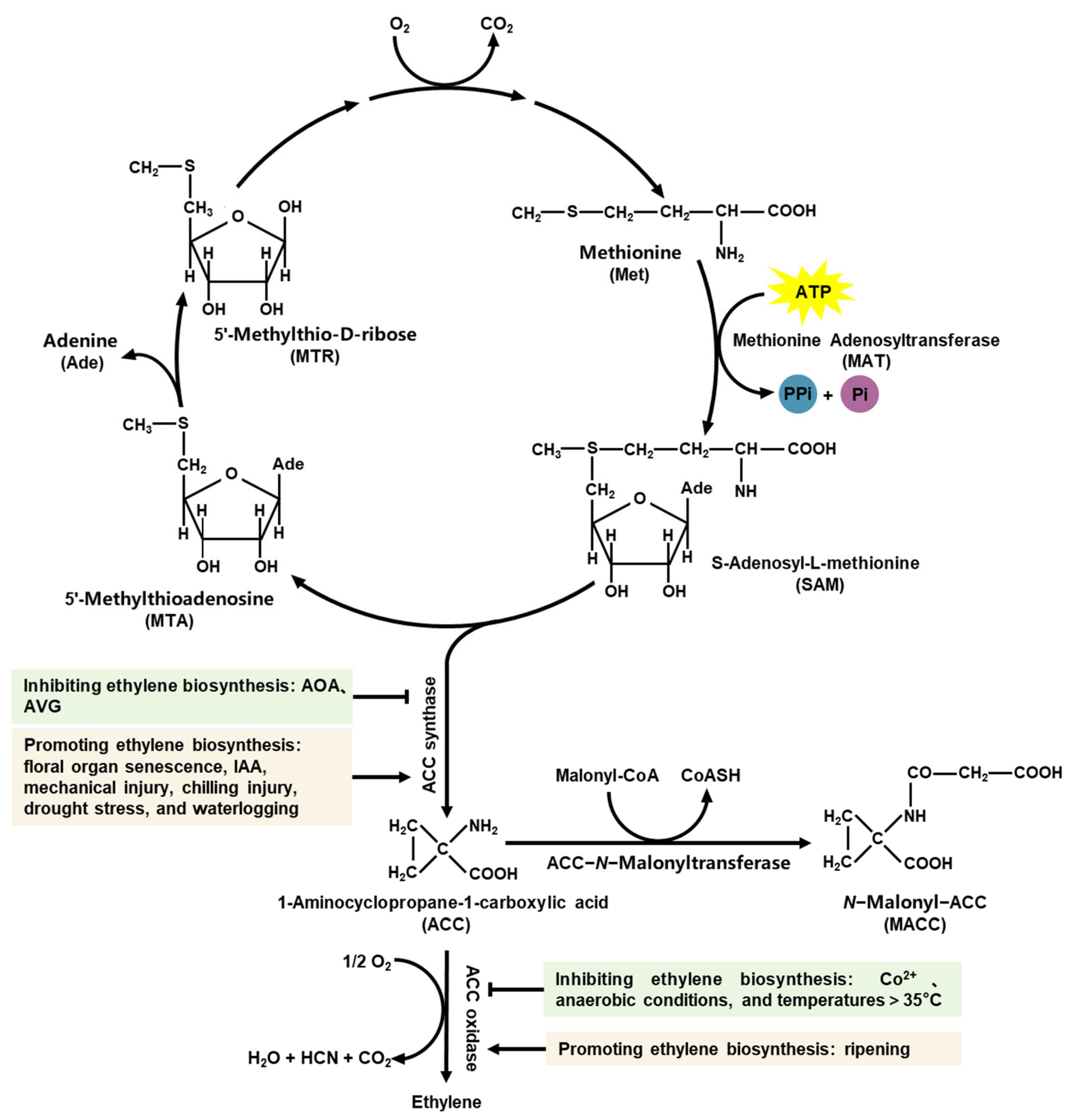
2.1. Ethylene Biosynthetic Pathway
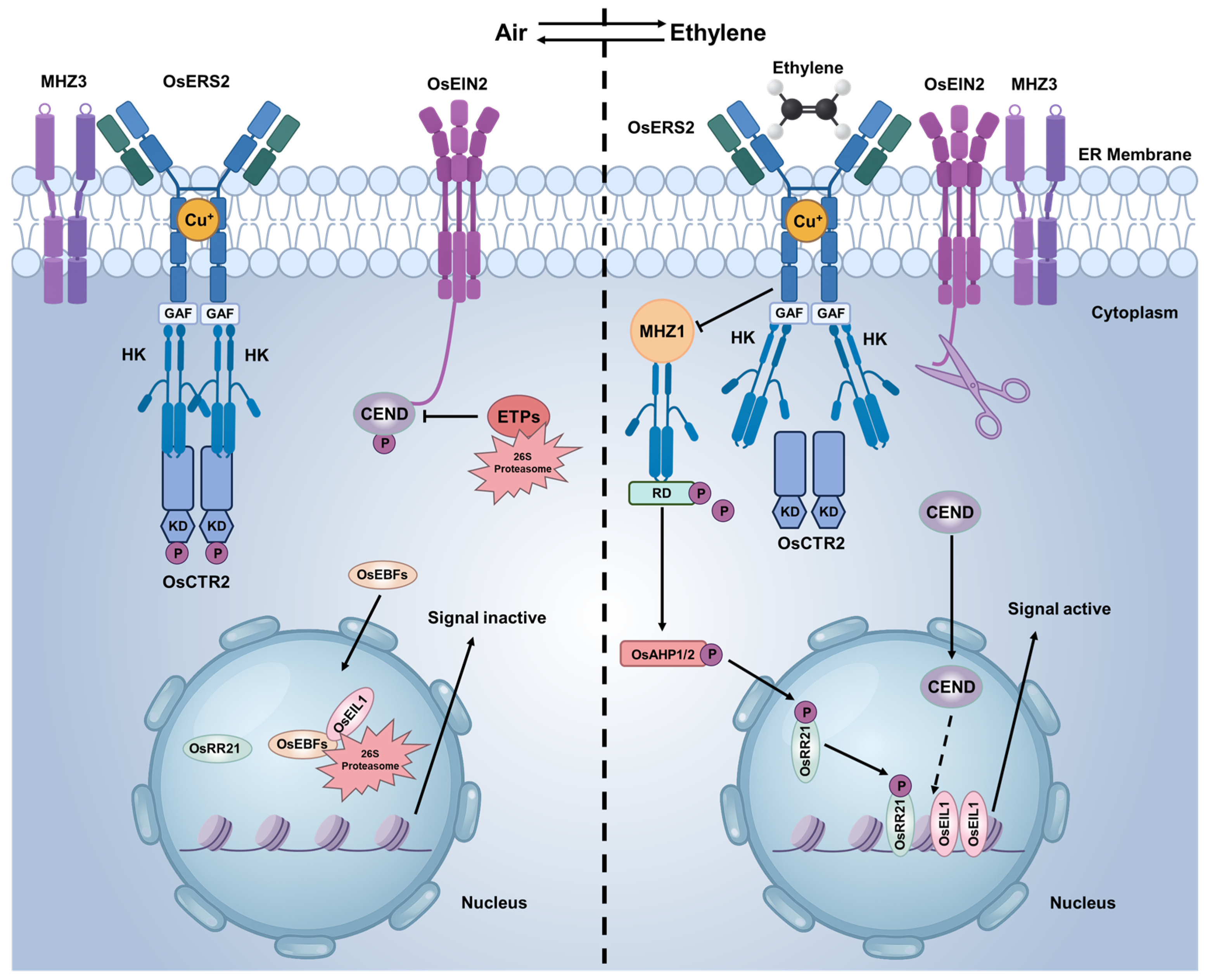
2.2. Ethylene Signaling Pathway
3. Ethylene Modulation of Root System Development
3.1. Root Primary Growth
3.2. Root Secondary Growth
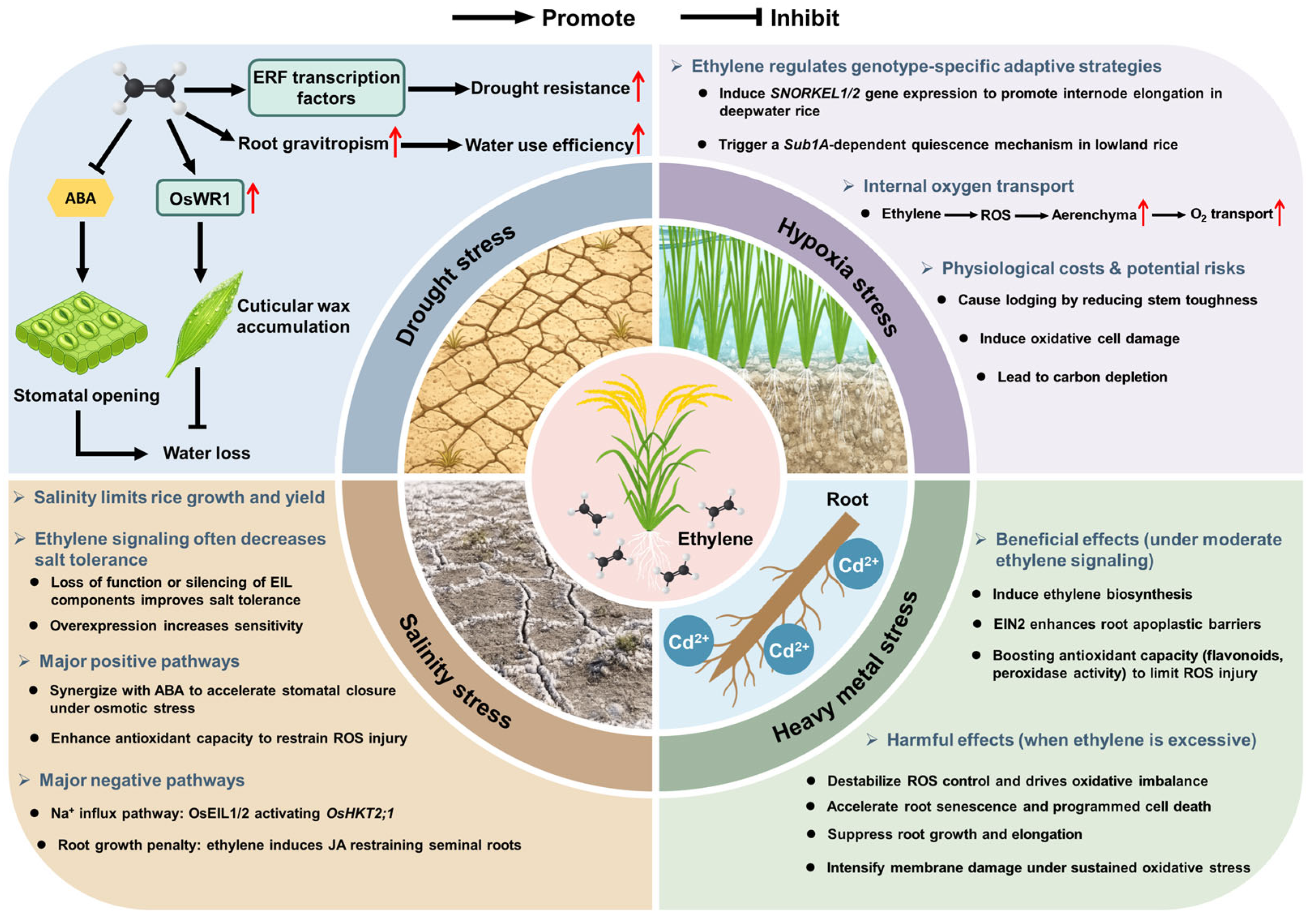
4. The Role of Ethylene in Rice Stress Responses
4.1. Drought Stress
4.2. Hypoxia Stress
4.3. Salinity Stress
4.4. Heavy Metal Stress
5. Ethylene-Mediated Root and Microbe Interactions
5.1. Rhizobacteria
5.2. Rhizosphere Fungi
6. Challenges and Prospects
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zhang, J.; Zhu, Y.; Yu, L.; Yang, M.; Zou, X.; Yin, C.; Lin, Y. Research advances in cadmium uptake, transport and resistance in rice (Oryza sativa L.). Cells 2022, 11, 569. [Google Scholar] [CrossRef]
- Bin Rahman, A.N.M.R.; Zhang, J. Trends in rice research: 2030 and beyond. Food Energy Secur. 2023, 12, e390. [Google Scholar] [CrossRef]
- Takehisa, H.; Sato, Y.; Igarashi, M.; Abiko, T.; Antonio, B.A.; Kamatsuki, K.; Minami, H.; Namiki, N.; Inukai, Y.; Nakazono, M.; et al. Genome-wide transcriptome dissection of the rice root system: Implications for developmental and physiological functions. Plant J. 2012, 69, 126–140. [Google Scholar] [CrossRef]
- Tardieu, F.; Davies, W.J. Integration of hydraulic and chemical signalling in the control of stomatal conductance and water status of droughted plants. Plant Cell Environ. 1993, 16, 341–349. [Google Scholar] [CrossRef]
- Liu, L.; Zhao, L.; Liu, Y.; Zhu, Y.; Chen, S.; Yang, L.; Li, X.; Chen, W.; Xu, Z.; Xu, P.; et al. Transcription factor OsWRKY72 controls rice leaf angle by regulating LAZY1-mediated shoot gravitropism. Plant Physiol. 2024, 195, 1586–1600. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Cheng, S. Root genetic research, an opportunity and challenge to rice improvement. Field Crops Res. 2014, 165, 111–124. [Google Scholar] [CrossRef]
- Zhang, Y.; Berman, A.; Shani, E. Plant hormone transport and localization: Signaling molecules on the move. Annu. Rev. Plant Biol. 2023, 74, 453–479. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.F.; Hoffman, N.E. Ethylene biosynthesis and its regulation in higher plants. Annu. Rev. Plant Physiol. 1984, 35, 155–189. [Google Scholar] [CrossRef]
- Ma, B.; Yin, C.C.; He, S.J.; Lu, X.; Zhang, W.K.; Lu, T.G.; Chen, S.Y.; Zhang, J.S. Ethylene-induced inhibition of root growth requires abscisic acid function in rice (Oryza sativa L.) seedlings. PLoS Genet. 2014, 10, e1004701. [Google Scholar] [CrossRef]
- Huang, G.; Kilic, A.; Karady, M.; Zhang, J.; Mehra, P.; Song, X.; Sturrock, C.J.; Zhu, W.; Qin, H.; Hartman, S.; et al. Ethylene inhibits rice root elongation in compacted soil via ABA- and auxin-mediated mechanisms. Proc. Natl. Acad. Sci. USA 2022, 119, e2201072119. [Google Scholar] [CrossRef]
- Yang, C.; Lu, X.; Ma, B.; Chen, S.Y.; Zhang, J.S. Ethylene signaling in rice and Arabidopsis: Conserved and diverged aspects. Mol. Plant 2015, 8, 495–505. [Google Scholar] [CrossRef]
- Yau, C.P. Differential expression of three genes encoding an ethylene receptor in rice during development, and in response to indole-3-acetic acid and silver ions. J. Exp. Bot. 2004, 55, 547–556. [Google Scholar] [CrossRef]
- Khattak, A.A.; Huang, Y.; Afzal, M.; Wang, X. MHZ3: A key regulator of ethylene signaling in rice. aBIOTECH 2025, 6, 133–138. [Google Scholar] [CrossRef]
- Zarembinski, T.I.; Theologis, A. Anaerobiosis and plant growth hormones induce two genes encoding 1-aminocyclopropane-1-carboxylate synthase in rice (Oryza sativa L.). Mol. Biol. Cell 1993, 4, 363–373. [Google Scholar] [CrossRef]
- Iwai, T.; Miyasaka, A.; Seo, S.; Ohashi, Y. Contribution of ethylene biosynthesis for resistance to blast fungus infection in young rice plants. Plant Physiol. 2006, 142, 1202–1215. [Google Scholar] [CrossRef]
- Pattyn, J.; Vaughan-Hirsch, J.; Van de Poel, B. The regulation of ethylene biosynthesis: A complex multilevel control circuitry. New Phytol. 2021, 229, 770–782. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Zhong, S.; Grierson, D. Recent advances in ethylene research. J. Exp. Bot. 2009, 60, 3311–3336. [Google Scholar] [CrossRef] [PubMed]
- Keunen, E.; Schellingen, K.; Vangronsveld, J.; Cuypers, A. Ethylene and metal stress: Small molecule, big impact. Front. Plant Sci. 2016, 7, 23. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.B.; Yang, S.F. Auxin-induced ethylene production and its inhibition by aminoethyoxyvinylglycine and cobalt ion. Plant Physiol. 1979, 64, 1074–1077. [Google Scholar] [CrossRef]
- Ramonell, K.M.; McClure, G.; Musgrave, M.E. Oxygen control of ethylene biosynthesis during seed development in Arabidopsis thaliana (L.) Heynh. Plant Cell Environ. 2002, 25, 793–801. [Google Scholar] [CrossRef]
- Biggs, M.S.; Woodson, W.R.; Handa, A.K. Biochemical basis of high-temperature inhibition of ethylene biosynthesis in ripening tomato fruits. Physiol. Plant. 1988, 72, 572–578. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, W.; Yin, Z.; Wen, C.K. Rice CONSTITUTIVE TRIPLE-RESPONSE2 is involved in the ethylene-receptor signalling and regulation of various aspects of rice growth and development. J. Exp. Bot. 2013, 64, 4863–4875. [Google Scholar] [CrossRef]
- Wuriyanghan, H.; Zhang, B.; Cao, W.H.; Ma, B.; Lei, G.; Liu, Y.F.; Wei, W.; Wu, H.J.; Chen, L.J.; Chen, H.W.; et al. The ethylene receptor ETR2 delays floral transition and affects starch accumulation in rice. Plant Cell 2009, 21, 1473–1494. [Google Scholar] [CrossRef]
- Li, X.K.; Huang, Y.H.; Zhao, R.; Cao, W.Q.; Lu, L.; Han, J.Q.; Zhou, Y.; Zhang, X.; Wu, W.A.; Tao, J.J.; et al. Membrane protein MHZ3 regulates the on-off switch of ethylene signaling in rice. Nat. Commun. 2024, 15, 5987. [Google Scholar] [CrossRef]
- Ma, B.; Zhou, Y.; Chen, H.; He, S.J.; Huang, Y.H.; Zhao, H.; Lu, X.; Zhang, W.K.; Pang, J.H.; Chen, S.Y.; et al. Membrane protein MHZ3 stabilizes OsEIN2 in rice by interacting with its Nramp-like domain. Proc. Natl. Acad. Sci. USA 2018, 115, 2520–2525. [Google Scholar] [CrossRef]
- Zhao, H.; Yin, C.; Ma, B.; Chen, S.; Zhang, J. Ethylene signaling in rice and Arabidopsis: New regulators and mechanisms. J. Integr. Plant Biol. 2021, 63, 102–125. [Google Scholar] [CrossRef]
- Zhao, H.; Duan, K.X.; Ma, B.; Yin, C.C.; Hu, Y.; Tao, J.J.; Huang, Y.H.; Cao, W.Q.; Chen, H.; Yang, C.; et al. Histidine kinase MHZ1/OsHK1 interacts with ethylene receptors to regulate root growth in rice. Nat. Commun. 2020, 11, 518. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Ma, B.; He, S.J.; Xiong, Q.; Duan, K.X.; Yin, C.C.; Chen, H.; Lu, X.; Chen, S.Y.; Zhang, J.S. MAOHUZI6/ETHYLENE INSENSITIVE3-LIKE1 and ETHYLENE INSENSITIVE3-LIKE2 regulate ethylene response of roots and coleoptiles and negatively affect salt tolerance in rice. Plant Physiol. 2015, 169, 148–165. [Google Scholar] [CrossRef] [PubMed]
- Li, X.K.; Yin, C.C.; Tao, J.J.; Chen, S.Y.; Zhao, H.; Zhang, J.S.; Li, X.K.; Yin, C.C.; Tao, J.J.; Chen, S.Y.; et al. Ethylene signaling in rice and Arabidopsis: From the perspective of protein complexes. Plant Horm. 2025, 1, e009. [Google Scholar] [CrossRef]
- Ma, B.; He, S.J.; Duan, K.X.; Yin, C.C.; Chen, H.; Yang, C.; Xiong, Q.; Song, Q.X.; Lu, X.; Chen, H.W.; et al. Identification of rice ethylene-response mutants and characterization of MHZ7/OSEIN2 in distinct ethylene response and yield trait regulation. Mol. Plant 2013, 6, 1830–1848. [Google Scholar] [CrossRef]
- Xu, K.; Xu, X.; Fukao, T.; Canlas, P.; Maghirang-Rodriguez, R.; Heuer, S.; Ismail, A.M.; Bailey-Serres, J.; Ronald, P.C.; Mackill, D.J. Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 2006, 442, 705–708. [Google Scholar] [CrossRef] [PubMed]
- Qin, H.; Pandey, B.K.; Li, Y.; Huang, G.; Wang, J.; Quan, R.; Zhou, J.; Zhou, Y.; Miao, Y.; Zhang, D.; et al. Orchestration of ethylene and gibberellin signals determines primary root elongation in rice. Plant Cell 2022, 34, 1273–1288. [Google Scholar] [CrossRef]
- Morgan, P.W.; Drew, M.C. Ethylene and plant responses to stress. Physiol. Plant. 1997, 100, 620–630. [Google Scholar] [CrossRef]
- Qin, H.; Zhang, Z.; Wang, J.; Chen, X.; Wei, P.; Huang, R. The activation of OsEIL1 on YUC8 transcription and auxin biosynthesis is required for ethylene-inhibited root elongation in rice early seedling development. PLoS Genet. 2017, 13, e1006955. [Google Scholar] [CrossRef]
- Yu, S.; Zheng, S.; Ning, J.; Shi, Y.; Guo, D.; Luo, R.; Xiao, G.; Saleem, S.; Ali, A.; Zhou, H.; et al. The rice OsCBL3-OsCIPK31 module regulates root development via abscisic acid and auxin signaling pathways. Crop J. 2025, 13, 694–704. [Google Scholar] [CrossRef]
- Zou, X.; Liu, L.; Hu, Z.; Wang, X.; Zhu, Y.; Zhang, J.; Li, X.; Kang, Z.; Lin, Y.; Yin, C. Salt-induced inhibition of rice seminal root growth is mediated by ethylene-jasmonate interaction. J. Exp. Bot. 2021, 72, 5656–5672. [Google Scholar] [CrossRef]
- Qin, H.; He, L.; Huang, R. The coordination of ethylene and other hormones in primary root development. Front. Plant Sci. 2019, 10, 874. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Xiong, Q.; Yin, C.; Ma, B.; Chen, S.; Zhang, J. Ethylene biosynthesis, signaling, and crosstalk with other hormones in rice. Small Methods 2020, 4, 1900278. [Google Scholar] [CrossRef]
- Mashiguchi, K.; Tanaka, K.; Sakai, T.; Sugawara, S.; Kawaide, H.; Natsume, M.; Hanada, A.; Yaeno, T.; Shirasu, K.; Yao, H.; et al. The main auxin biosynthesis pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 18512–18517. [Google Scholar] [CrossRef]
- Růžička, K.; Ljung, K.; Vanneste, S.; Podhorská, R.; Beeckman, T.; Friml, J.; Benková, E. Ethylene regulates root growth through effects on auxin biosynthesis and transport-dependent auxin distribution. Plant Cell 2007, 19, 2197–2212. [Google Scholar] [CrossRef]
- Yin, C.C.; Ma, B.; Collinge, D.P.; Pogson, B.J.; He, S.J.; Xiong, Q.; Duan, K.X.; Chen, H.; Yang, C.; Lu, X.; et al. Ethylene responses in rice roots and coleoptiles are differentially regulated by a carotenoid isomerase-mediated abscisic acid pathway. Plant Cell 2015, 27, 1061–1081. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Chen, Z.; Gao, J.; Gong, Z. Abscisic acid inhibits root growth in Arabidopsis through ethylene biosynthesis. Plant J. 2014, 79, 44–55. [Google Scholar] [CrossRef]
- Clark, L.J.; Whalley, W.R.; Barraclough, P.B. How do roots penetrate strong soil? Plant Soil 2003, 255, 93–104. [Google Scholar] [CrossRef]
- Zhou, Y.; Gao, Y.H.; Zhang, B.C.; Yang, H.L.; Tian, Y.B.; Huang, Y.H.; Yin, C.C.; Tao, J.J.; Wei, W.; Zhang, W.K.; et al. CELLULOSE SYNTHASE-LIKE C proteins modulate cell wall establishment during ethylene-mediated root growth inhibition in rice. Plant Cell 2024, 36, 3751–3769. [Google Scholar] [CrossRef]
- Yu, P.; Gutjahr, C.; Li, C.; Hochholdinger, F. Genetic control of lateral root formation in cereals. Trends Plant Sci. 2016, 21, 951–961. [Google Scholar] [CrossRef]
- Muday, G.K.; Rahman, A.; Binder, B.M. Auxin and ethylene: Collaborators or competitors? Trends Plant Sci. 2012, 17, 181–195. [Google Scholar] [CrossRef] [PubMed]
- Steffens, B.; Rasmussen, A. The physiology of adventitious roots. Plant Physiol. 2016, 170, 603–617. [Google Scholar] [CrossRef]
- Zhao, Y.; Cheng, S.; Song, Y.; Huang, Y.; Zhou, S.; Liu, X.; Zhou, D.X. The interaction between rice ERF3 and WOX11 promotes crown root development by regulating gene expression involved in cytokinin signaling. Plant Cell 2015, 27, 2469–2483. [Google Scholar] [CrossRef]
- Song, L.; Yu, H.; Dong, J.; Che, X.; Jiao, Y.; Liu, D. The molecular mechanism of ethylene-mediated root hair development induced by phosphate starvation. PLoS Genet. 2016, 12, e1006194. [Google Scholar] [CrossRef] [PubMed]
- Moon, S.; Cho, L.H.; Kim, Y.J.; Gho, Y.S.; Jeong, H.Y.; Hong, W.J.; Lee, C.; Park, H.; Jwa, N.S.; Dangol, S.; et al. RSL class II transcription factors guide the nuclear localization of RHL1 to regulate root hair development. Plant Physiol. 2019, 179, 558–568. [Google Scholar] [CrossRef]
- Xiong, H.; Yu, J.; Miao, J.; Li, J.; Zhang, H.; Wang, X.; Liu, P.; Zhao, Y.; Jiang, C.; Yin, Z.; et al. Natural variation in OsLG3 increases drought tolerance in rice by inducing ROS scavenging. Plant Physiol. 2018, 178, 451–467. [Google Scholar] [CrossRef] [PubMed]
- Kong, X.; Xiong, Y.; Song, X.; Wadey, S.; Yu, S.; Rao, J.; Lale, A.; Lombardi, M.; Fusi, R.; Bhosale, R.; et al. Ethylene regulates auxin-mediated root gravitropic machinery and controls root angle in cereal crops. Plant Physiol. 2024, 195, 1969–1980. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wan, L.; Zhang, L.; Zhang, Z.; Zhang, H.; Quan, R.; Zhou, S.; Huang, R. An ethylene response factor OsWR1 responsive to drought stress transcriptionally activates wax synthesis related genes and increases wax production in rice. Plant Mol. Biol. 2012, 78, 275–288. [Google Scholar] [CrossRef]
- Yang, J.C.; Zhang, J.H.; Ye, Y.X.; Wang, Z.Q.; Zhu, Q.S.; Liu, L.J. Involvement of abscisic acid and ethylene in the responses of rice grains to water stress during filling. Plant Cell Environ. 2004, 27, 1055–1064. [Google Scholar] [CrossRef]
- Young, T.E.; Meeley, R.B.; Gallie, D.R. ACC synthase expression regulates leaf performance and drought tolerance in maize. Plant J. 2004, 40, 813–825. [Google Scholar] [CrossRef]
- Huang, S.; Wang, H.; Liu, S.; Lu, S.; Hua, J.; Zou, B. Ethylene antagonizes ABA and inhibits stomatal closure and chilling tolerance in rice. J. Exp. Bot. 2025, 76, 5011–5024. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.C.; Liu, K.; Zhang, S.F.; Wang, X.M.; Wang, Z.Q.; Liu, L.J. Hormones in rice spikelets in responses to water stress during meiosis. Acta Agron. Sin. 2008, 34, 111–118. [Google Scholar] [CrossRef]
- Peng, Y.; Zhou, Z.; Tong, R.; Hu, X.; Du, K. Anatomy and ultrastructure adaptations to soil flooding of two full-sib poplar clones differing in flood-tolerance. Flora 2017, 233, 90–98. [Google Scholar] [CrossRef]
- Hartman, S.; Liu, Z.; van Veen, H.; Vicente, J.; Reinen, E.; Martopawiro, S.; Zhang, H.; van Dongen, N.; Bosman, F.; Bassel, G.W.; et al. Ethylene-mediated nitric oxide depletion pre-adapts plants to hypoxia stress. Nat. Commun. 2019, 10, 4020. [Google Scholar] [CrossRef]
- Weits, D.A.; Giuntoli, B.; Kosmacz, M.; Parlanti, S.; Hubberten, H.M.; Riegler, H.; Hoefgen, R.; Perata, P.; van Dongen, J.T.; Licausi, F. Plant cysteine oxidases control the oxygen-dependent branch of the N-end-rule pathway. Nat. Commun. 2014, 5, 3425. [Google Scholar] [CrossRef]
- Yamauchi, T.; Yoshioka, M.; Fukazawa, A.; Mori, H.; Nishizawa, N.K.; Tsutsumi, N.; Yoshioka, H.; Nakazono, M. An NADPH oxidase RBOH functions in rice roots during lysigenous aerenchyma formation under oxygen-deficient conditions. Plant Cell 2017, 29, 775–790. [Google Scholar] [CrossRef]
- Vriezen, W.H.; Zhou, Z.; Van Der Straeten, D. Regulation of submergence-induced enhanced shoot elongation in Oryza sativa L. Ann. Bot. 2003, 91, 263–270. [Google Scholar] [CrossRef] [PubMed]
- Steffens, B.; Sauter, M. Epidermal cell death in rice is confined to cells with a distinct molecular identity and is mediated by ethylene and H2O2 through an autoamplified signal pathway. Plant Cell 2009, 21, 184–196. [Google Scholar] [CrossRef]
- Fukao, T.; Yeung, E.; Bailey-Serres, J. The submergence tolerance regulator SUB1A mediates crosstalk between submergence and drought tolerance in rice. Plant Cell 2011, 23, 412–427. [Google Scholar] [CrossRef] [PubMed]
- Qin, H.; Wang, J.; Chen, X.; Wang, F.; Peng, P.; Zhou, Y.; Miao, Y.; Zhang, Y.; Gao, Y.; Qi, Y.; et al. Rice OsDOF15 contributes to ethylene-inhibited primary root elongation under salt stress. New Phytol. 2019, 223, 798–813. [Google Scholar] [CrossRef] [PubMed]
- Sackey, O.K.; Feng, N.; Mohammed, Y.Z.; Dzou, C.F.; Zheng, D.; Zhao, L.; Shen, X. A comprehensive review on rice responses and tolerance to salt stress. Front. Plant Sci. 2025, 16, 1561280. [Google Scholar] [CrossRef]
- Steffens, B. The role of ethylene and ROS in salinity, heavy metal, and flooding responses in rice. Front. Plant Sci. 2014, 5, 685. [Google Scholar] [CrossRef]
- Liang, S.; Xiong, W.; Yin, C.; Xie, X.; Jin, Y.; Zhang, S.; Yang, B.; Ye, G.; Chen, S.; Luan, W. Overexpression of OsARD1 improves submergence, drought, and salt tolerances of seedling through the enhancement of ethylene synthesis in rice. Front. Plant Sci. 2019, 10, 1088. [Google Scholar] [CrossRef]
- Angon, P.B.; Islam, M.S.; Kc, S.; Das, A.; Anjum, N.; Poudel, A.; Suchi, S.A. Sources, effects and present perspectives of heavy metals contamination: Soil, plants and human food chain. Heliyon 2024, 10, e28357. [Google Scholar] [CrossRef]
- Saini, S.; Kaur, N.; Pati, P.K. Phytohormones: Key players in the modulation of heavy metal stress tolerance in plants. Ecotoxicol. Environ. Saf. 2021, 223, 112578. [Google Scholar] [CrossRef]
- Haider, F.U.; Liqun, C.; Coulter, J.A.; Cheema, S.A.; Wu, J.; Zhang, R.; Wenjun, M.; Farooq, M. Cadmium toxicity in plants: Impacts and remediation strategies. Ecotoxicol. Environ. Saf. 2021, 211, 111887. [Google Scholar] [CrossRef]
- Chen, H.; Zhang, Q.; Lv, W.; Yu, X.; Zhang, Z. Ethylene positively regulates Cd tolerance via reactive oxygen species scavenging and apoplastic transport barrier formation in rice. Environ. Pollut. 2022, 302, 119063. [Google Scholar] [CrossRef] [PubMed]
- Tao, Q.; Liu, J.; Zhang, K.; Yan, M.; Li, M.; Wu, Y.; Wang, C.; Li, B. Ethylene-mediated root endodermal barrier development in impeding Cd radial transport and accumulation in rice (Oryza sativa L.). Plant Physiol. Biochem. 2025, 219, 109313. [Google Scholar] [CrossRef]
- Azhar, W.; Khan, A.R.; Salam, A.; Ulhassan, Z.; Qi, J.; Shah, G.; Liu, Y.; Chunyan, Y.; Yang, S.; Gan, Y. Ethylene accelerates copper oxide nanoparticle-induced toxicity at physiological, biochemical, and ultrastructural levels in rice seedlings. Environ. Sci. Pollut. Res. 2023, 30, 26137–26149. [Google Scholar] [CrossRef] [PubMed]
- Bal, H.B.; Nayak, L.; Das, S.; Adhya, T.K. Isolation of ACC deaminase producing PGPR from rice rhizosphere and evaluating their plant growth promoting activity under salt stress. Plant Soil 2013, 366, 93–105. [Google Scholar] [CrossRef]
- Sodhi, G.K.; Saxena, S. Plant growth promotion and abiotic stress mitigation in rice using endophytic fungi: Advances made in the last decade. Environ. Exp. Bot. 2023, 209, 105312. [Google Scholar] [CrossRef]
- Harbort, C.J.; Hashimoto, M.; Inoue, H.; Niu, Y.; Guan, R.; Rombolà, A.D.; Kopriva, S.; Voges, M.J.E.E.E.; Sattely, E.S.; Garrido-Oter, R.; et al. Root-secreted coumarins and the microbiota interact to improve iron nutrition in Arabidopsis. Cell Host Microbe 2020, 28, 825–837.e6. [Google Scholar] [CrossRef]
- Halshoy, H.S.; Braim, S.A.; Hama, J.R. Phytohormonal regulation of root exudation: Mechanisms and rhizosphere function. Plant Signal. Behav. 2025, 20, 2587486. [Google Scholar] [CrossRef]
- Badri, D.V.; Vivanco, J.M. Regulation and function of root exudates. Plant Cell Environ. 2009, 32, 666–681. [Google Scholar] [CrossRef]
- Bais, H.P.; Weir, T.L.; Perry, L.G.; Gilroy, S.; Vivanco, J.M. The role of root exudates in rhizosphere interactions with plants and other organisms. Annu. Rev. Plant Biol. 2006, 57, 233–266. [Google Scholar] [CrossRef]
- Dimkpa, C.; Weinand, T.; Asch, F. Plant-rhizobacteria interactions alleviate abiotic stress conditions. Plant Cell Environ. 2009, 32, 1682–1694. [Google Scholar] [CrossRef]
- Barnawal, D.; Maji, D.; Bharti, N.; Chanotiya, C.S.; Kalra, A. ACC deaminase-containing bacillus subtilis reduces stress ethylene-induced damage and improves mycorrhizal colonization and rhizobial nodulation in trigonella foenum-graecum under drought stress. J. Plant Growth Regul. 2013, 32, 809–822. [Google Scholar] [CrossRef]
- Glick, B.R. Plant growth-promoting bacteria: Mechanisms and applications. Scientifica 2012, 2012, 963401. [Google Scholar] [CrossRef]
- Van Wees, S.C.M.; Van der Ent, S.; Pieterse, C.M. Plant immune responses triggered by beneficial microbes. Curr. Opin. Plant Biol. 2008, 11, 443–448. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Qiu, Y.L. Phylogenetic distribution and evolution of mycorrhizas in land plants. Mycorrhiza 2006, 16, 299–363. [Google Scholar] [CrossRef] [PubMed]
- Kalamulla, R.; Karunarathna, S.C.; Tibpromma, S.; Galappaththi, M.C.A.; Suwannarach, N.; Stephenson, S.L.; Asad, S.; Salem, Z.S.; Yapa, N. Arbuscular mycorrhizal fungi in sustainable agriculture. Sustainability 2022, 14, 12250. [Google Scholar] [CrossRef]
- Das, D.; Varshney, K.; Ogawa, S.; Torabi, S.; Huettl, R.; Nelson, D.C.; Gutjahr, C. Ethylene promotes SMAX1 accumulation to inhibit arbuscular mycorrhiza symbiosis. Nat. Commun. 2025, 16, 2025. [Google Scholar] [CrossRef]
- Carbonnel, S.; Das, D.; Varshney, K.; Kolodziej, M.C.; Villaécija-Aguilar, J.A.; Gutjahr, C. The karrikin signaling regulator SMAX1 controls Lotus japonicus root and root hair development by suppressing ethylene biosynthesis. Proc. Natl. Acad. Sci. USA 2020, 117, 21757–21765. [Google Scholar] [CrossRef]
- Helliwell, E.E.; Wang, Q.; Yang, Y. Transgenic rice with inducible ethylene production exhibits broad-spectrum disease resistance to the fungal pathogens Agnaporthe oryzae and Rhizoctonia solani. Plant Biotechnol. J. 2013, 11, 33–42. [Google Scholar] [CrossRef]
- Yang, C.; Li, W.; Cao, J.; Meng, F.; Yu, Y.; Huang, J.; Jiang, L.; Liu, M.; Zhang, Z.; Chen, X.; et al. Activation of ethylene signaling pathways enhances disease resistance by regulating ROS and phytoalexin production in rice. Plant J. Cell Mol. Biol. 2017, 89, 338–353. [Google Scholar] [CrossRef]
- Zhai, K.; Liang, D.; Li, H.; Jiao, F.; Yan, B.; Liu, J.; Lei, Z.; Huang, L.; Gong, X.; Wang, X.; et al. NLRs guard metabolism to coordinate pattern- and effector-triggered immunity. Nature 2022, 601, 245–251. [Google Scholar] [CrossRef] [PubMed]
| Search Phase | Primary Databases or Sources | Representative Keyword Combinations | Key Screening and Inclusion Criteria |
|---|---|---|---|
| Phase 1: Broad Identification | Web of Science, Scopus, PubMed, Google Scholar | (Oryza sativa OR rice) AND ethylene AND (root system architecture OR root development) | Relevance to core topic; exclusion of non-research articles (e.g., non-peer-reviewed reports). |
| Phase 2: Targeted Expansion | RiceXPro, Publisher Portals (e.g., ScienceDirect) | Core keywords extended with: stress (drought, salinity, hypoxia), root-microbe interactions, ACS/ACO, signaling pathway | Peer-reviewed original research; must include experimental data on ethylene’s role in rice roots. |
| Phase 3: Final Curation | Cross-check across all above sources | Combined use of core and extended keywords | 1. Central focus on ethylene in rice root biology; 2. Publication period: 1979–2025; 3. Emphasis on influential studies (assessed by citation count as a heuristic). |
| Stress Type | Core Role of Ethylene | Key Molecular Mechanisms | Impact on Root Architecture and Physiology | Reference |
|---|---|---|---|---|
| Drought | Dual role; beneficial at low levels, harmful when excessive. | Positive: Induces drought-responsive TFs (OsLG3); promotes wax synthesis (OsWR1). Negative: Antagonizes ABA signaling; triggers senescence programs. | Adaptive: 1. Promotes a compact, deeply anchored RSA, improving water uptake; 2. Reduces transpirational water loss; 3. Facilitates grain filling under moderate stress. Detrimental: 1. Exacerbates water loss; 2. Leads to loss of photosynthetic capacity; 3. Significantly increases spikelet sterility. | [51,52,53,54,55,56,57] |
| Hypoxia | Core adaptive signal; survival trade-offs occur at high levels. | Positive: Stabilizes ERF-VII TFs (e.g., SK1/SK2, Sub1A); activates RBOH for aerenchyma. Negative: Causes ROS overproduction; suppresses photosynthesis. | Adaptive: 1. Forms lysigenous aerenchyma, enhancing internal O2 transport; 2. Promotes internode elongation (escape) or triggers quiescence (endurance). Detrimental: 1. Causes plant lodging due to reduced stem toughness; 2. Induces oxidative cell damage; 3. Leads to carbon depletion. | [26,59,60,61,62,63,64] |
| Salinity | Primarily negative; positive modulation occurs in specific pathways. | Positive: Synergizes with ABA; activates antioxidants; OsDOF15 suppresses OsACS1. Negative: OsEIL1/2 upregulate OsHKT2;1 (Na+ uptake); induces JA. | Adaptive: 1. Alleviates osmotic stress; 2. Mitigates oxidative damage; 3. Maintains root growth via suppressed ethylene biosynthesis. Detrimental: 1. Exacerbates ionic toxicity; 2. Impairs root growth and salt tolerance. | [28,36,65,66,67,68] |
| Heavy Metal | Early positive, late negative; dose- and time-dependent. | Positive: Upregulates ACS/ACO; EIN2 enhances apoplastic barriers (Casparian strip). Negative: Disrupts ROS homeostasis; promotes cell death. | Adaptive: 1. Strengthens apoplastic barriers, reducing Cd translocation to shoots; 2. Alleviates oxidative damage. Detrimental: 1. Inhibits root elongation; 2. Exacerbates oxidative stress damage. | [72,73,74] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2026 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Zhang, N.; Lv, X.; Yan, Y.; Meng, Q.; Wang, C.; Jing, W.; Zhang, Y.; Xiao, Z.; Zhang, H. Physiological and Molecular Mechanisms of Ethylene in Sculpting Rice Root System Architecture. Agronomy 2026, 16, 355. https://doi.org/10.3390/agronomy16030355
Zhang N, Lv X, Yan Y, Meng Q, Wang C, Jing W, Zhang Y, Xiao Z, Zhang H. Physiological and Molecular Mechanisms of Ethylene in Sculpting Rice Root System Architecture. Agronomy. 2026; 16(3):355. https://doi.org/10.3390/agronomy16030355
Chicago/Turabian StyleZhang, Nan, Xinping Lv, Yu Yan, Qinghao Meng, Chaorui Wang, Wenjiang Jing, Ying Zhang, Zhilin Xiao, and Hao Zhang. 2026. "Physiological and Molecular Mechanisms of Ethylene in Sculpting Rice Root System Architecture" Agronomy 16, no. 3: 355. https://doi.org/10.3390/agronomy16030355
APA StyleZhang, N., Lv, X., Yan, Y., Meng, Q., Wang, C., Jing, W., Zhang, Y., Xiao, Z., & Zhang, H. (2026). Physiological and Molecular Mechanisms of Ethylene in Sculpting Rice Root System Architecture. Agronomy, 16(3), 355. https://doi.org/10.3390/agronomy16030355







