Spectral Estimation of Nitrogen Content in Cotton Leaves Under Coupled Nitrogen and Phosphorus Conditions
Abstract
1. Introduction
2. Materials and Methods
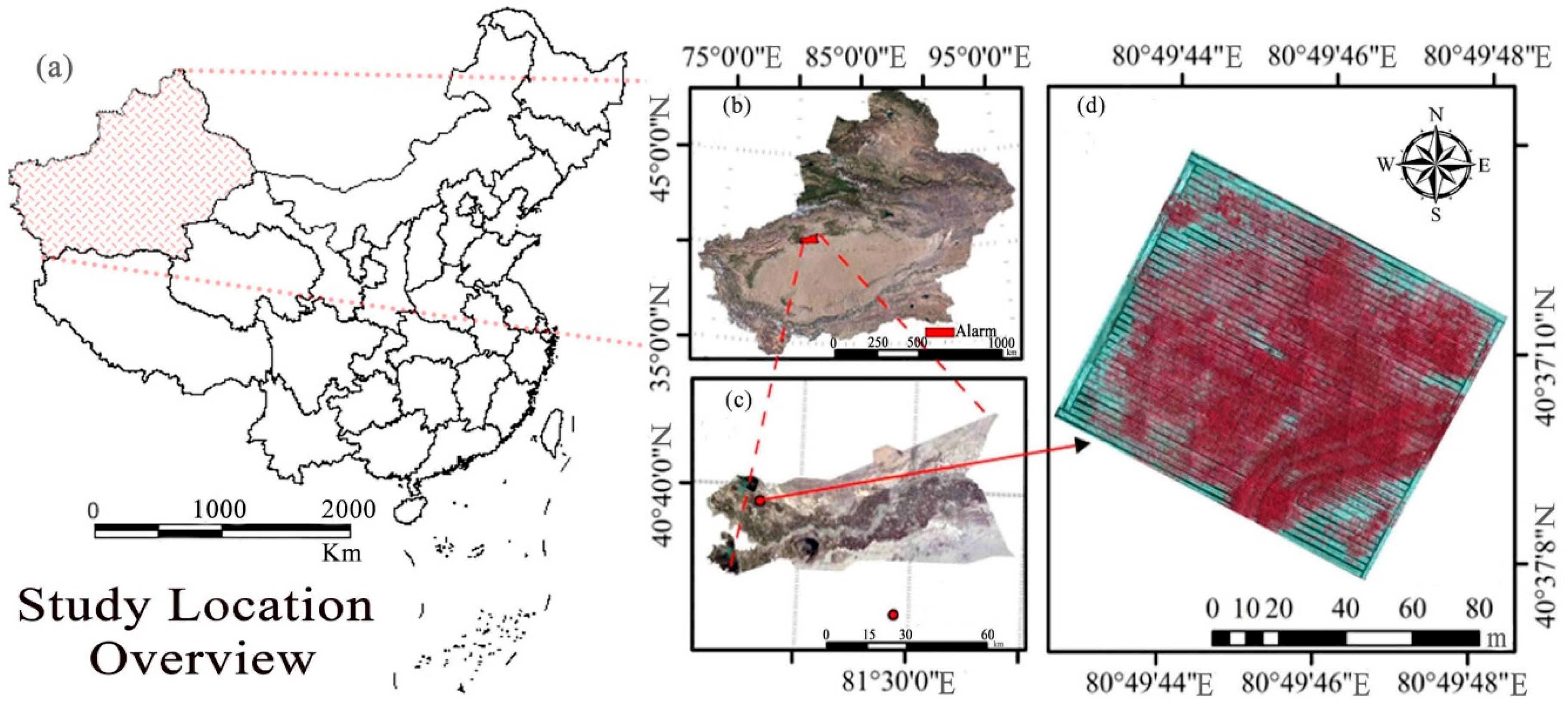
2.1. Overview of the Study Area
2.2. Experimental Design
2.3. Data Acquisition
2.3.1. Collection and Determination of Cotton Samples
2.3.2. Determination of the Spectral Data of the Cotton Leaf
2.4. Data Processing and Analysis
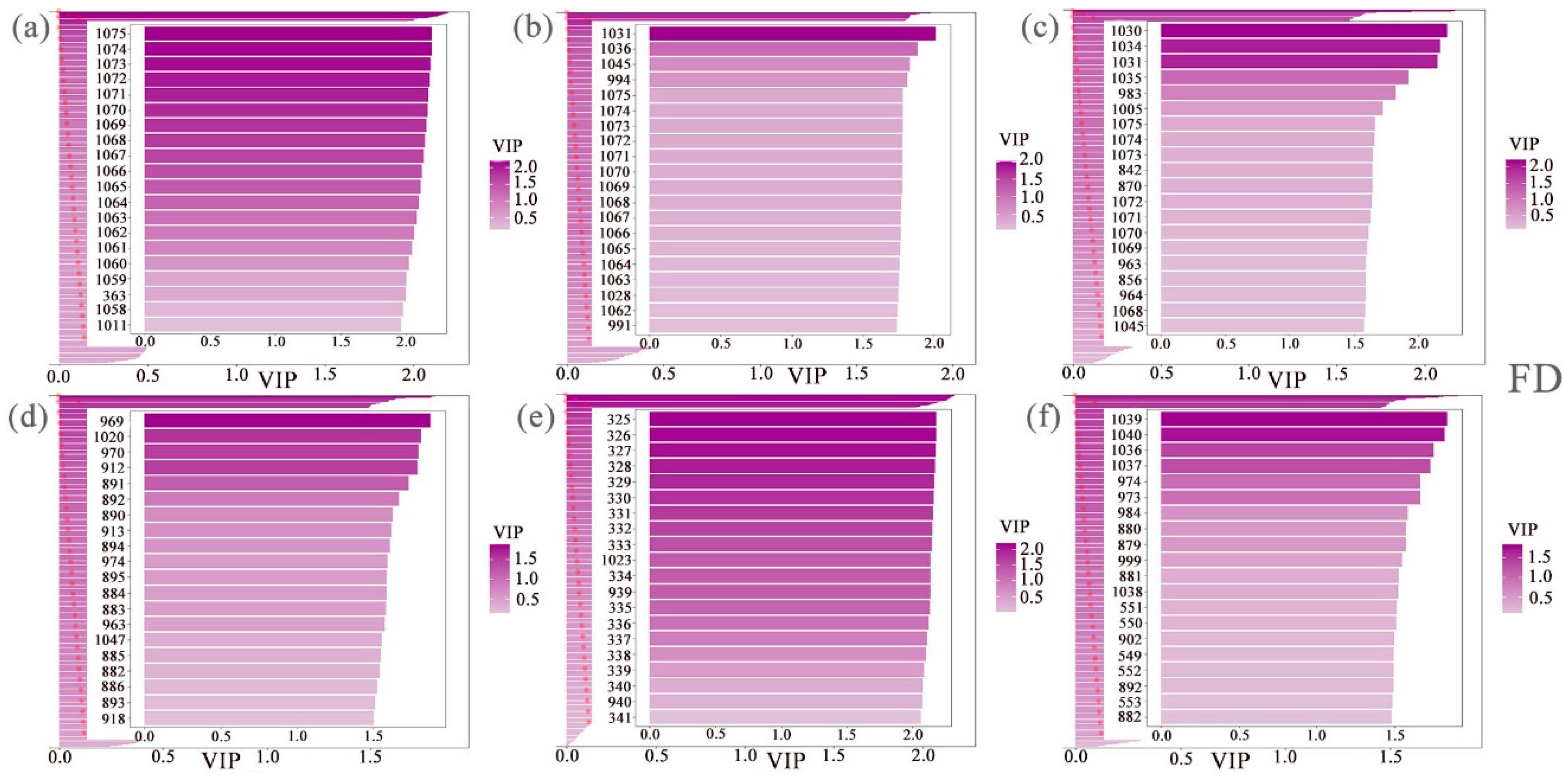
2.4.1. Screening Feature Band
2.4.2. Model Construction and Accuracy Checking
3. Results
3.1. Cotton LNCs Variations at Different Growth Stages Under Different Nitrogen and Phosphorus Coupling Conditions
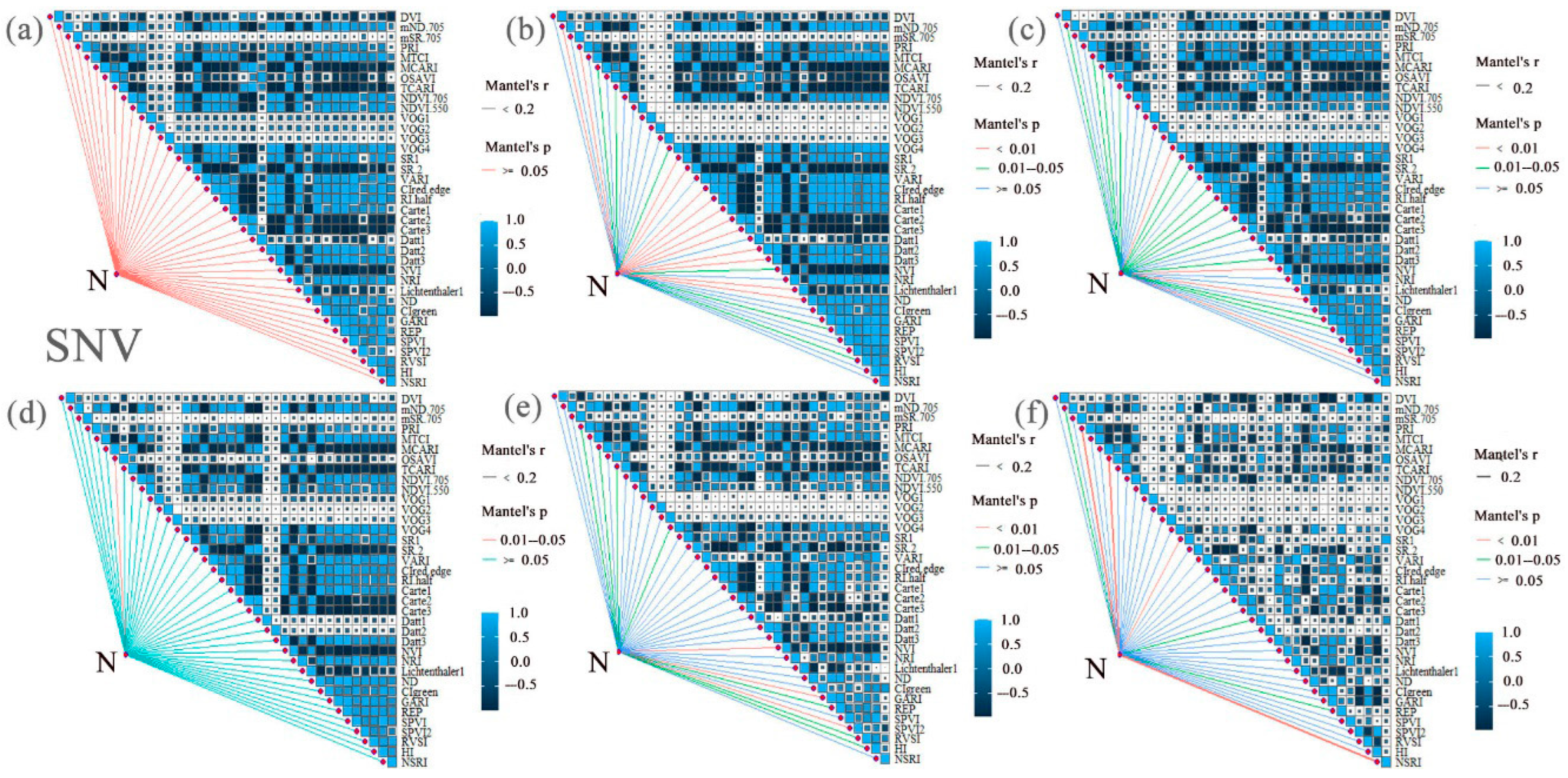
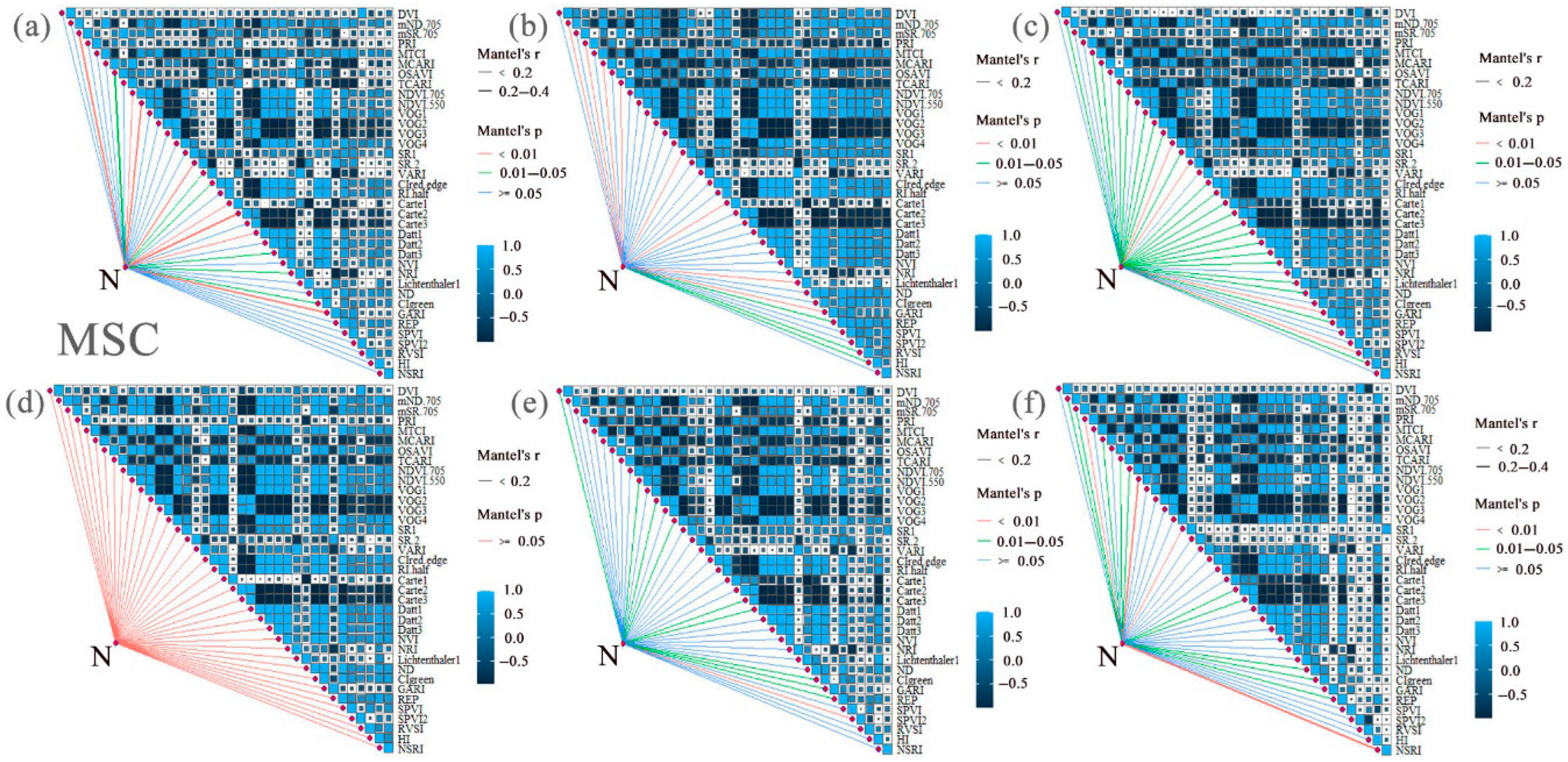
3.2. Correlation Analysis and Extraction of LNCs and SIs from Cotton Under Different Nitrogen and Phosphorus Coupling Conditions
3.3. Extraction of the Spectral Reflectance of Cotton Leaves Under Different Nitrogen and Phosphorus Coupling Conditions Via PLS–DA
3.4. Modeling of Cotton LNCs via Hyperspectral Technology
4. Discussion
4.1. The Influence of Nitrogen-Phosphorus Coupling on LNCs in Cotton
4.2. Potential of Spectral Pretreatment Techniques for Crop LNC Estimation
4.3. Advantages of Spectral Fusion in Estimating Crop LNCs
5. Conclusions
- (1)
- Under the conditions of nitrogen and phosphorus coupling, cotton LNC predictions performed better under N3P1, N3P0, N2P2, N2P2, N0P3, and N3P2 at the squaring stage, initial bloom stage, peak bloom stage, initial boll stage, peak boll stage, and boll opening stage, respectively, showing a trend of first increasing and then decreasing, and the LNCs reached a maximum at the peak bloom stage.
- (2)
- The results revealed that SVM–MSC in the squaring stage, SVM–FD in the initial bloom stage, SVM–FD in the peak bloom stage, SVM–FD in the initial boll stage, RF–SNV in the peak boll stage, and SVM–FD in the boll opening stage could be used as LNC recognition models for cotton. FD showed the best performance compared with the other three treatments, and the SVM model had a higher R2 value and lower RMSE value than the RF model.
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Dai, J.; Kong, X.; Zhang, D.; Li, W.; Dong, H. Technologies and theoretical basis of light and simplified cotton cultivation in China. Field Crops Res. 2017, 214, 142–148. [Google Scholar] [CrossRef]
- Ata-Ul-Karim, S.T.; Cang, L.; Wang, Y.; Zhou, D. Effects of soil properties, nitrogen application, plant phenology, and their interactions on plant uptake of cadmium in wheat. J. Hazard. Mater. 2020, 384, 121452. [Google Scholar] [CrossRef]
- Ma, J.; He, P.; Xu, X.; He, W.; Liu, Y.; Yang, F.; Chen, F.; Li, S.; Tu, S.; Jin, J.; et al. Temporal and spatial changes in soil available phosphorus in China (1990–2012). Field Crops Res. 2016, 192, 13–20. [Google Scholar] [CrossRef]
- Lemercier, B.; Gaudin, L.; Walter, C.; Aurousseau, P.; Arrouays, D.; Schvartz, C.; Saby, N.P.A.; Follain, S.; Abrassart, J. Soil phosphorus monitoring at the regional level by means of a soil test database. Soil Use Manag. 2008, 24, 131–138. [Google Scholar] [CrossRef]
- Sonobe, R.; Hirono, Y. Carotenoid Content Estimation in Tea Leaves Using Noisy Reflectance Data. Remote Sens. 2023, 15, 4303. [Google Scholar] [CrossRef]
- Falcioni, R.; Antunes, W.C.; Demattê, J.A.M.; Nanni, M.R. A Novel Method for Estimating Chlorophyll and Carotenoid Concentrations in Leaves: A Two Hyperspectral Sensor Approach. Sensors 2023, 23, 3843. [Google Scholar] [CrossRef]
- Yang, Z.; Tian, J.; Zhang, L.; Zan, O.; Yan, X.; Feng, K. Spectral detection of leaf carbon and nitrogen as a proxy for remote assessment of photosynthetic capacity for wheat and maize under nitrogen stress. Comput. Electron. Agric. 2024, 224, 109174. [Google Scholar] [CrossRef]
- Li, D.; Hu, Q.; Zhang, J.; Dian, Y.; Hu, C.; Zhou, J. Leaf Nitrogen and Phosphorus Variation and Estimation of Citrus Tree under Two Labor-Saving Cultivation Modes Using Hyperspectral Data. Remote Sens. 2024, 16, 3261. [Google Scholar] [CrossRef]
- Li, J.; Shi, L.; Mo, X.; Hu, X.; Su, C.; Han, J.; Deng, X.; Du, S.; Li, S. Self-correcting deep learning for estimating rice leaf nitrogen concentration with mobile phone images. Comput. Electron. Agric. 2024, 227, 109497. [Google Scholar] [CrossRef]
- Hu, W.; Tang, W.; Li, C.; Wu, J.; Liu, H.; Wang, C.; Luo, X.; Tang, R. Handling the Challenges of Small-Scale Labeled Data and Class Imbalances in Classifying the N and K Statuses of Rubber Leaves Using Hyperspectroscopy Techniques. Plant Phenomics 2024, 6, 0154. [Google Scholar] [CrossRef] [PubMed]
- Furlanetto, R.H.; Crusiol, L.G.T.; Gonçalves, J.V.F.; Nanni, M.R.; Junior, A.d.O.; de Oliveira, F.A.; Sibaldelli, R.N.R. Machine learning as a tool to predict potassium concentration in soybean leaf using hyperspectral data. Precis. Agric. 2023, 24, 2264–2292. [Google Scholar] [CrossRef]
- Jin, X.; Xu, L.; Feng, H.; Wang, K.; Niu, J.; Su, X.; Chen, L.; Zheng, H.; Huang, J. Optimizing Multidimensional Spectral Indices and Ensemble Learning Methods for Estimating Nitrogen Content in Torreya grandis Leaves Based on UAV Hyperspectral. Forests 2024, 16, 40. [Google Scholar] [CrossRef]
- Wang, J.; Li, Z.; Jin, X.; Liang, G.; Struik, P.C.; Gu, J.; Zhou, Y. Phenotyping flag leaf nitrogen content in rice using a three-band spectral index. Comput. Electron. Agric. 2019, 162, 475–481. [Google Scholar] [CrossRef]
- Yang, S.; Li, J.; Li, J.; Zhang, X.; Ma, C.; Liu, Z.; Ren, M. Estimating the Canopy Nitrogen Content in Maize by Using the Transform-Based Dynamic Spectral Indices and Random Forest. Sustainability 2024, 16, 8011. [Google Scholar] [CrossRef]
- Wang, J.; Wang, H.; Tian, T.; Cui, J.; Shi, X.; Song, J.; Li, T.; Li, W.; Zhong, M.; Zhang, W. Construction of spectral index based on multi-angle spectral data for estimating cotton leaf nitrogen concentration. Comput. Electron. Agric. 2022, 201, 107328. [Google Scholar] [CrossRef]
- Lu, M.; Wang, H.; Xu, J.; Wei, Z.; Li, Y.; Hu, J.; Tian, S. A Vis/NIRS device for evaluating leaf nitrogen content using K-means algorithm and feature extraction methods. Comput. Electron. Agric. 2024, 225, 109301. [Google Scholar] [CrossRef]
- Tan, J.; Ding, J.; Wang, Z.; Han, L.; Wang, X.; Li, Y.; Zhang, Z.; Meng, S.; Cai, W.; Hong, Y. Estimating soil salinity in mulched cotton fields using UAV-based hyperspectral remote sensing and a Seagull Optimization Algorithm-Enhanced Random Forest Model. Comput. Electron. Agric. 2024, 221, 109017. [Google Scholar] [CrossRef]
- Yang, H.; Yin, H.; Li, F.; Hu, Y.; Yu, K. Machine learning models fed with optimized spectral indices to advance crop nitrogen monitoring. Field Crops Res. 2023, 293, 108844. [Google Scholar] [CrossRef]
- Qin, S.; Ding, Y.; Zhou, T.; Zhai, M.; Zhang, Z.; Fan, M.; Lv, X.; Zhang, Z.; Zhang, L. “Image-Spectral” fusion monitoring of small cotton samples nitrogen content based on improved deep forest. Comput. Electron. Agric. 2024, 221, 109002. [Google Scholar] [CrossRef]
- Sun, Q.; Gu, X.; Chen, L.; Qu, X.; Zhang, S.; Zhou, J.; Pan, Y. Hyperspectral estimation of maize (Zea mays L.) yield loss under lodging stress. Field Crops Res. 2023, 302, 109042. [Google Scholar] [CrossRef]
- Wang, J.; Tian, T.; Wang, H.; Cui, J.; Zhu, Y.; Zhang, W.; Tong, X.; Zhou, T.; Yang, Z.; Sun, J. Estimating cotton leaf nitrogen by combining the bands sensitive to nitrogen concentration and oxidase activities using hyperspectral imaging. Comput. Electron. Agric. 2021, 189, 106390. [Google Scholar] [CrossRef]
- Flynn, K.C.; Witt, T.W.; Baath, G.S.; Chinmayi, H.K.; Smith, D.R.; Gowda, P.H.; Ashworth, A.J. Hyperspectral reflectance and machine learning for multi-site monitoring of cotton growth. Smart Agric. Technol. 2024, 9, 100536. [Google Scholar] [CrossRef]
- Yin, C.; Lv, X.; Zhang, L.; Ma, L.; Wang, H.; Zhang, L.; Zhang, Z. Hyperspectral UAV Images at Different Altitudes for Monitoring the Leaf Nitrogen Content in Cotton Crops. Remote Sens. 2022, 14, 2576. [Google Scholar] [CrossRef]
- Shu, M.; Zhu, J.; Yang, X.; Gu, X.; Li, B.; Ma, Y. A spectral decomposition method for estimating the leaf nitrogen status of maize by UAV-based hyperspectral imaging. Comput. Electron. Agric. 2023, 212, 108100. [Google Scholar] [CrossRef]
- Chen, R.; Liu, W.; Yang, H.; Jin, X.; Yang, G.; Zhou, Y.; Zhang, C.; Han, S.; Meng, Y.; Zhai, C.; et al. A novel framework to assess apple leaf nitrogen content: Fusion of hyperspectral reflectance and phenology information through deep learning. Comput. Electron. Agric. 2024, 219, 108816. [Google Scholar] [CrossRef]
- Shao, Y.; Yang, W.; Wang, J.; Lu, Z.; Zhang, M.; Chen, D. Cotton Disease Recognition Method in Natural Environment Based on Convolutional Neural Network. Agriculture 2024, 14, 1577. [Google Scholar] [CrossRef]
- Aierken, N.; Yang, B.; Li, Y.; Jiang, P.; Pan, G.; Li, S. A review of unmanned aerial vehicle based remote sensing and machine learning for cotton crop growth monitoring. Comput. Electron. Agric. 2024, 227, 109601. [Google Scholar] [CrossRef]
- Karmakar, P.; Teng, S.W.; Murshed, M.; Pang, S.; Li, Y.; Lin, H. Crop monitoring by multimodal remote sensing: A review. Remote Sens. Appl. Soc. Environ. 2024, 33, 101093. [Google Scholar] [CrossRef]
- Stein, M.; Bargoti, S.; Underwood, J. Image Based Mango Fruit Detection, Localisation and Yield Estimation Using Multiple View Geometry. Sensors 2016, 16, 1915. [Google Scholar] [CrossRef]
- Savitzky, A.; Golay, M.J.E. Smoothing and Differentiation of Data by Simplified Least Squares Procedures. Anal. Chem. 1964, 36, 1627–1639. [Google Scholar] [CrossRef]
- Chong, I.; Jun, C. Performance of some variable selection methods when multicollinearity is present. Chemom. Intell. Lab. Syst. 2004, 78, 103–112. [Google Scholar] [CrossRef]
- Montero, D.; Aybar, C.; Mahecha, M.D.; Martinuzzi, F.; Söchting, M.; Wieneke, S. A standardized catalogue of spectral indices to advance the use of remote sensing in Earth system research. Sci. Data 2023, 10, 197. [Google Scholar] [CrossRef]
- Richardson, A.J.; Wiegand, A. Distinguishing vegetation from soil background information. Photogramm. Eng. Remote Sens. 1977, 43, 1541–1552. [Google Scholar]
- Sims, D.A.; Gamon, J.A. Relationships between leaf pigment content and spectral reflectance across a wide range of species, leaf structures and developmental stages. Remote Sens. Environ. 2002, 81, 337–354. [Google Scholar] [CrossRef]
- Gamon, J.A.; Peñuelas, J.; Field, C.B. A narrow-waveband spectral index that tracks diurnal changes in photosynthetic efficiency. Remote Sens. Environ. 1992, 41, 35–44. [Google Scholar] [CrossRef]
- Dash, J.; Curran, P.J. The MERIS terrestrial chlorophyll index. Int. J. Remote Sens. 2004, 25, 5403–5413. [Google Scholar] [CrossRef]
- Rondeaux, G.; Steven, M.; Baret, F. Optimization of soil-adjusted vegetation index. Remote Sens. Environ. 1996, 24, 109–127. [Google Scholar] [CrossRef]
- Haboudane, D.; Miller, J.R.; Tremblay, N.; Zarco-Tejada, P.J.; Dextraze, L. Integrated narrow-band vegetation indices for prediction of crop chlorophyll content for application to precision agriculture. Remote Sens. Environ. 2002, 81, 416–426. [Google Scholar] [CrossRef]
- Gitelson, A.; Merzlyak, M.N. Spectral Reflectance Changes Associated with Autumn Senescence of Aesculus hippocastanum L. and Acer platanoides L. Leaves. Spectral Features and Relation to Chlorophyll Estimation. J. Plant Physiol. 1994, 143, 286–292. [Google Scholar] [CrossRef]
- Zarco-Tejada, P.J.; Miller, J.R.; Noland, T.L.; Mohammed, G.H.; Sampson, P.H. Scaling-up and model inversion methods with narrowband optical indices for chlorophyll content estimation in closed forest canopies with hyperspectral data. IEEE Trans. Geosci. Remote Sens. 2001, 39, 1491–1507. [Google Scholar] [CrossRef]
- Wang, F.; Yan, Z.; Zhao, X.; Guo, X.; Zhou, Y.; Guo, J. Parameter selection of partial least squares model for hyperspectral estimation of chlorophyll content in camellia oleifera leaves. Acta Agric. Univ. Jiangxiensis 2022, 44, 86–96. [Google Scholar] [CrossRef]
- Schlerf, M.; Atzberger, C.; Hill, J. Remote sensing of forest biophysical variables using HyMap imaging spectrometer data. Remote Sens. Environ. 2004, 95, 177–194. [Google Scholar] [CrossRef]
- Liang, L.; Di, L.; Zhang, L.; Deng, M.; Qin, Z.; Zhao, S.; Lin, H. Estimation of crop LAI using hyperspectral vegetation indices and a hybrid inversion method. Remote Sens. Environ. 2015, 165, 123–134. [Google Scholar] [CrossRef]
- Hong, S.; Zhang, Z.; Zhang, L.; Ma, L.; Hai, X.; Wang, Z.; Zhang, H.; Lv, X. Hyperspectral estimation model of chlorophyll content in cotton canopy leaves under drip irrigation at different growths. Cotton Sci. 2019, 31, 138–146. [Google Scholar] [CrossRef]
- Fu, K.; Zhang, W.; Cao, H.; Zhu, Y.; Waili, S.; Zhang, W.; Feng, C. Research progress on crop diseases and insect pests monitoring based on spectrum. J. Agric. Sci. Technol. 2014, 16, 90–98. [Google Scholar] [CrossRef]
- Sun, L.; Cheng, L. Analysis of spectral response of vegetation leaf biochemical components. Spectrosc. Spectr. Anal. 2010, 30, 3031–3035. [Google Scholar] [CrossRef]
- Chen, S.; Chen, Y.; Ren, F.; Zheng, Y. Estimation of maize chlorophyll content based on spectral index. J. Xinyang Norm. Univ. Nat. Sci. Ed. 2021, 34, 225–229. [Google Scholar]
- Naidu, R.A.; Perry, E.M.; Pierce, F.J.; Mekuria, T. The potential of spectral reflectance technique for the detection of Grapevine leafroll-associated virus-3 in two red-berried wine grape cultivars. Comput. Electron. Agric. 2009, 66, 38–45. [Google Scholar] [CrossRef]
- Mahlein, A.-K.; Rumpf, T.; Welke, P.; Dehne, H.-W.; Plümer, L.; Steiner, U.; Oerke, E.-C. Development of spectral indices for detecting and identifying plant diseases. Remote Sens. Environ. 2013, 128, 21–30. [Google Scholar] [CrossRef]
- Liu, L.; Huang, W.; Pu, R.; Wang, J. Detection of Internal Leaf Structure Deterioration Using a New Spectral Ratio Index in the Near-Infrared Shoulder Region. J. Integr. Agric. 2014, 13, 760–769. [Google Scholar] [CrossRef]
- Hong, Y.; Liu, Y.; Chen, Y.; Liu, Y.; Yu, L.; Liu, Y.; Cheng, H. Application of fractional-order derivative in the quantitative estimation of soil organic matter content through visible and near-infrared spectroscopy. Geoderma 2019, 337, 758–769. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Lin, L.I.-K. A concordance correlation coefficient to evaluate reproducibility. Biometrics 1989, 45, 255–268. [Google Scholar] [CrossRef]
- Peng, L.; Xin, N.; Lv, X.; Li, N.; Li, F.; Geng, L.; Chen, H.; Lai, N. Inversion of nitrogen and phosphorus contents in cotton leaves based on the Gaussian mixture model and differences in hyperspectral features of UAV. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2025, 327, 125419. [Google Scholar] [CrossRef]
- Luo, H.; Wang, Q.; Zhang, J.; Wang, L.; Li, Y.; Yang, G. Minimum fertilization at the appearance of the first flower benefits cotton nutrient utilization of nitrogen, phosphorus and potassium. Sci. Rep. 2020, 10, 6815. [Google Scholar] [CrossRef]
- Arneberg, R.; Rajalahti, T.; Flikka, K.; Berven, F.S.; Kroksveen, A.C.; Berle, M.; Myhr, K.M.; Vedeler, C.A.; Ulvik, R.J.; Kvalheim, O.M. Pretreatment of mass spectral profiles: Application to proteomic data. Anal. Chem. 2007, 79, 7014–7026. [Google Scholar] [CrossRef]
- Nguyen, T.H.; Lee, B. Assessment of rice leaf growth and nitrogen status by hyperspectral canopy reflectance and partial least square regression. Eur. J. Agron. 2006, 24, 349–356. [Google Scholar] [CrossRef]
- Li, L.; Lu, J.; Wang, S.; Ma, Y.; Wei, Q.; Li, X.; Cong, R.; Ren, T. Methods for estimating leaf nitrogen concentration of winter oilseed rape (Brassica napus L.) using in situ leaf spectroscopy. Ind. Crops Prod. 2016, 91, 194–204. [Google Scholar] [CrossRef]
- Liu, L.; Wang, J.; Bian, H.; Abdalla, A.N. Near-Infrared Spectroscopy Combined With Support Vector Machine Model to Realize Quality Control of Ginkgolide Production. IEEE Photonics J. 2024, 16, 6600608. [Google Scholar] [CrossRef]
- Chen, D.; Chen, Y.; Xue, D. 1-D and 2-D digital fractional-order Savitzky–Golay differentiator. Signal Image Video Process. 2012, 6, 503–511. [Google Scholar] [CrossRef]
- Hong, Y.; Chen, Y.; Yu, L.; Liu, Y.; Liu, Y.; Zhang, Y.; Liu, Y.; Cheng, H. Combining Fractional Order Derivative and Spectral Variable Selection for Organic Matter Estimation of Homogeneous Soil Samples by VIS–NIR Spectroscopy. Remote Sens. 2018, 10, 479. [Google Scholar] [CrossRef]
- Kahraman, S.; Bacher, R. A comprehensive review of hyperspectral data fusion with LIDAR and SAR data. Annu. Rev. Control. 2021, 51, 236–253. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, Y.; Wen, M.; Lu, Y.; Ma, F. Accuracy Comparison of Estimation on Cotton Leaf and Plant Nitrogen Content Based on UAV Digital Image under Different Nutrition Treatments. Agronomy 2023, 13, 1686. [Google Scholar] [CrossRef]
- Rajaei, A.; Abiri, E.; Helfroush, M.S. Self-supervised spectral super-resolution for a fast hyperspectral and multispectral image fusion. Sci. Rep. 2024, 14, 29820. [Google Scholar] [CrossRef] [PubMed]
- Yu, P.; Yang, T.; Chen, S.; Kuo, C.; Tseng, H.W. Comparison of random forests and support vector machine for real-time radar-derived rainfall forecasting. J. Hydrol. 2017, 552, 92–104. [Google Scholar] [CrossRef]
- Abe, B.T.; Olugbara, O.O.; Marwala, T. Experimental comparison of support vector machines with random forests for hyperspectral image land cover classification. J. Earth Syst. Sci. 2014, 123, 779–790. [Google Scholar] [CrossRef]
- Bawa, A.; Samanta, S.; Himanshu, S.K.; Singh, J.; Kim, J.J.; Zhang, T.; Chang, A.; Jung, J.; DeLaune, P.; Bordovsky, J.; et al. A support vector machine and image processing based approach for counting open cotton bolls and estimating lint yield from UAV imagery. Smart Agric. Technol. 2023, 3, 100140. [Google Scholar] [CrossRef]
- Ma, Y.; Chen, H.; Zhao, G.; Wang, Z.; Wang, D. Spectral Index Fusion for Salinized Soil Salinity Inversion Using Sentinel-2A and UAV Images in a Coastal Area. IEEE Access 2020, 8, 159595–159608. [Google Scholar] [CrossRef]
- Guan, S.; Fukami, K.; Matsunaka, H.; Okami, M.; Tanaka, R.; Nakano, H.; Sakai, T.; Nakano, K.; Ohdan, H.; Takahashi, K. Assessing Correlation of High-Resolution NDVI with Fertilizer Application Level and Yield of Rice and Wheat Crops Using Small UAVs. Remote Sens. 2019, 11, 112. [Google Scholar] [CrossRef]
- Sultana, R.S.; Ali, A.; Ahmad, A.; Mubeen, M.; Zia-Ul-Haq, M.; Ahmad, S.; Ercisli, S.; Jaafar, Z.E.H. Normalized Difference Vegetation Index as a Tool for Wheat Yield Estimation: A Case Study from Faisalabad, Pakistan. Sci. World J. 2014, 2014, 725326. [Google Scholar] [CrossRef]



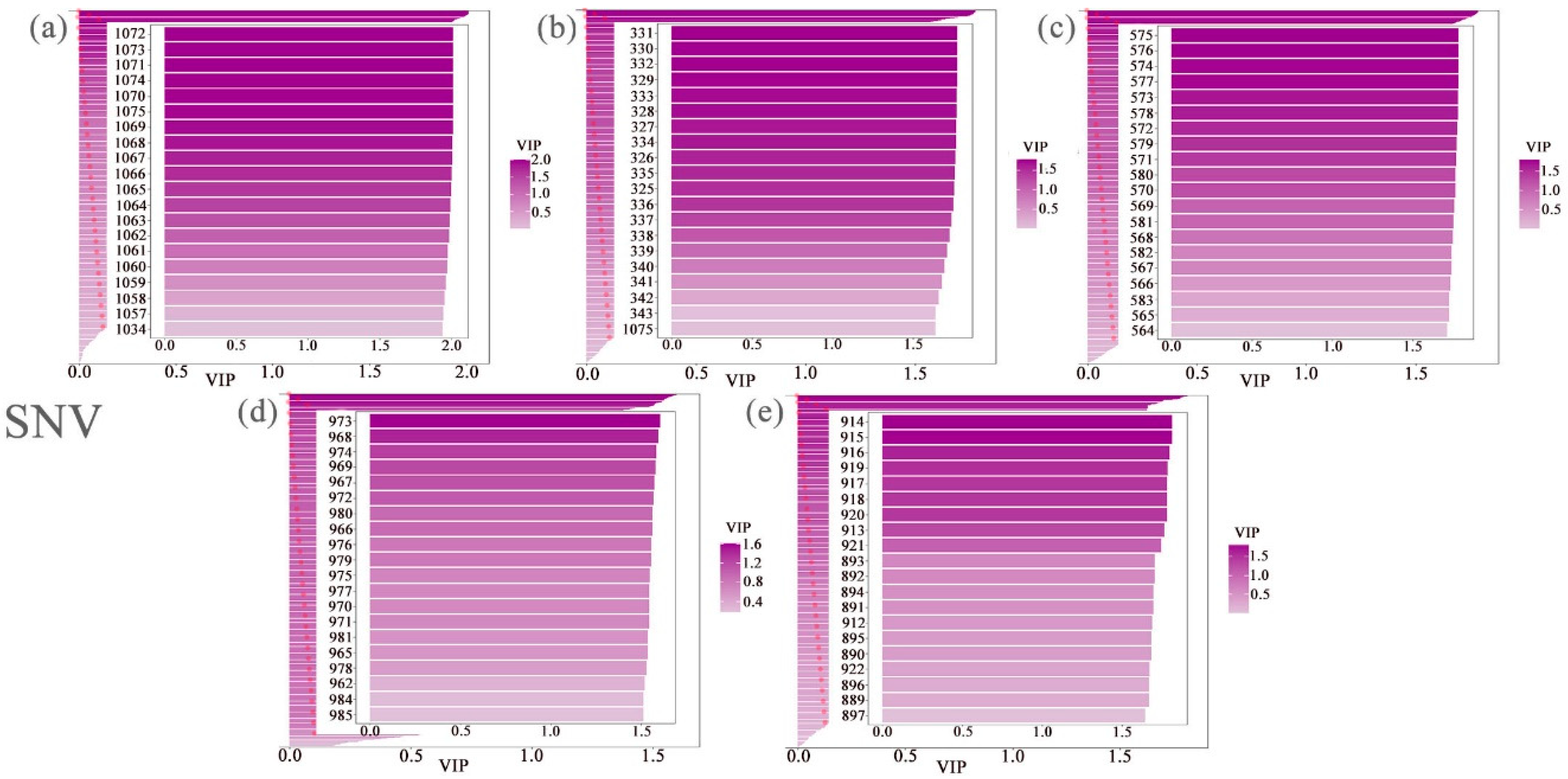
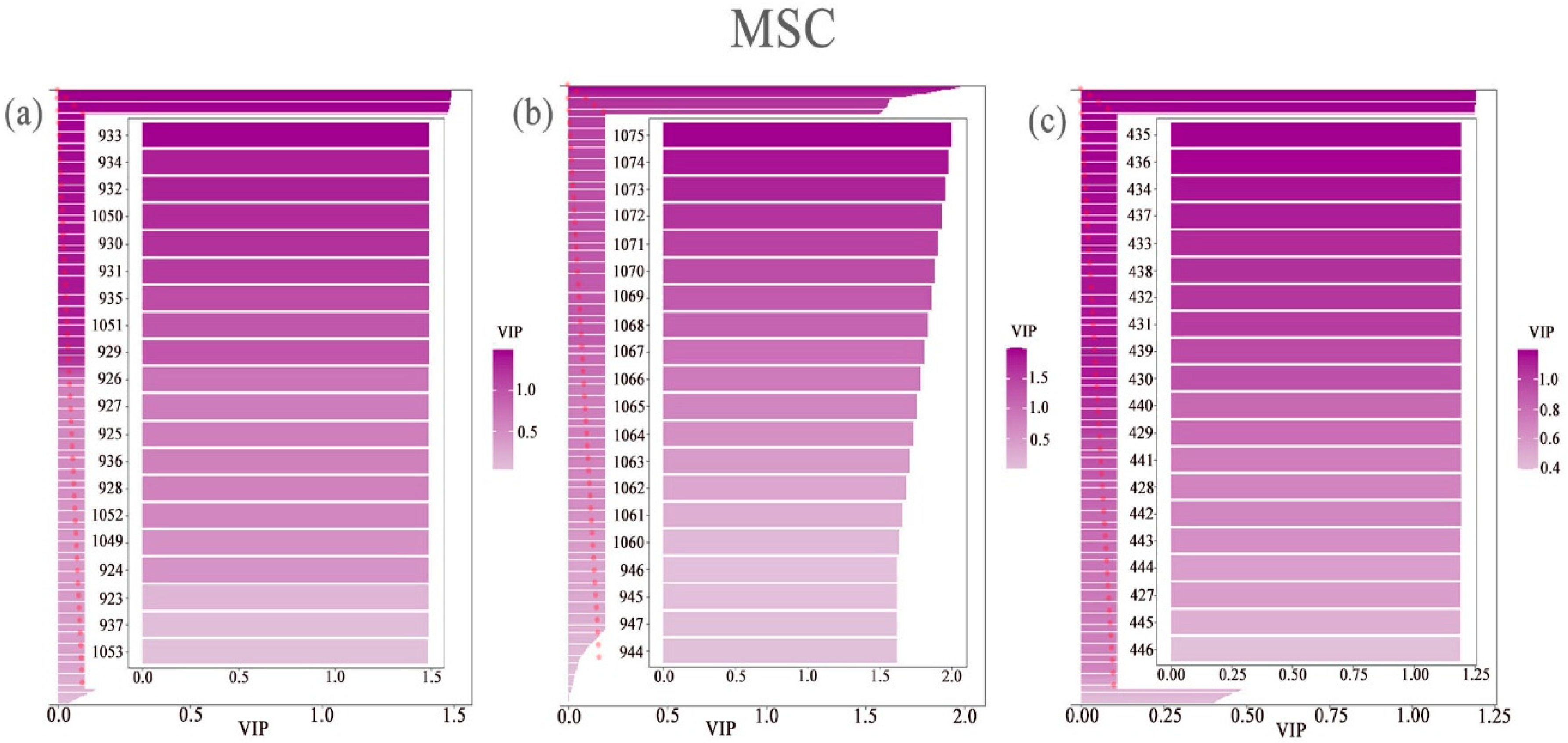
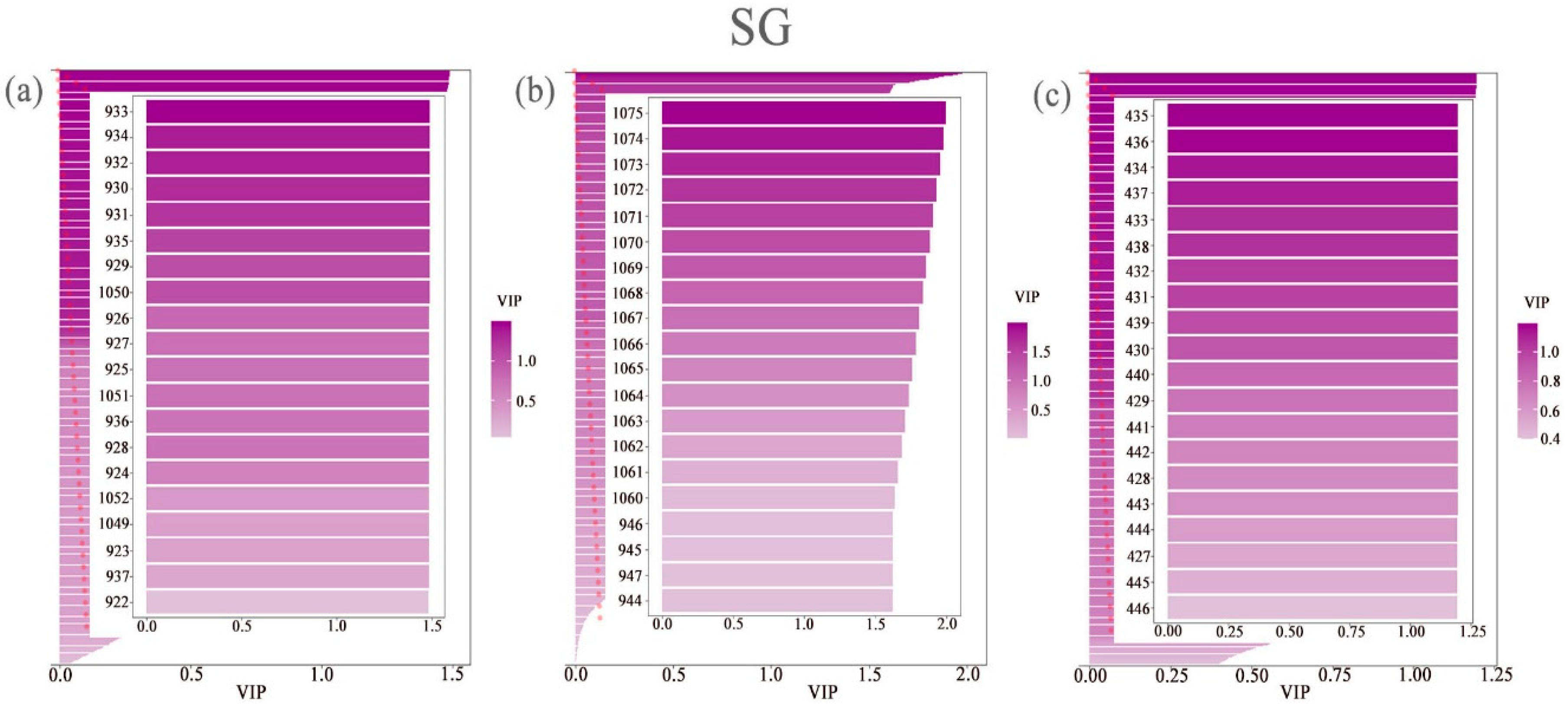

| Spectral Index | Formula | Document Source |
|---|---|---|
| DVI | R 800−R 680 | Richardson et al. [33] |
| mND705 | (R750 − R705)/(R750 + R705 − 2 × R445) | Sims et al. [34] |
| mSR705 | (R750 − R445)/(R705 − R445) | Sims et al. [34] |
| PRI | (R570 − R531)/(R570 + R531) | Gamon et al. [35] |
| MTCI | (R750 − R710)/(R710 − R680) | Dash et al. [36] |
| MCARI | [(R700 − R670) − 0.2 × (R700 − R550)] × (R700/R 670) | Dash et al. [36] |
| OSAVI | (1 + 0.16) × (R800 − R670)/(R800 + R670 + 0.16) | Rondeaux et al. [37] |
| TCARI | 3 × [(R700 − R670) − 0.2 × (R700 − R550)] × (R700/R670) | Haboudane et al. [38] |
| NDVI705 | (R750 − R705)/(R750 + R705) | Gitelson et al. [39] |
| NDVI550 | (R750 − R550)/(R750 + R550) | Gamon et al. [35] |
| VOG1 | R740/R720 | Zarco-Tejada et al. [40] |
| VOG2 | (R734 − R 747)/(R715 + R 726) | Zarco-Tejada et al. [40] |
| VOG3 | (R734 − R747)/(R715 + R 720) | Zarco-Tejada et al. [40] |
| VOG4 | R780/R740 | Wang et al. [41] |
| SR1 | R985/R745 | Schlerf et al. [42] |
| SR2 | R675/R700 | Liang et al. [43] |
| VARI | (R555 − R680)/(R580 + R680 + R480) | Wang et al. [41] |
| CIred edge | R750/R705 − 1 | Hong et al. [44] |
| RI-half | R747/R708 | Hong et al. [44] |
| Carte1 | R695/R420 | Liang et al. [43] |
| Carte2 | R695/R760 | Liang et al. [43] |
| Carte3 | R710/R760 | Liang et al. [43] |
| Datt1 | (R850 − R710)/(R850 + R680) | Liang et al. [43] |
| Datt2 | R850/R710 | Liang et al. [43] |
| Datt3 | R754/R704 | Liang et al. [43] |
| NVI | (R777 − R747)/R673 | Liang et al. [43] |
| NRI | (R570 − R670)/(R570 + R670) | Fu et al. [45] |
| Lichtenthaler | R440/R690 | Sun et al. [46] |
| ND | (R935 − R705)/(R935 + R705) | Sun et al. [46] |
| CIgreen | R800/R550 − 1 | Sun et al. [46] |
| GARI | [R800 − [R550 − 1.7 × (R470 − R670)]]/ [R800 + [R550 − 1.7 × (R470 + R670)]] | Chen et al. [47] |
| REP | 700 + 40 × [(R670 + R780)/2-R700]/(R740 − R700) | Liang et al. [43] |
| SPVI | 0.4 × 3.7 × (R670 − R550) − 1.2 × |R530 − R670| | Liang et al. [43] |
| SPVI2 | 0.4 × 3.7 × (R800 − R670) − 1.2 × |R550 − R670| | Liang et al. [43] |
| RVSI | (R714 + R752)/2 − R733 | Naidu et al. [48] |
| HI | (R534 − R698)/(R534 + R698) − 0.5R704 | Mahlein et al. [49] |
| NSRI | R890/R780 | Liu et al. [50] |
| SP | Stage | Spectral Index |
|---|---|---|
| FD | Squaring | / |
| Initial bloom | mND705, PRI, MCARI, TCARI, SRI, Lichtenthaler1, GARI | |
| Peak bloom | VOG4, SR2, GARI, RVSI | |
| Initial boll | / | |
| Peak boll | SPVI2 | |
| Boll opening | PRI, NDVI550, SR1, NSRI | |
| SNV | Squaring | / |
| Initial bloom | mND705, PRI, SP1, SR2, Clred edge, RI-half, Carte1, Carte2, Carte3, Datt2, Datt3, Lichtenthaler1, ND | |
| Peak bloom | VOG4, NVI, ND, RVSI | |
| Initial boll | SPVI2 | |
| Peak boll | NVI, GARI, SPVI | |
| Boll opening | PRI, OSAVI, SR1, NSRI | |
| MSC | Squaring | mSR705, NDVI705, NDVI550, SR2, Clarte1, Carte2, Datt1, GARI |
| Initial bloom | mND705, PRI, MCARI, TCARI, SR1, Carte1, Lichtenthaler1, GARI | |
| Peak bloom | VOG4, SR2, GARI, RVSI | |
| Initial boll | / | |
| Peak boll | SPVI2 | |
| Boll opening | PRI, NDVI550, SR1, NSRI | |
| SG | Squaring | NDVI705, NDVI550, SR2, Clarte1, Carte2, Datt1, GARI |
| Initial bloom | mND705, PRI, MCARI, TCARI, SR1, Carte1, Lichtenthaler1, GARI | |
| Peak bloom | VOG4, SR2, GARI, RVSI | |
| Initial boll | / | |
| Peak boll | SPVI2 | |
| Boll opening | PRI, NDVI550, SR1, NSRI |
| Model | SP | Stage | Training Set | Validation Set | ||||
|---|---|---|---|---|---|---|---|---|
| R2 | RMSE | LCCC | R2 | RMSE | LCCC | |||
| SVM | FD | Squaring | 0.6358 | 0.1250 | 0.3051 | 0.5800 | 0.1732 | 0.6979 |
| Initial bloom | 0.9960 | 0.0084 | 0.9943 | 0.6270 | 0.1412 | 0.4561 | ||
| Peak bloom | 0.9850 | 0.0073 | 0.9891 | 0.7360 | 0.0870 | 0.4290 | ||
| Initial boll | 0.9777 | 0.0015 | 0.9822 | 0.7281 | 0.1168 | 0.4648 | ||
| Peak boll | 0.6915 | 0.0638 | 0.7000 | 0.6318 | 0.0211 | 0.1653 | ||
| Boll opening | 0.9815 | 0.0064 | 0.9670 | 0.6211 | 0.0012 | 0.7766 | ||
| SNV | Squaring | 0.6044 | 0.0256 | 0.6188 | 0.5935 | 0.1045 | 0.4497 | |
| Initial bloom | 0.8901 | 0.1046 | 0.9003 | 0.6118 | 0.1695 | 0.7158 | ||
| Peak bloom | 0.6778 | 0.0392 | 0.7191 | 0.6271 | 0.0526 | 0.6981 | ||
| Initial boll | 0.7265 | 0.0054 | 0.6657 | 0.6605 | 0.0807 | 0.3650 | ||
| Peak boll | 0.7127 | 0.0068 | 0.6966 | 0.6410 | 0.0620 | 0.6323 | ||
| Boll opening | 0.5405 | 0.1257 | 0.4323 | 0.5579 | 0.0434 | 0.2579 | ||
| MSC | Squaring | 0.9596 | 0.0120 | 0.9719 | 0.5059 | 0.0023 | 0.6443 | |
| Initial bloom | 0.9087 | 0.0396 | 0.9474 | 0.6025 | 0.0131 | 0.7245 | ||
| Peak bloom | 0.5678 | 0.0313 | 0.6049 | 0.6099 | 0.0677 | 0.3734 | ||
| Initial boll | 0.6025 | 0.0314 | 0.6863 | 0.6944 | 0.0126 | 0.1865 | ||
| Peak boll | 0.5372 | 0.1246 | 0.5569 | 0.5848 | 0.0401 | 0.6055 | ||
| Boll opening | 0.6589 | 0.2829 | 0.6603 | 0.6459 | 0.0957 | 0.5395 | ||
| SG | Squaring | 0.9121 | 0.0138 | 0.9256 | 0.5572 | 0.1594 | 0.5931 | |
| Initial bloom | 0.9311 | 0.0085 | 0.9132 | 0.5790 | 0.0443 | 0.4599 | ||
| Peak bloom | 0.6389 | 0.0158 | 0.7386 | 0.5757 | 0.1312 | 0.5850 | ||
| Initial boll | 0.6903 | 0.0719 | 0.7439 | 0.6080 | 0.0093 | 0.3196 | ||
| Peak boll | 0.5399 | 0.1143 | 0.6116 | 0.6290 | 0.0106 | 0.6947 | ||
| Boll opening | 0.6709 | 0.0537 | 0.6179 | 0.6090 | 0.0599 | 0.5930 | ||
| RF | FD | Squaring | 0.6709 | 0.0562 | 0.6052 | 0.5153 | 0.1484 | 0.5373 |
| Initial bloom | 0.8137 | 0.1412 | 0.6575 | 0.6036 | 0.1427 | 0.6518 | ||
| Peak bloom | 0.8382 | 0.0023 | 0.7445 | 0.7002 | 0.0637 | 0.4286 | ||
| Initial boll | 0.7720 | 0.1704 | 0.5237 | 0.5730 | 0.0589 | 0.7182 | ||
| Peak boll | 0.7400 | 0.1157 | 0.5017 | 0.6444 | 0.0294 | 0.5554 | ||
| Boll opening | 0.7865 | 0.1214 | 0.5110 | 0.6101 | 0.0804 | 0.6816 | ||
| SNV | Squaring | 0.6047 | 0.0432 | 0.4451 | 0.5576 | 0.1334 | 0.5384 | |
| Initial bloom | 0.7810 | 0.0160 | 0.7063 | 0.6194 | 0.0486 | 0.7120 | ||
| Peak bloom | 0.7272 | 0.0183 | 0.5269 | 0.6605 | 0.0237 | 0.6530 | ||
| Initial boll | 0.7406 | 0.0534 | 0.5477 | 0.6444 | 0.0844 | 0.5421 | ||
| Peak boll | 0.8003 | 0.0551 | 0.6737 | 0.6532 | 0.0846 | 0.6309 | ||
| Boll opening | 0.6721 | 0.2389 | 0.4867 | 0.6027 | 0.0146 | 0.6787 | ||
| MSC | Squaring | 0.7697 | 0.1599 | 0.6164 | 0.6848 | 0.0151 | 0.6272 | |
| Initial bloom | 0.7661 | 0.0193 | 0.7428 | 0.6697 | 0.0734 | 0.3307 | ||
| Peak bloom | 0.6130 | 0.0163 | 0.5172 | 0.5719 | 0.0355 | 0.2448 | ||
| Initial boll | 0.6716 | 0.0402 | 0.6047 | 0.6214 | 0.0030 | 0.5684 | ||
| Peak boll | 0.6322 | 0.0130 | 0.6243 | 0.5141 | 0.1211 | 0.5907 | ||
| Boll opening | 0.7051 | 0.0026 | 0.5818 | 0.6387 | 0.0186 | 0.3511 | ||
| SG | Squaring | 0.7620 | 0.0434 | 0.7122 | 0.6989 | 0.0122 | 0.8213 | |
| Initial bloom | 0.7776 | 0.0928 | 0.7099 | 0.6097 | 0.0569 | 0.5353 | ||
| Peak bloom | 0.6558 | 0.0008 | 0.5082 | 0.5022 | 0.0546 | 0.2203 | ||
| Initial boll | 0.5888 | 0.1348 | 0.4363 | 0.5637 | 0.1058 | 0.3990 | ||
| Peak boll | 0.5390 | 0.0190 | 0.5498 | 0.5149 | 0.0648 | 0.6034 | ||
| Boll opening | 0.6987 | 0.0987 | 0.5627 | 0.5250 | 0.1271 | 0.5165 | ||
| Model | SP | Stage | Training Set | Validation Set | ||||
|---|---|---|---|---|---|---|---|---|
| R2 | RMSE | LCCC | R2 | RMSE | LCCC | |||
| RF | FD | Squaring | 0.6709 | 0.0562 | 0.6052 | 0.5153 | 0.1484 | 0.5373 |
| Initial bloom | 0.8137 | 0.1412 | 0.6575 | 0.6036 | 0.1427 | 0.6518 | ||
| Peak bloom | 0.8382 | 0.0023 | 0.7445 | 0.7002 | 0.0637 | 0.4286 | ||
| Initial boll | 0.7720 | 0.1704 | 0.5237 | 0.5730 | 0.0589 | 0.7182 | ||
| Peak boll | 0.7400 | 0.1157 | 0.5017 | 0.6444 | 0.0294 | 0.5554 | ||
| Boll opening | 0.7865 | 0.1214 | 0.5110 | 0.6101 | 0.0804 | 0.6816 | ||
| SNV | Squaring | 0.6047 | 0.0432 | 0.4451 | 0.5576 | 0.1334 | 0.5384 | |
| Initial bloom | 0.7810 | 0.0160 | 0.7063 | 0.6194 | 0.0486 | 0.7120 | ||
| Peak bloom | 0.7272 | 0.0183 | 0.5269 | 0.6605 | 0.0237 | 0.6530 | ||
| Initial boll | 0.7406 | 0.0534 | 0.5477 | 0.6444 | 0.0844 | 0.5421 | ||
| Peak boll | 0.8003 | 0.0551 | 0.6737 | 0.6532 | 0.0846 | 0.6309 | ||
| Boll opening | 0.6721 | 0.2389 | 0.4867 | 0.6027 | 0.0146 | 0.6787 | ||
| MSC | Squaring | 0.7697 | 0.1599 | 0.6164 | 0.6848 | 0.0151 | 0.6272 | |
| Initial bloom | 0.7661 | 0.0193 | 0.7428 | 0.6697 | 0.0734 | 0.3307 | ||
| Peak bloom | 0.6130 | 0.0163 | 0.5172 | 0.5719 | 0.0355 | 0.2448 | ||
| Initial boll | 0.6716 | 0.0402 | 0.6047 | 0.6214 | 0.0030 | 0.5684 | ||
| Peak boll | 0.6322 | 0.0130 | 0.6243 | 0.5141 | 0.1211 | 0.5907 | ||
| Boll opening | 0.7051 | 0.0026 | 0.5818 | 0.6387 | 0.0186 | 0.3511 | ||
| SG | Squaring | 0.7620 | 0.0434 | 0.7122 | 0.6989 | 0.0122 | 0.8213 | |
| Initial bloom | 0.7776 | 0.0928 | 0.7099 | 0.6097 | 0.0569 | 0.5353 | ||
| Peak bloom | 0.6558 | 0.0008 | 0.5082 | 0.5022 | 0.0546 | 0.2203 | ||
| Initial boll | 0.5888 | 0.1348 | 0.4363 | 0.5637 | 0.1058 | 0.3990 | ||
| Peak boll | 0.5390 | 0.0190 | 0.5498 | 0.5149 | 0.0648 | 0.6034 | ||
| Boll opening | 0.6987 | 0.0987 | 0.5627 | 0.5250 | 0.1271 | 0.5165 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qiao, S.; Fu, W.; Wang, J.; An, X.; Li, F.; Liu, W.; Cai, C. Spectral Estimation of Nitrogen Content in Cotton Leaves Under Coupled Nitrogen and Phosphorus Conditions. Agronomy 2025, 15, 1701. https://doi.org/10.3390/agronomy15071701
Qiao S, Fu W, Wang J, An X, Li F, Liu W, Cai C. Spectral Estimation of Nitrogen Content in Cotton Leaves Under Coupled Nitrogen and Phosphorus Conditions. Agronomy. 2025; 15(7):1701. https://doi.org/10.3390/agronomy15071701
Chicago/Turabian StyleQiao, Shunyu, Wenjin Fu, Jiaqiang Wang, Xiaolong An, Fuqing Li, Weiyang Liu, and Chongfa Cai. 2025. "Spectral Estimation of Nitrogen Content in Cotton Leaves Under Coupled Nitrogen and Phosphorus Conditions" Agronomy 15, no. 7: 1701. https://doi.org/10.3390/agronomy15071701
APA StyleQiao, S., Fu, W., Wang, J., An, X., Li, F., Liu, W., & Cai, C. (2025). Spectral Estimation of Nitrogen Content in Cotton Leaves Under Coupled Nitrogen and Phosphorus Conditions. Agronomy, 15(7), 1701. https://doi.org/10.3390/agronomy15071701






