Genome-Wide Identification of the bHLH Gene Family in Kiwifruit (Actinidia chinensis) and the Responses of AcbHLH84 and AcbHLH97 Under Drought Stress
Abstract
1. Introduction
2. Materials and Methods
2.1. Gene Family Identification and Physicochemical Property Analysis
2.2. Phylogenetic Analysis
2.3. Gene Structure and Conserved Motif Analysis
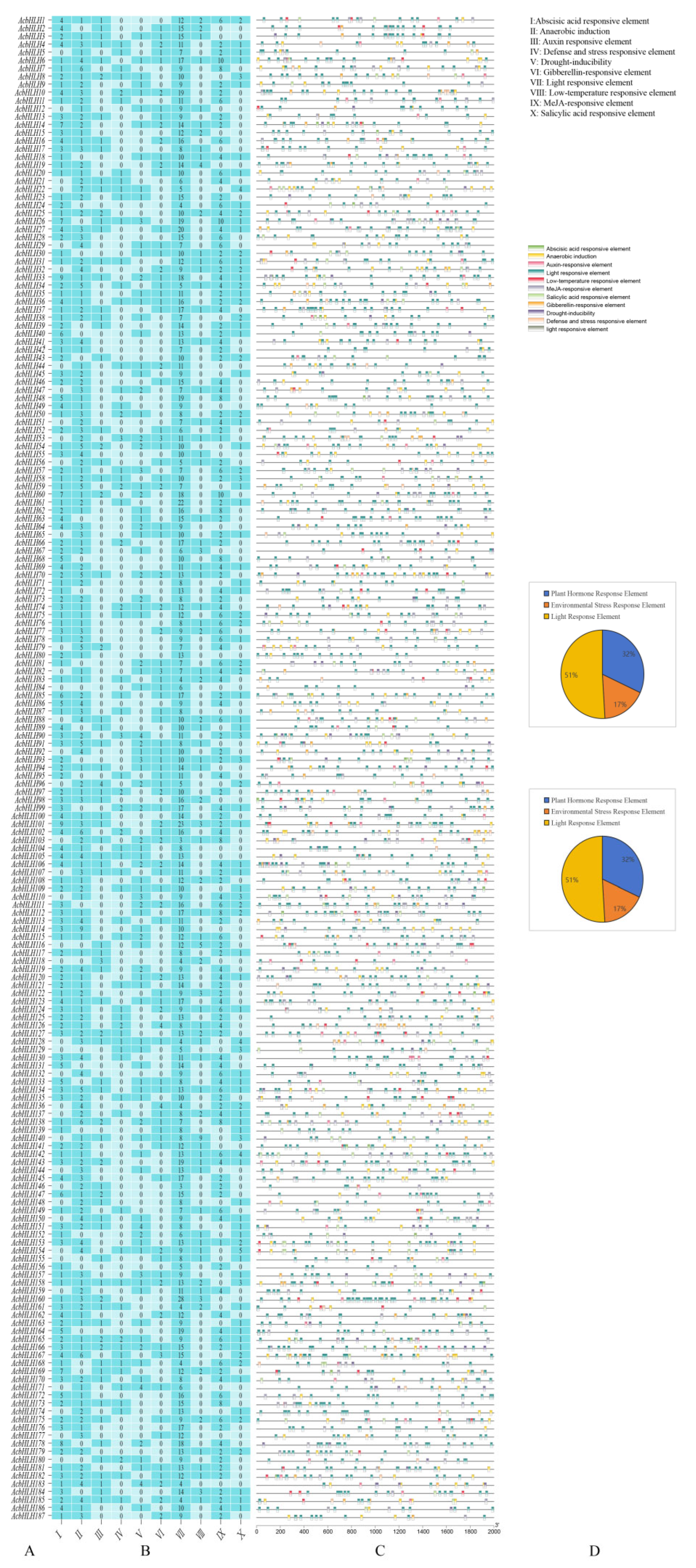
2.4. Analysis of Cis-Acting Elements
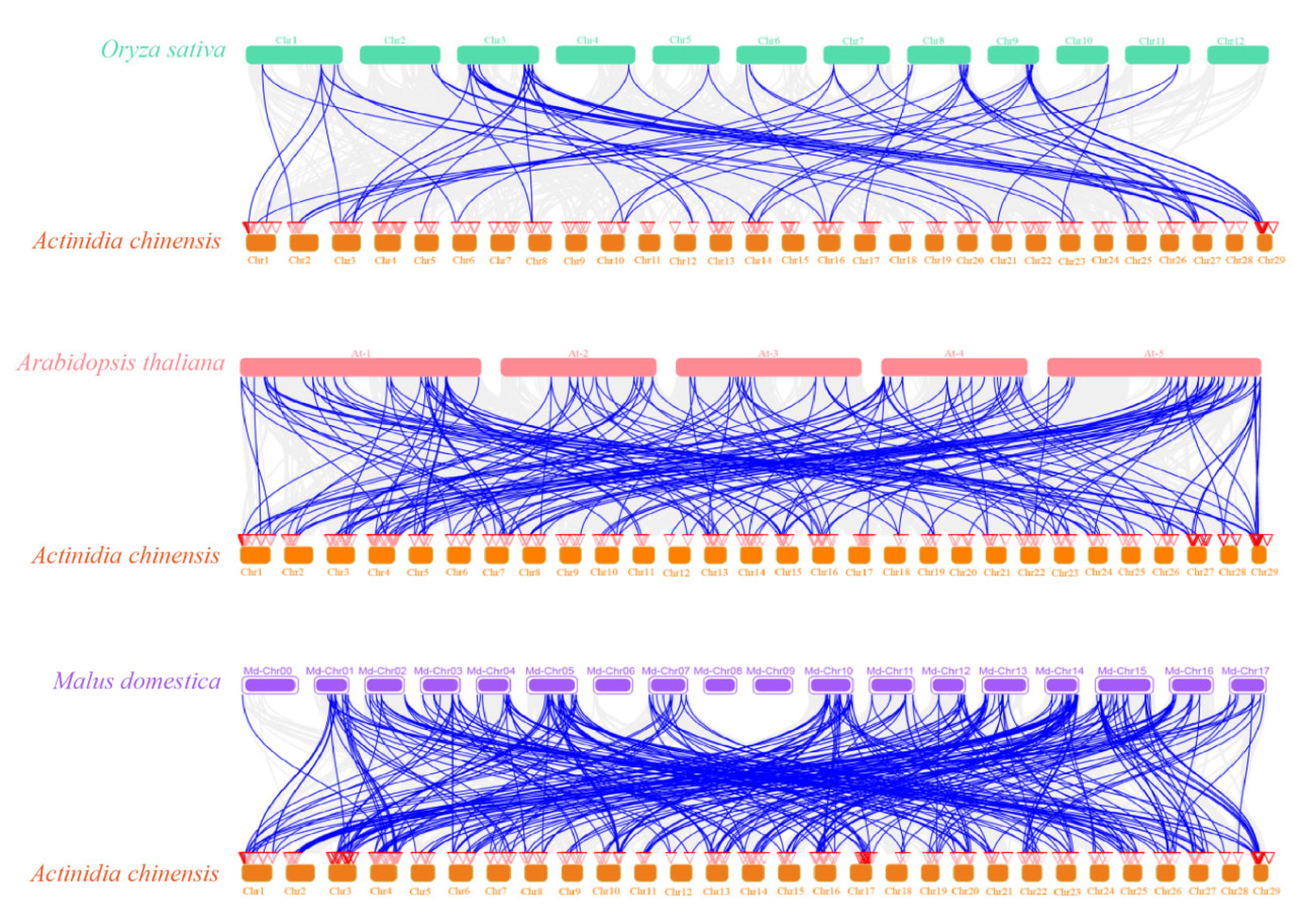
2.5. Chromosome Localization and Collinearity Analysis
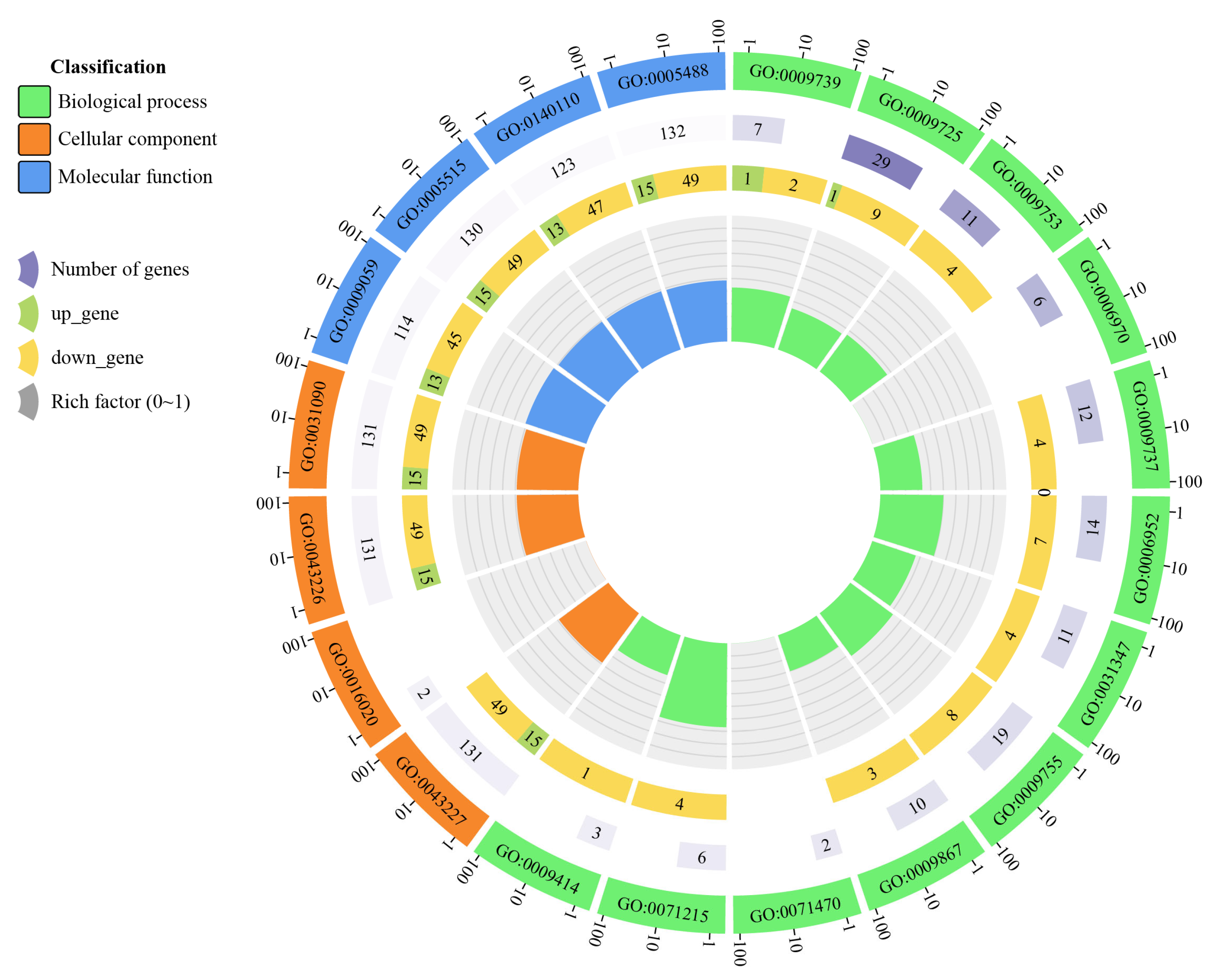
2.6. GO Function and KEGG Pathway Enrichment Analysis
2.7. Drought Stress Treatment, Transcriptome Sequencing, and Hormone Expression Determination of Kiwifruit Leaves
2.8. qRT-PCR Analysis
2.9. Statistics
3. Results
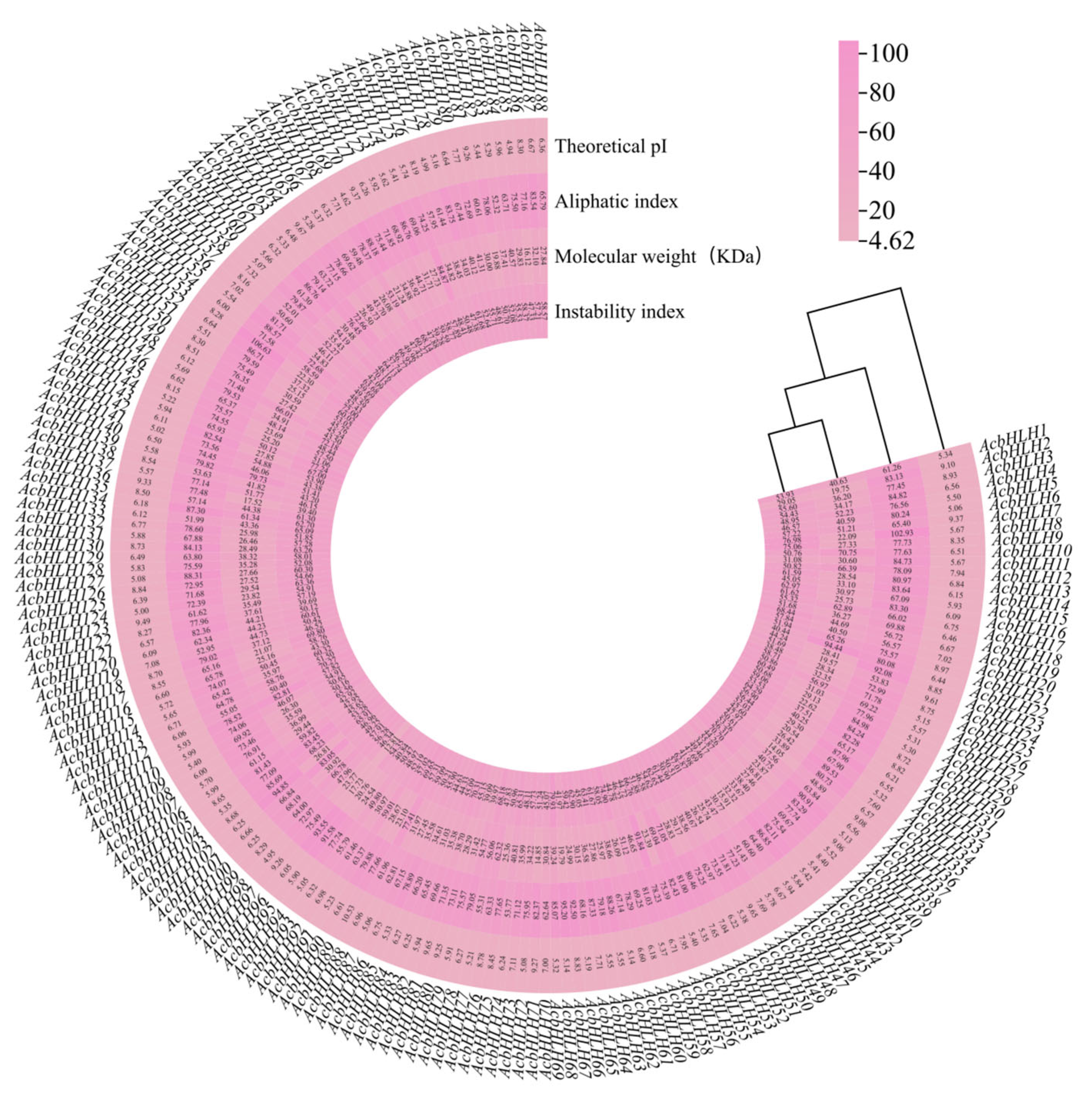
3.1. Identification and Physicochemical Properties of the bHLH Gene Family in Kiwifruit
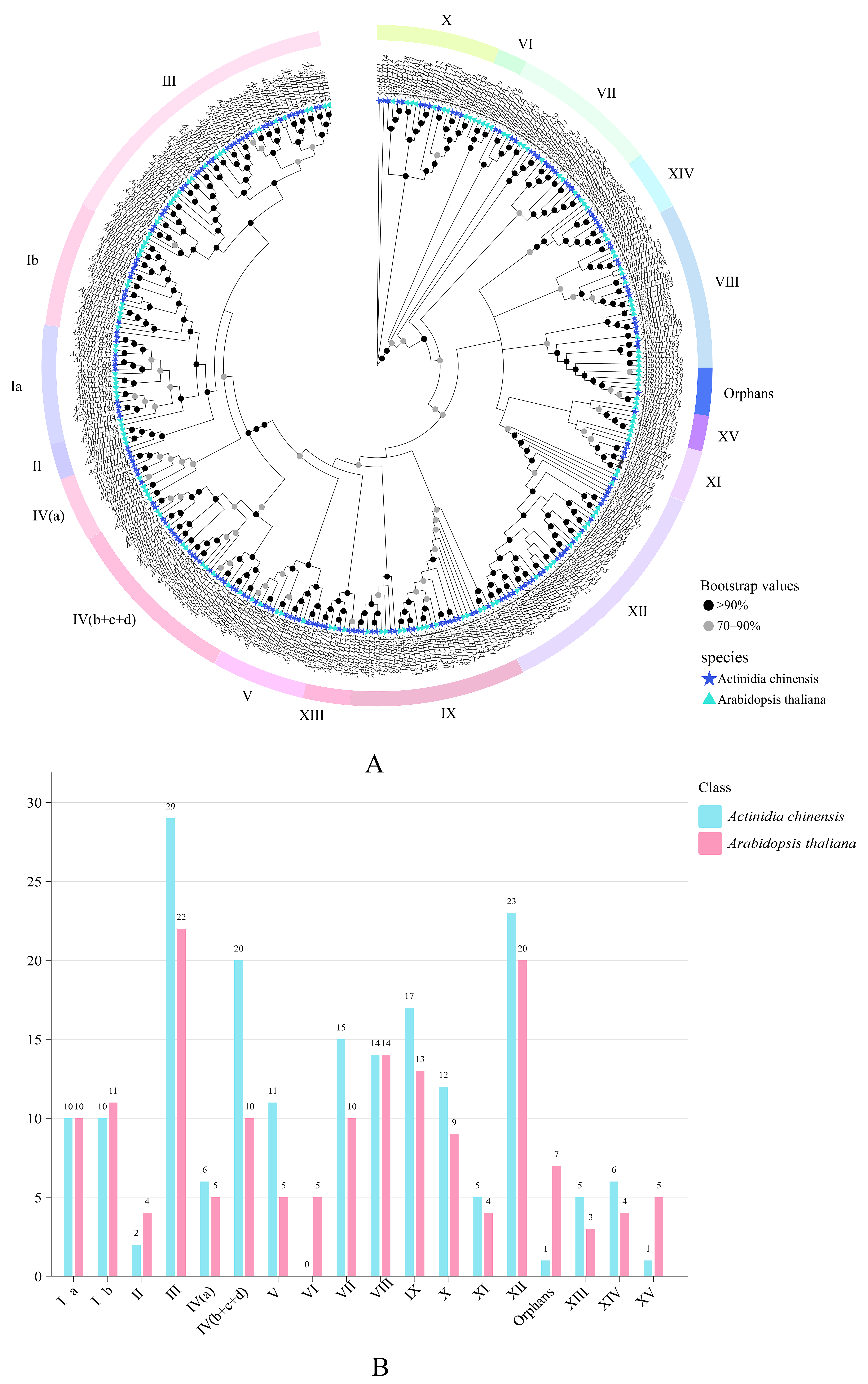
3.2. Phylogeny of the AcbHLH Gene Family in Kiwifruit
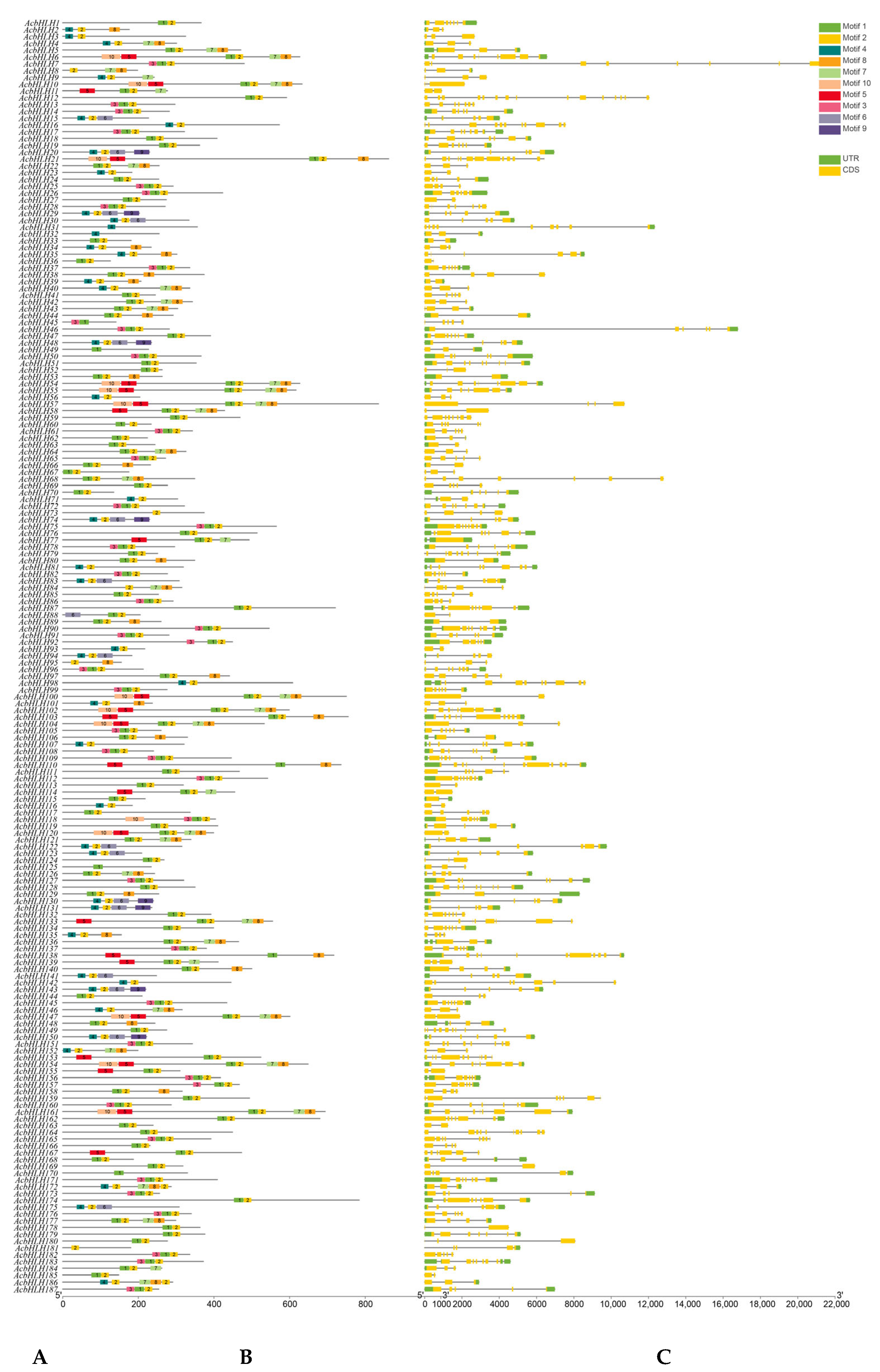
3.3. Gene Structure and Conserved Motif Analysis of the AcbHLH Gene
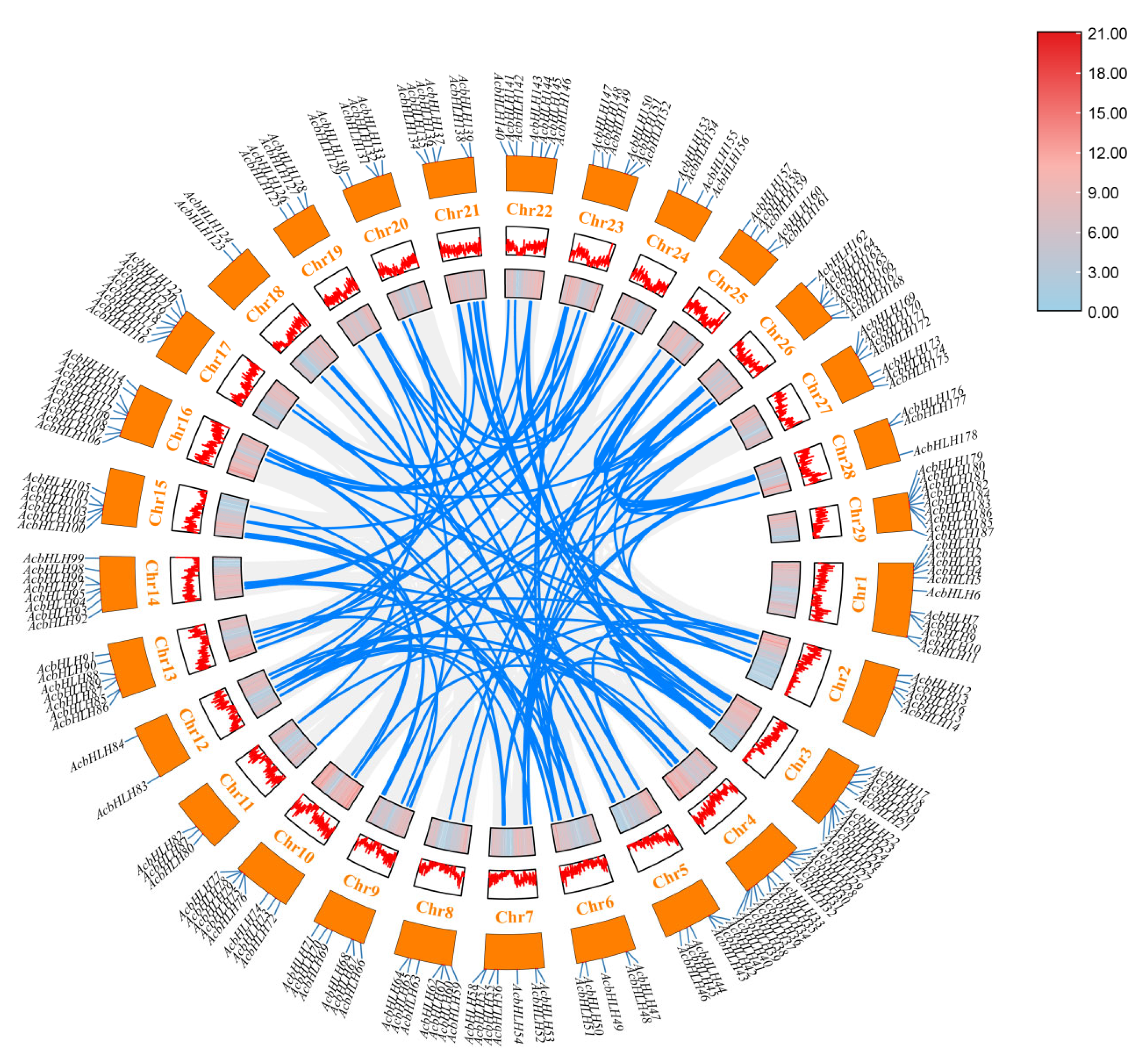
3.4. Chromosomal Mapping and Duplication Gene Analysis of AcbHLH Genes in Kiwifruit
3.5. Analysis of Cis-Acting Elements of AcbHLH Gene Family Promoters in Kiwifruit
3.6. GO Functional Enrichment Analysis
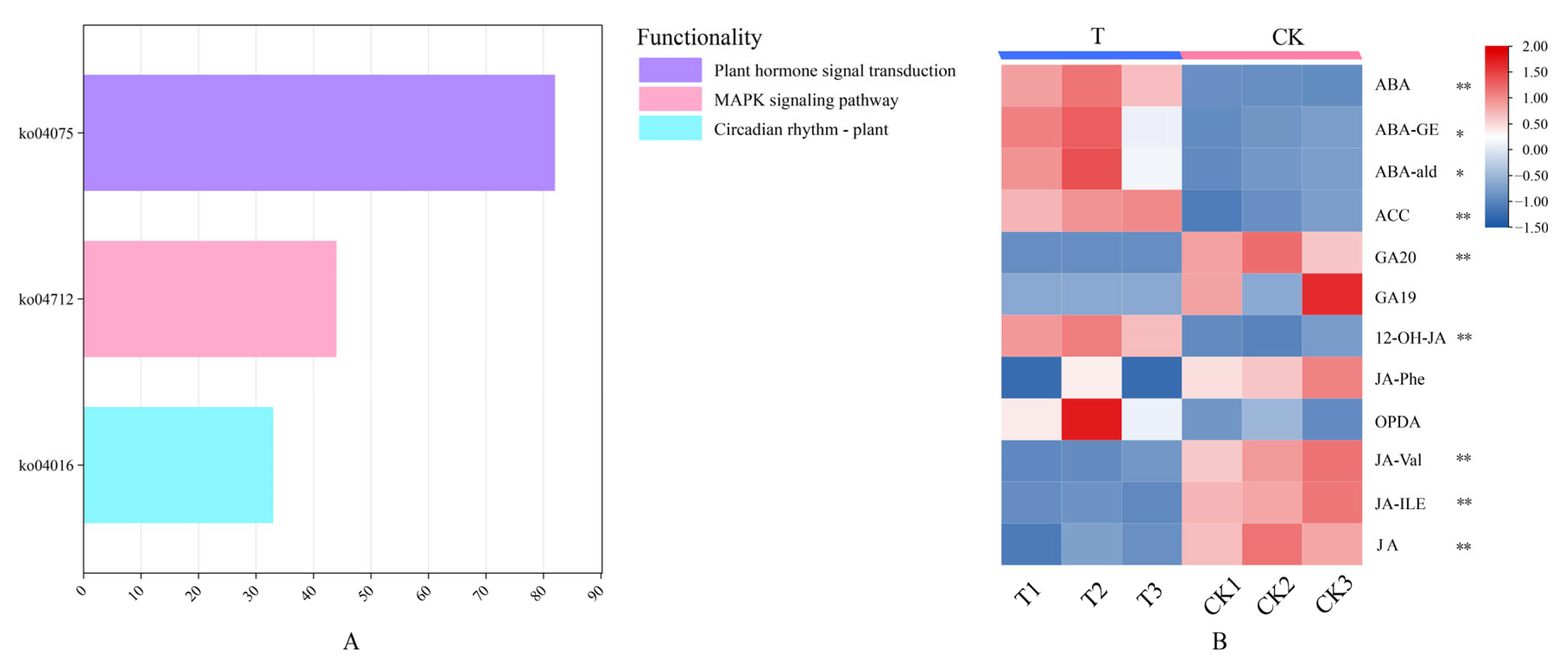
3.7. KEGG Enrichment of the AcbHLH Gene Family and Metabolite Analysis of Kiwifruit Under Drought Stress
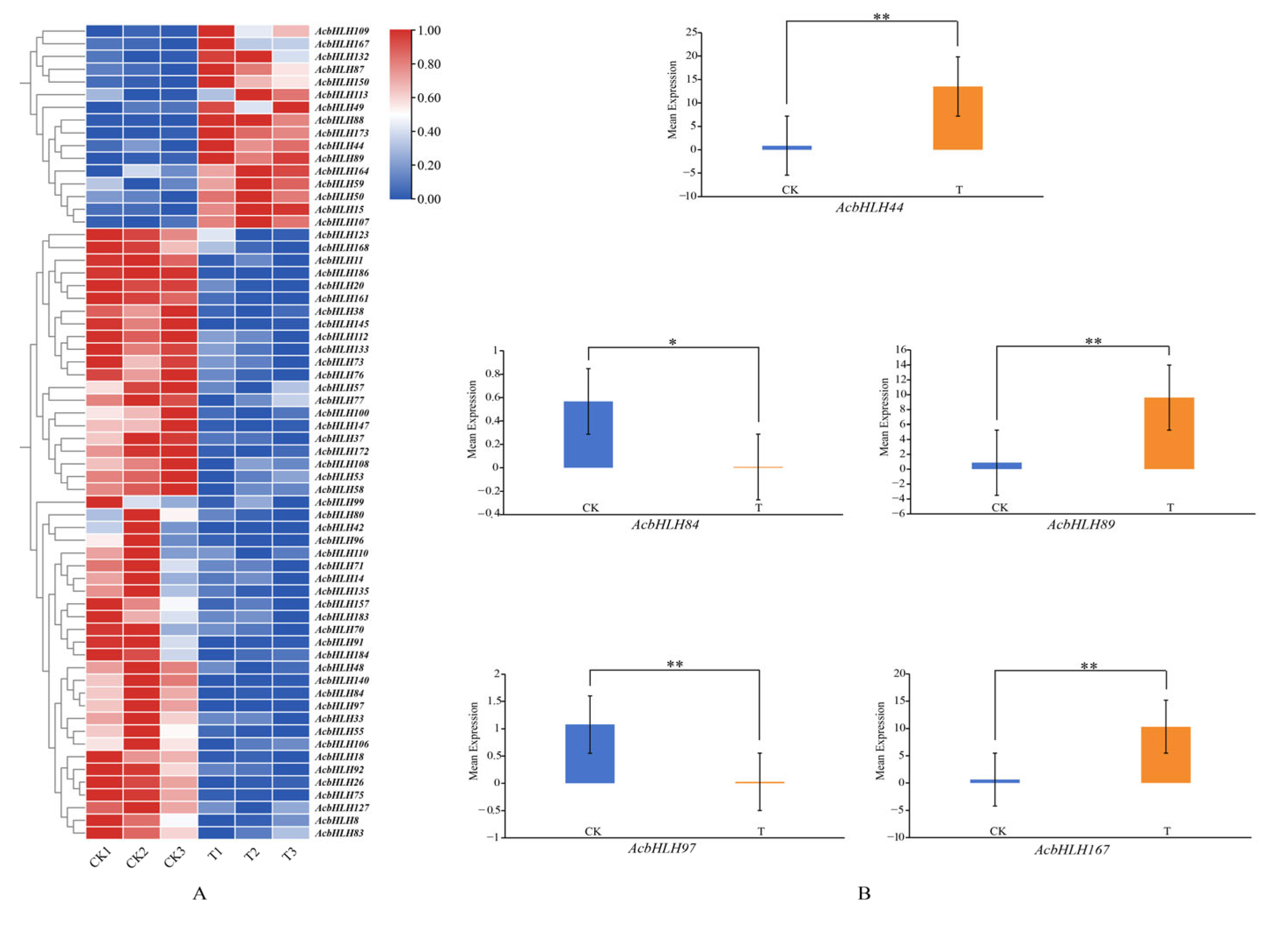
3.8. Expression Pattern and qRT-PCR Analysis of bHLH Genes in Kiwifruit Under Drought Stress
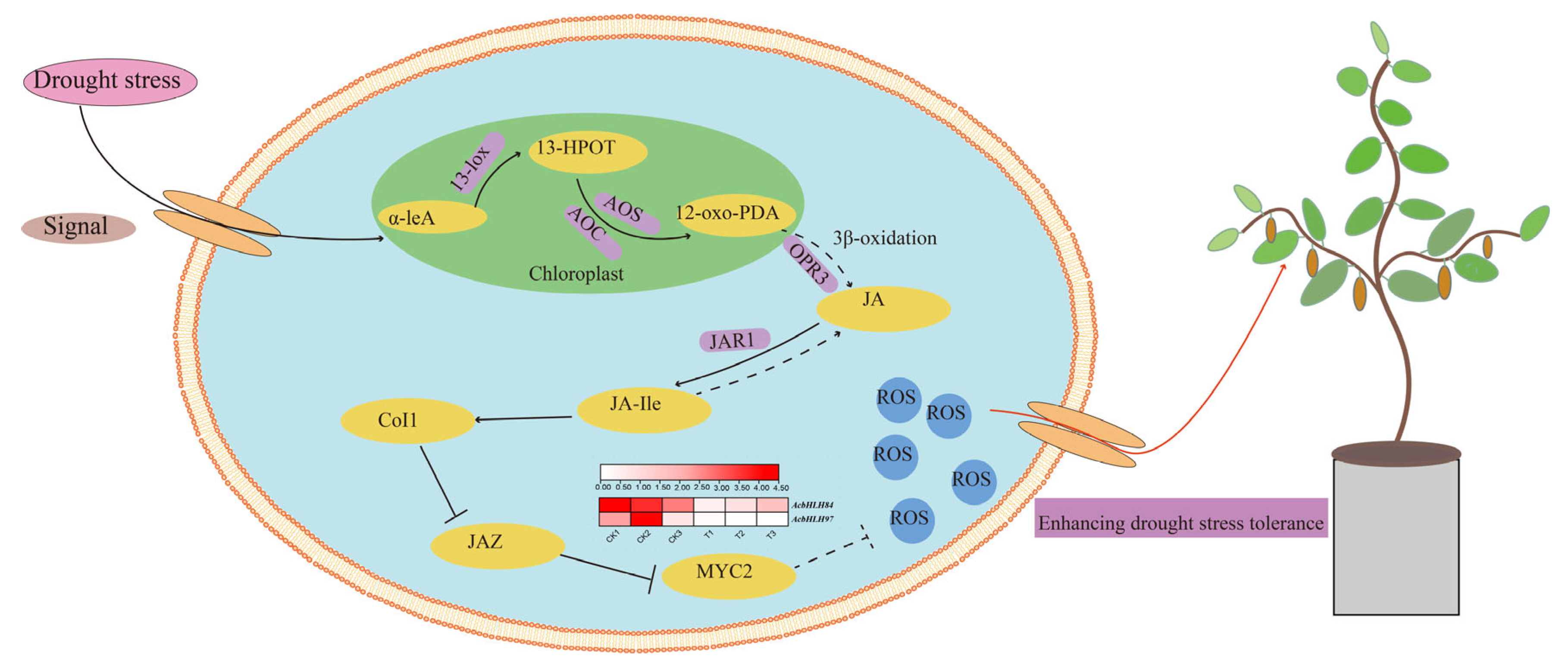
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Sequence ID | Gene Name | Number of Amino Acids (aa) | Molecular Weight (kDa) | Theoretical pI | Instability Index | Aliphatic Index | Subcellular Localization |
|---|---|---|---|---|---|---|---|
| Actinidia26970 | AcbHLH1 | 366 | 40.63 | 5.34 | 53.93 | 61.26 | Nucleus |
| Actinidia01875 | AcbHLH2 | 176 | 19.75 | 9.10 | 29.05 | 83.13 | Nucleus |
| Actinidia16554 | AcbHLH3 | 325 | 36.20 | 8.93 | 55.60 | 77.45 | Nucleus |
| Actinidia23610 | AcbHLH4 | 301 | 34.17 | 6.56 | 54.43 | 84.82 | Nucleus |
| Actinidia31749 | AcbHLH5 | 471 | 52.23 | 5.50 | 48.95 | 76.56 | Nucleus |
| Actinidia10014 | AcbHLH6 | 627 | 40.59 | 5.06 | 46.57 | 80.24 | Nucleus |
| Actinidia32939 | AcbHLH7 | 480 | 51.21 | 9.37 | 57.22 | 65.40 | Nucleus |
| Actinidia37008 | AcbHLH8 | 198 | 22.09 | 5.67 | 76.98 | 102.93 | Nucleus |
| Actinidia33194 | AcbHLH9 | 242 | 27.33 | 8.35 | 75.06 | 77.73 | Nucleus |
| Actinidia07970 | AcbHLH10 | 633 | 70.75 | 6.51 | 50.76 | 77.63 | Nucleus |
| Actinidia07995 | AcbHLH11 | 277 | 30.60 | 5.67 | 31.08 | 84.73 | Nucleus |
| Actinidia21691 | AcbHLH12 | 592 | 66.39 | 7.94 | 50.82 | 78.09 | Nucleus |
| Actinidia25535 | AcbHLH13 | 297 | 33.10 | 6.15 | 45.05 | 83.64 | Nucleus |
| Actinidia04941 | AcbHLH14 | 282 | 30.97 | 5.93 | 62.97 | 67.09 | Nucleus |
| Actinidia04904 | AcbHLH15 | 227 | 25.73 | 6.09 | 61.62 | 83.30 | Nucleus |
| Actinidia04791 | AcbHLH16 | 573 | 62.89 | 6.75 | 55.32 | 66.02 | Nucleus |
| Actinidia08901 | AcbHLH17 | 322 | 36.27 | 6.46 | 51.68 | 69.88 | Nucleus |
| Actinidia35022 | AcbHLH18 | 408 | 44.69 | 6.67 | 68.44 | 56.72 | Nucleus |
| Actinidia35292 | AcbHLH19 | 362 | 40.50 | 7.02 | 57.84 | 56.57 | Nucleus |
| Actinidia35313 | AcbHLH20 | 230 | 65.26 | 8.97 | 51.94 | 75.57 | Nucleus |
| Actinidia35482 | AcbHLH21 | 862 | 94.44 | 6.44 | 40.44 | 80.08 | Nucleus |
| Actinidia02421 | AcbHLH22 | 255 | 28.41 | 8.85 | 44.24 | 92.08 | Nucleus |
| Actinidia12937 | AcbHLH23 | 183 | 19.57 | 9.61 | 51.69 | 53.83 | Nucleus |
| Actinidia17488 | AcbHLH24 | 254 | 28.34 | 8.75 | 38.48 | 72.99 | Nucleus |
| Actinidia34363 | AcbHLH25 | 292 | 32.35 | 5.15 | 58.71 | 71.78 | Nucleus |
| Actinidia34350 | AcbHLH26 | 423 | 56.97 | 5.57 | 50.86 | 69.22 | Nucleus |
| Actinidia33692 | AcbHLH27 | 274 | 31.03 | 5.31 | 60.49 | 77.96 | Nucleus |
| Actinidia04493 | AcbHLH28 | 271 | 29.13 | 5.30 | 50.68 | 84.98 | Nucleus |
| Actinidia04455 | AcbHLH29 | 203 | 22.62 | 8.72 | 59.06 | 84.24 | Nucleus |
| Actinidia02716 | AcbHLH30 | 334 | 37.51 | 8.82 | 53.53 | 82.28 | Nucleus |
| Actinidia26274 | AcbHLH31 | 356 | 40.25 | 6.21 | 64.29 | 65.17 | Nucleus |
| Actinidia38158 | AcbHLH32 | 255 | 29.30 | 6.55 | 56.90 | 87.96 | Nucleus |
| Actinidia33915 | AcbHLH33 | 181 | 20.54 | 5.32 | 56.44 | 67.90 | Nucleus |
| Actinidia13536 | AcbHLH34 | 234 | 26.42 | 7.60 | 52.03 | 89.53 | Nucleus |
| Actinidia13535 | AcbHLH35 | 302 | 33.89 | 6.57 | 42.6 | 80.73 | Nucleus |
| Actinidia29085 | AcbHLH36 | 126 | 14.05 | 9.08 | 47.97 | 48.89 | Nucleus |
| Actinidia29185 | AcbHLH37 | 322 | 37.56 | 6.56 | 50.61 | 63.84 | Nucleus |
| Actinidia00959 | AcbHLH38 | 374 | 40.75 | 5.13 | 53.26 | 90.91 | Nucleus |
| Actinidia03094 | AcbHLH39 | 207 | 23.87 | 9.06 | 56.31 | 83.29 | Nucleus |
| Actinidia03158 | AcbHLH40 | 336 | 36.81 | 5.52 | 70.7 | 77.74 | Nucleus |
| Actinidia36367 | AcbHLH41 | 245 | 27.46 | 8.40 | 44.26 | 69.67 | Nucleus |
| Actinidia36394 | AcbHLH42 | 343 | 38.40 | 5.41 | 55.87 | 75.54 | Nucleus |
| Actinidia36395 | AcbHLH43 | 304 | 33.67 | 5.42 | 34.17 | 82.11 | Nucleus |
| Actinidia16614 | AcbHLH44 | 292 | 32.32 | 5.84 | 49.46 | 86.85 | Nucleus |
| Actinidia07746 | AcbHLH45 | 141 | 15.91 | 5.94 | 47.69 | 64.40 | Nucleus |
| Actinidia04303 | AcbHLH46 | 282 | 30.77 | 6.67 | 41.98 | 60.60 | Nucleus |
| Actinidia09941 | AcbHLH47 | 391 | 43.47 | 5.78 | 62.87 | 51.43 | Nucleus |
| Actinidia09958 | AcbHLH48 | 235 | 25.74 | 7.69 | 57.01 | 77.23 | Nucleus |
| Actinidia05467 | AcbHLH49 | 227 | 26.54 | 9.65 | 50.51 | 71.81 | Nucleus |
| Actinidia09371 | AcbHLH50 | 366 | 40.67 | 5.38 | 50.9 | 73.55 | Nucleus |
| Actinidia09432 | AcbHLH51 | 353 | 38.56 | 6.22 | 61.44 | 62.97 | Nucleus |
| Actinidia07680 | AcbHLH52 | 263 | 29.17 | 7.04 | 62.65 | 75.25 | Nucleus |
| Actinidia07635 | AcbHLH53 | 263 | 28.83 | 7.65 | 54.62 | 80.46 | Nucleus |
| Actinidia20908 | AcbHLH54 | 627 | 71.05 | 5.35 | 46.58 | 81 | Nucleus |
| Actinidia14565 | AcbHLH55 | 617 | 69.04 | 5.4 | 51.88 | 82.43 | Nucleus |
| Actinidia14495 | AcbHLH56 | 204 | 23.39 | 7.95 | 66.76 | 75.39 | Nucleus |
| Actinidia20248 | AcbHLH57 | 835 | 91.84 | 6.71 | 44.72 | 78.23 | Nucleus |
| Actinidia20274 | AcbHLH58 | 428 | 46.65 | 5.37 | 46.22 | 81.03 | Nucleus |
| Actinidia01660 | AcbHLH59 | 469 | 51.12 | 6.18 | 44.78 | 69.25 | Nucleus |
| Actinidia38064 | AcbHLH60 | 234 | 26.09 | 6.6 | 74.9 | 78.29 | Nucleus |
| Actinidia38100 | AcbHLH61 | 343 | 38.66 | 5.14 | 58.05 | 67.14 | Nucleus |
| Actinidia29648 | AcbHLH62 | 224 | 25.97 | 5.55 | 61.67 | 88.26 | Nucleus |
| Actinidia29264 | AcbHLH63 | 244 | 27.86 | 5.55 | 63.41 | 79.18 | Nucleus |
| Actinidia04410 | AcbHLH64 | 326 | 36.58 | 7.71 | 45.05 | 87.33 | Nucleus |
| Actinidia04383 | AcbHLH65 | 272 | 30.15 | 5.19 | 62.9 | 68.16 | Nucleus |
| Actinidia28828 | AcbHLH66 | 232 | 24.99 | 8.83 | 59.42 | 92.5 | Nucleus |
| Actinidia38451 | AcbHLH67 | 175 | 19.79 | 5.14 | 41.97 | 95.2 | Nucleus |
| Actinidia38499 | AcbHLH68 | 349 | 39.24 | 5.32 | 79.93 | 85.07 | Nucleus |
| Actinidia22793 | AcbHLH69 | 277 | 30.84 | 7 | 45.07 | 62.64 | Nucleus |
| Actinidia27112 | AcbHLH70 | 135 | 14.85 | 9.27 | 51.24 | 82.37 | Nucleus |
| Actinidia35869 | AcbHLH71 | 304 | 34.22 | 5.08 | 57.73 | 75.95 | Nucleus |
| Actinidia36836 | AcbHLH72 | 322 | 35.99 | 7.11 | 48.11 | 71.12 | Nucleus |
| Actinidia19012 | AcbHLH73 | 374 | 40.81 | 6.24 | 65.31 | 53.77 | Nucleus |
| Actinidia10704 | AcbHLH74 | 230 | 25.36 | 8.45 | 50.96 | 77.65 | Nucleus |
| Actinidia23778 | AcbHLH75 | 565 | 62.32 | 8.78 | 47.83 | 63.33 | Nucleus |
| Actinidia23742 | AcbHLH76 | 514 | 56.06 | 5.21 | 68.18 | 55.31 | Nucleus |
| Actinidia21175 | AcbHLH77 | 493 | 54.77 | 6.27 | 39.97 | 79.05 | Nucleus |
| Actinidia21152 | AcbHLH78 | 296 | 31.42 | 5.91 | 39.09 | 75.57 | Nucleus |
| Actinidia21104 | AcbHLH79 | 251 | 28.29 | 9.25 | 55.15 | 73.11 | Nucleus |
| Actinidia13678 | AcbHLH80 | 349 | 38.70 | 9.65 | 70.31 | 71.35 | Nucleus |
| Actinidia19903 | AcbHLH81 | 319 | 35.38 | 5.94 | 55.09 | 69.66 | Nucleus |
| Actinidia19843 | AcbHLH82 | 277 | 31.03 | 6.25 | 44.33 | 65.45 | Nucleus |
| Actinidia17442 | AcbHLH83 | 308 | 34.61 | 6.27 | 44.63 | 66.2 | Nucleus |
| Actinidia11941 | AcbHLH84 | 315 | 35.58 | 5.33 | 55.9 | 78.89 | Nucleus |
| Actinidia00404 | AcbHLH85 | 253 | 27.45 | 6.75 | 50.21 | 67.15 | Nucleus |
| Actinidia00378 | AcbHLH86 | 292 | 31.97 | 5.06 | 59.09 | 62.81 | Nucleus |
| Actinidia17735 | AcbHLH87 | 721 | 77.41 | 6.96 | 57.31 | 61.96 | Nucleus |
| Actinidia16869 | AcbHLH88 | 205 | 23.10 | 10.53 | 69.65 | 77.66 | Nucleus |
| Actinidia16799 | AcbHLH89 | 260 | 28.67 | 6.61 | 57.02 | 79.88 | Nucleus |
| Actinidia13413 | AcbHLH90 | 546 | 59.01 | 5.23 | 52.19 | 63.37 | Nucleus |
| Actinidia01170 | AcbHLH91 | 281 | 30.97 | 6.98 | 48.97 | 61.46 | Nucleus |
| Actinidia06881 | AcbHLH92 | 449 | 49.80 | 6.32 | 50.47 | 55.79 | Nucleus |
| Actinidia07184 | AcbHLH93 | 217 | 24.54 | 5.05 | 40.84 | 77.74 | Nucleus |
| Actinidia37637 | AcbHLH94 | 183 | 20.76 | 5.9 | 54.35 | 91.58 | Nucleus |
| Actinidia33058 | AcbHLH95 | 155 | 17.73 | 6.05 | 12.81 | 93.55 | Nucleus |
| Actinidia18317 | AcbHLH96 | 213 | 23.77 | 9.26 | 46.34 | 75.49 | Nucleus |
| Actinidia18311 | AcbHLH97 | 441 | 47.96 | 4.95 | 56.28 | 72.97 | Nucleus |
| Actinidia10438 | AcbHLH98 | 608 | 66.78 | 8.29 | 57.91 | 64 | Nucleus |
| Actinidia20625 | AcbHLH99 | 276 | 30.92 | 6.25 | 49.25 | 68.19 | Nucleus |
| Actinidia14806 | AcbHLH100 | 750 | 83.05 | 6.66 | 46.96 | 66.84 | Nucleus |
| Actinidia27418 | AcbHLH101 | 237 | 26.81 | 6.25 | 65.91 | 94.85 | Nucleus |
| Actinidia27380 | AcbHLH102 | 599 | 68.23 | 8.68 | 53.28 | 85.69 | Nucleus |
| Actinidia27296 | AcbHLH103 | 755 | 83.45 | 5.35 | 45.65 | 77.09 | Nucleus |
| Actinidia27284 | AcbHLH104 | 533 | 59.82 | 8.65 | 42.95 | 81.43 | Nucleus |
| Actinidia14904 | AcbHLH105 | 260 | 29.44 | 5.99 | 42.92 | 61.15 | Nucleus |
| Actinidia38397 | AcbHLH106 | 330 | 36.99 | 5.7 | 65.56 | 76.91 | Nucleus |
| Actinidia31927 | AcbHLH107 | 321 | 35.59 | 6 | 50.16 | 73.46 | Nucleus |
| Actinidia08197 | AcbHLH108 | 240 | 26.30 | 5.4 | 50.65 | 69.92 | Nucleus |
| Actinidia38336 | AcbHLH109 | 446 | 46.07 | 5.99 | 54.91 | 74.06 | Nucleus |
| Actinidia09561 | AcbHLH110 | 736 | 82.81 | 5.93 | 57.82 | 78.52 | Nucleus |
| Actinidia09499 | AcbHLH111 | 467 | 50.40 | 6.06 | 70.27 | 55.05 | Nucleus |
| Actinidia09453 | AcbHLH112 | 542 | 58.76 | 6.71 | 51.5 | 64.78 | Nucleus |
| Actinidia30501 | AcbHLH113 | 319 | 35.97 | 5.65 | 60.32 | 65.42 | Nucleus |
| Actinidia24957 | AcbHLH114 | 455 | 50.45 | 5.72 | 43.3 | 74.07 | Nucleus |
| Actinidia06642 | AcbHLH115 | 218 | 25.16 | 6.6 | 58.76 | 65.78 | Nucleus |
| Actinidia06533 | AcbHLH116 | 184 | 21.07 | 8.55 | 69.8 | 65.16 | Nucleus |
| Actinidia21519 | AcbHLH117 | 337 | 37.12 | 8.7 | 46.52 | 79.02 | Nucleus |
| Actinidia21564 | AcbHLH118 | 404 | 44.73 | 7.08 | 50.42 | 52.95 | Nucleus |
| Actinidia33486 | AcbHLH119 | 410 | 44.23 | 6.09 | 60.61 | 62.34 | Nucleus |
| Actinidia28094 | AcbHLH120 | 399 | 44.21 | 6.57 | 50.12 | 82.36 | Nucleus |
| Actinidia28011 | AcbHLH121 | 339 | 37.61 | 8.27 | 39.69 | 77.96 | Nucleus |
| Actinidia31235 | AcbHLH122 | 315 | 35.49 | 9.49 | 57.19 | 61.62 | Nucleus |
| Actinidia39893 | AcbHLH123 | 209 | 23.82 | 5 | 54.91 | 72.39 | Nucleus |
| Actinidia39559 | AcbHLH124 | 268 | 29.54 | 6.39 | 63.36 | 71.68 | Nucleus |
| Actinidia08446 | AcbHLH125 | 234 | 27.52 | 8.84 | 54.66 | 72.95 | Nucleus |
| Actinidia07548 | AcbHLH126 | 243 | 27.66 | 5.08 | 60.3 | 88.31 | Nucleus |
| Actinidia20294 | AcbHLH127 | 320 | 35.28 | 5.83 | 52.08 | 75.59 | Nucleus |
| Actinidia20396 | AcbHLH128 | 350 | 38.32 | 6.49 | 58.01 | 63.8 | Nucleus |
| Actinidia10972 | AcbHLH129 | 254 | 28.49 | 8.73 | 63.26 | 84.13 | Nucleus |
| Actinidia12492 | AcbHLH130 | 240 | 26.46 | 5.88 | 57.28 | 67.88 | Nucleus |
| Actinidia05126 | AcbHLH131 | 236 | 25.98 | 6.77 | 51.85 | 78.6 | Nucleus |
| Actinidia05158 | AcbHLH132 | 392 | 43.36 | 6.12 | 65.09 | 51.99 | Nucleus |
| Actinidia00747 | AcbHLH133 | 555 | 61.34 | 6.18 | 62.7 | 87.3 | Nucleus |
| Actinidia05553 | AcbHLH134 | 399 | 44.38 | 8.5 | 61.3 | 57.14 | Nucleus |
| Actinidia05642 | AcbHLH135 | 155 | 17.52 | 9.33 | 39.4 | 77.48 | Nucleus |
| Actinidia05815 | AcbHLH136 | 465 | 51.77 | 5.57 | 46.15 | 77.14 | Nucleus |
| Actinidia31050 | AcbHLH137 | 380 | 41.82 | 8.54 | 43.2 | 53.63 | Nucleus |
| Actinidia12888 | AcbHLH138 | 717 | 79.73 | 5.58 | 51.41 | 79.82 | Nucleus |
| Actinidia33765 | AcbHLH139 | 411 | 46.06 | 6.5 | 43.38 | 74.45 | Nucleus |
| Actinidia37726 | AcbHLH140 | 500 | 54.88 | 5.02 | 53.9 | 73.56 | Nucleus |
| Actinidia17543 | AcbHLH141 | 248 | 27.85 | 6.11 | 67 | 82.54 | Nucleus |
| Actinidia17610 | AcbHLH142 | 445 | 50.12 | 5.94 | 77.24 | 65.93 | Nucleus |
| Actinidia11340 | AcbHLH143 | 220 | 25.20 | 5.22 | 51.06 | 74.55 | Nucleus |
| Actinidia11564 | AcbHLH144 | 210 | 23.69 | 8.15 | 57.5 | 75.57 | Nucleus |
| Actinidia13834 | AcbHLH145 | 434 | 48.14 | 6.62 | 48.44 | 65.37 | Nucleus |
| Actinidia31866 | AcbHLH146 | 316 | 34.91 | 5.69 | 72.18 | 79.53 | Nucleus |
| Actinidia25964 | AcbHLH147 | 601 | 66.01 | 6.12 | 43.5 | 71.48 | Nucleus |
| Actinidia29708 | AcbHLH148 | 244 | 27.42 | 8.51 | 45.24 | 76.35 | Nucleus |
| Actinidia02969 | AcbHLH149 | 275 | 30.59 | 8.3 | 45.76 | 75.49 | Nucleus |
| Actinidia40062 | AcbHLH150 | 222 | 25.15 | 5.51 | 53.02 | 79.59 | Nucleus |
| Actinidia40097 | AcbHLH151 | 343 | 37.32 | 6.64 | 60.02 | 86.71 | Nucleus |
| Actinidia40189 | AcbHLH152 | 199 | 22.30 | 8.28 | 72 | 106.63 | Nucleus |
| Actinidia22194 | AcbHLH153 | 524 | 58.59 | 6 | 52.43 | 71.58 | Nucleus |
| Actinidia02003 | AcbHLH154 | 649 | 72.68 | 5.54 | 48.59 | 88.57 | Nucleus |
| Actinidia28511 | AcbHLH155 | 310 | 34.83 | 7.02 | 49.56 | 81.71 | Nucleus |
| Actinidia20683 | AcbHLH156 | 417 | 46.11 | 8.16 | 59.69 | 50.6 | Nucleus |
| Actinidia16303 | AcbHLH157 | 467 | 52.27 | 7.32 | 63.68 | 52.01 | Nucleus |
| Actinidia24146 | AcbHLH158 | 316 | 35.43 | 5.07 | 42.09 | 79.87 | Nucleus |
| Actinidia30102 | AcbHLH159 | 494 | 54.19 | 5.66 | 70.15 | 61.3 | Nucleus |
| Actinidia09094 | AcbHLH160 | 287 | 30.48 | 6.32 | 48.16 | 86.76 | Nucleus |
| Actinidia09144 | AcbHLH161 | 694 | 76.45 | 5.33 | 64.13 | 79.14 | Nucleus |
| Actinidia11692 | AcbHLH162 | 680 | 72.66 | 6.48 | 57.74 | 63.72 | Nucleus |
| Actinidia16479 | AcbHLH163 | 239 | 26.50 | 9.67 | 56.77 | 77.15 | Nucleus |
| Actinidia16476 | AcbHLH164 | 449 | 49.73 | 5.28 | 66.95 | 78.66 | Nucleus |
| Actinidia16406 | AcbHLH165 | 392 | 43.70 | 5.37 | 49.96 | 69.62 | Nucleus |
| Actinidia17228 | AcbHLH166 | 231 | 26.08 | 6.32 | 47.42 | 59.48 | Nucleus |
| Actinidia29906 | AcbHLH167 | 473 | 53.19 | 7.71 | 60.27 | 78.37 | Nucleus |
| Actinidia29957 | AcbHLH168 | 187 | 21.24 | 4.62 | 68.14 | 88.18 | Nucleus |
| Actinidia23288 | AcbHLH169 | 318 | 34.88 | 9.37 | 47.88 | 75.44 | Nucleus |
| Actinidia01096 | AcbHLH170 | 330 | 36.92 | 6.26 | 59.28 | 71.85 | Nucleus |
| Actinidia10226 | AcbHLH171 | 409 | 44.71 | 5.92 | 59.59 | 68.92 | Nucleus |
| Actinidia10353 | AcbHLH172 | 287 | 31.71 | 5.62 | 58.77 | 86.76 | Nucleus |
| Actinidia01558 | AcbHLH173 | 256 | 27.73 | 5.41 | 57.89 | 69.06 | Nucleus |
| Actinidia22852 | AcbHLH174 | 784 | 84.87 | 5.74 | 48.41 | 74.25 | Nucleus |
| Actinidia22974 | AcbHLH175 | 308 | 34.82 | 8.19 | 50.48 | 57.95 | Nucleus |
| Actinidia08250 | AcbHLH176 | 340 | 38.45 | 4.99 | 47.38 | 61.44 | Nucleus |
| Actinidia19604 | AcbHLH177 | 299 | 34.03 | 5.16 | 61.68 | 83.75 | Nucleus |
| Actinidia32124 | AcbHLH178 | 363 | 40.12 | 6.64 | 52.64 | 67.44 | Nucleus |
| Actinidia22518 | AcbHLH179 | 376 | 41.31 | 7.77 | 55.71 | 72.69 | Nucleus |
| Actinidia22640 | AcbHLH180 | 277 | 30.00 | 9.26 | 48.61 | 60.61 | Nucleus |
| Actinidia22645 | AcbHLH181 | 180 | 19.88 | 5.44 | 50.7 | 78.06 | Nucleus |
| Actinidia22688 | AcbHLH182 | 336 | 37.41 | 5.29 | 52.08 | 52.32 | Nucleus |
| Actinidia19291 | AcbHLH183 | 372 | 40.57 | 5.96 | 55.53 | 63.71 | Nucleus |
| Actinidia19259 | AcbHLH184 | 262 | 29.83 | 4.94 | 58.37 | 75.5 | Nucleus |
| Actinidia30810 | AcbHLH185 | 148 | 16.12 | 8.3 | 47.33 | 77.16 | Nucleus |
| Actinidia30799 | AcbHLH186 | 291 | 32.10 | 6.67 | 57.21 | 83.54 | Nucleus |
| Actinidia15954 | AcbHLH187 | 254 | 27.84 | 6.36 | 58.57 | 65.79 | Nucleus |
| ID | Classification | GO Terms | All Gene | Up_Gene | Down_Gene |
|---|---|---|---|---|---|
| GO:0009739 | Biological process | Response to gibberellin | 7 | 1 | 2 |
| GO:0009725 | Biological process | Response to hormone | 29 | 1 | 9 |
| GO:0009753 | Biological process | Response to jasmonic acid | 11 | 4 | |
| GO:0006970 | Biological process | Response to osmotic stress | 6 | 0 | |
| GO:0009737 | Biological process | Response to abscisic acid | 12 | 4 | |
| GO:0006952 | Biological process | Defense response | 14 | 7 | |
| GO:0031347 | Biological process | Regulation of defense response | 11 | 4 | |
| GO:0009755 | Biological process | Hormone-mediated signaling pathway | 19 | 8 | |
| GO:0009867 | Biological process | Jasmonic acid mediated signaling pathway | 10 | 3 | |
| GO:0071470 | Biological process | Cellular response to osmotic stress | 2 | 0 | |
| GO:0071215 | Biological process | Cellular response to abscisic acid stimulus | 6 | 4 | |
| GO:0009414 | Biological process | Response to water deprivation | 3 | 1 | |
| GO:0043227 | Cellular component | Membrane-bounded organelle | 131 | 15 | 49 |
| GO:0016020 | Cellular component | Membrane | 2 | ||
| GO:0043226 | Cellular component | Organelle | 131 | 15 | 49 |
| GO:0031090 | Cellular component | Organelle membrane | 131 | 15 | 49 |
| GO:0009059 | Molecular function | Macromolecule biosynthetic Process | 114 | 13 | 45 |
| GO:0005515 | Molecular function | Protein binding | 130 | 15 | 49 |
| GO:0140110 | Molecular function | Transcription regulator activity | 123 | 13 | 47 |
| GO:0005488 | Molecular function | Binding | 132 | 15 | 49 |
| ID | Count | Description |
|---|---|---|
| ko04075 | 82 | Plant hormone signal transduction |
| ko04016 | 33 | MAPK signaling pathway |
| ko04712 | 44 | Circadian rhythm—plant |
| Index | Compounds | T1 | T2 | T3 | CK1 | CK2 | CK3 |
|---|---|---|---|---|---|---|---|
| ABA | Abscisic acid | 1155.61 | 1322.69 | 1037.88 | 100.12 | 97.47 | 80.19 |
| ABA-GE | ABA-glucosyl ester | 33448.82 | 35722.10 | 23656.63 | 13126.39 | 14161.25 | 14810.04 |
| ABA-ald | Abscisic acid | 283.66 | 326.9766 | 202.40 | 93.87 | 107.26 | 111.77 |
| ACC | Ethylene | 68.08 | 72.88 | 74.33 | 28.50 | 32.62 | 35.84 |
| GA20 | Gibberellin | 0.0 | 0.0 | 0.0 | 6.53 | 7.77 | 5.60 |
| GA19 | Gibberellin | 0.0 | 0.0 | 0.0 | 2.45 | 0.0 | 3.76 |
| 12-OH-JA | Jasmonic acid | 3782.05 | 4012.92 | 3487.32 | 1378.49 | 1280.27 | 1596.73 |
| JA-Phe | Jasmonic acid | 0.0 | 0.26 | 0.0 | 0.27 | 0.30 | 0.37 |
| OPDA | Jasmonic acid | 500.31 | 883.45 | 422.35 | 162.25 | 253.69 | 132.85 |
| JA-Val | Jasmonic acid | 0.97 | 1.05 | 1.26 | 3.92 | 4.49 | 4.98 |
| JA-ILE | Jasmonic acid | 62.40 | 66.17 | 59.56 | 166.79 | 172.82 | 192.75 |
| JA | Jasmonic acid | 65.22 | 98.02 | 83.97 | 215.15 | 255.65 | 228.16 |
References
- Pan, L.; Ma, J.; Li, J.; Yin, B.; Fu, C. Advancses of salt stress-responsive transcription factors in plants. Sheng Wu Gong Cheng Xue Bao 2022, 38, 50–65. (In Chinese) [Google Scholar] [CrossRef] [PubMed]
- Riechmann, L.; Muyldermans, S. Single domain antibodies: Comparison of camel VH and camelised human VH domains. J. Immunol. Methods 1999, 231, 25–38. [Google Scholar] [CrossRef]
- Carretero-Paulet, L.; Galstyan, A.; Roig-Villanova, I.; Martinez-Garcia, J.F.; Bilbao-Castro, J.R.; Robertson, D.L. Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiol. 2010, 153, 1398–1412. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yang, S.; Sun, Q.W.; Chen, H.Y.; Yang, T.; Huang, Y. Genome-wide identification and expression analysis of the bHLH transcription factor family in Dendrobium cinereum. Biotechnol. Bull. 2024, 40, 203–218. [Google Scholar]
- Ludwig, S.R.; Habera, L.F.; Dellaporta, S.L.; Wessler, S.R. Lc, a Member of the Maize R Gene Family Responsible for Tissue-Specific Anthocyanin Production, Encodes a Protein Similar to Transcriptional Activators and Contains the Myc-Homology Region. Proc. Natl. Acad. Sci. USA 1989, 86, 7092–7096. [Google Scholar] [CrossRef] [PubMed]
- Heim, M.A.; Jakoby, M.; Werber, M.; Martin, C.; Weisshaar, B.; Bailey, P.C. The Basic Helix-Loop-Helix Transcription Factor Family in Plants: A Genome-Wide Study of Protein Structure and Functional Diversity. Mol. Biol. Evol. 2003, 20, 735–747. [Google Scholar] [CrossRef]
- Murre, C.; McCaw, P.S.; Baltimore, D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell 1989, 56, 777–783. [Google Scholar] [CrossRef]
- Li, G.; Jin, L.; Sheng, S. Genome-wide identification of bHLH transcription factor in Medicago sativa in response to cold stress. Genes 2022, 13, 2371. [Google Scholar] [CrossRef]
- Li, X.; Duan, X.; Jiang, H.; Sun, Y.; Tang, Y.; Yuan, Z.; Guo, J.; Liang, W.; Chen, L.; Yin, J.; et al. Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis. Plant Physiol. 2006, 141, 1167–1184. [Google Scholar] [CrossRef]
- Liu, Y.; Lin-Wang, K.; Espley, R.V.; Wang, L.; Li, Y.; Liu, Z.; Zhou, P.; Zeng, L.; Zhang, X.; Zhang, J.; et al. StMYB44 negatively regulates anthocyanin biosynthesis at high temperatures in tuber flesh of potato. J. Exp. Bot. 2019, 70, 3809–3824. [Google Scholar] [CrossRef]
- Kavas, M.; Baloglu, M.C.; Atabay, E.S.; Ziplar, U.T.; Dasgan, H.Y.; Ünver, T. Genome-wide characterization and expression analysis of common bean bHLH transcription factors in response to excess salt concentration. Mol. Genet. Genom. 2016, 291, 129–143. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Zhu, H.; Chen, D.; Li, Z.; Peng, R.; Yao, Q. A grape bHLH transcription factor gene, VvbHLH1, increases the accumulation of flavonoids and enhances salt and drought tolerance in transgenic Arabidopsis thaliana. Plant Cell Tissue Organ Cult. 2016, 125, 387–398. [Google Scholar] [CrossRef]
- Yang, J.; Gao, M.; Huang, L.; Wang, Y.; van Nocker, S.; Wan, R.; Guo, C.; Wang, X.; Gao, H. Identification and expression analysis of the apple (Malus × domestica) basic helix-loop-helix transcription factor family. Sci. Rep. 2017, 7, 28. [Google Scholar] [CrossRef]
- Chen, C.; Li, X.; Wen, K.; Yin, T.; Tian, P.; Zhao, K.; Zhang, L.; Zhou, X.; Liu, X.; Zhang, H. Identification of the bHLH transcription factor family in Orah mandarin and the response of CrbHLH46 to low-temperature stress. Plants 2025, 14, 882. [Google Scholar] [CrossRef] [PubMed]
- Buck, M.J.; Atchley, W.R. Phylogenetic analysis of plant basic helix-loop-helix proteins. J. Mol. Evol. 2003, 56, 742–750. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Li, X.; Han, P.; Liu, H.; Gong, J.; Zhou, W.; Shi, B.; Liu, A.; Xu, L. Genome-wide investigation of bHLH genes and expression analysis under different biotic and abiotic stresses in Helianthus annuus L. Int. J. Biol. Macromol. 2021, 189, 72–83. [Google Scholar] [CrossRef]
- Ji, X.; Nie, X.; Liu, Y.; Zheng, L.; Zhao, H.; Zhang, B.; Huo, L.; Wang, Y. A bHLH gene from Tamarix hispida improves abiotic stress tolerance by enhancing osmotic potential and decreasing reactive oxygen species accumulation. Tree Physiol. 2016, 36, 193–207. [Google Scholar] [CrossRef]
- Yang, Y.-Y.; Zheng, P.-F.; Ren, Y.-R.; Yao, Y.-X.; You, C.-X.; Wang, X.-F.; Hao, Y.-J. Apple MdSAT1 encodes a bHLHm1 transcription factor involved in salinity and drought responses. Planta 2021, 253, 46. [Google Scholar] [CrossRef]
- Liu, W.; Tai, H.; Li, S.; Gao, W.; Zhao, M.; Xie, C.; Li, W. bHLH122 Is important for drought and osmotic stress resistance in Arabidopsis and in the repression of aba catabolism. New Phytol. 2014, 201, 1192–1204. [Google Scholar] [CrossRef]
- Zhu, L.; Yin, T.; Zhang, M.; Yang, X.; Wu, J.; Cai, H.; Yang, N.; Li, X.; Wen, K.; Chen, D.; et al. Genome-wide identification and expression pattern analysis of the kiwifruit GRAS transcription factor family in response to salt stress. BMC Genom. 2024, 25, 12. [Google Scholar] [CrossRef]
- Ferguson, A.R.; Ferguson, L.R. Are kiwifruit really good for you? Acta Hortic. 2003, 610, 131–138. [Google Scholar] [CrossRef]
- Lee, Y.K.; Low, K.Y.; Siah, K.; Drummond, L.M.; Gwee, K.A. Kiwifruit (Actinidia deliciosa) changes intestinal microbial profile. Microb. Ecol. Health Dis. 2012, 23, 18572. [Google Scholar] [CrossRef] [PubMed]
- FAOSTAT: The United Nations Food and Agriculture Organisation of the United Nations (FAO). Available online: https://www.fao.org/ (accessed on 26 September 2024).
- Xia, H.; Ni, Z.; Hu, R.; Lin, L.; Deng, H.; Wang, J.; Tang, Y.; Sun, G.; Wang, X.; Li, H.; et al. Melatonin alleviates drought stress by a non-enzymatic and enzymatic antioxidative system in kiwifruit seedlings. Int. J. Mol. Sci. 2020, 21, 852. [Google Scholar] [CrossRef]
- Bao, W.W.; Chen, X.; Li, R.N.; Li, M.; Xie, C.; Dou, M.; Zhang, K.; Wang, J.; Gao, Z.; Liu, Z.; et al. Comprehensive assessment of drought resistance and recovery in kiwifruit genotypes using multivariate analysis. Plant J. 2024, 119, 100–114. [Google Scholar] [CrossRef]
- Liu, X.; Quan, W.; Bartels, D. Stress memory responses and seed priming correlate with drought tolerance in plants: An overview. Planta 2022, 255, 45. [Google Scholar] [CrossRef]
- Ndlovu, E.; van Staden, J.; Maphosa, M. Morpho-physiological effects of moisture, heat and combined stresses on Sorghum bicolor [Moench (L.)] and its acclimation mechanisms. Plant Stress 2021, 2, 100018. [Google Scholar] [CrossRef]
- Hussain, H.A.; Hussain, S.; Khaliq, A.; Ashraf, U.; Anjum, S.A.; Men, S.; Wang, L. Chilling and drought stresses in crop plants: Implications, cross talk, and potential management opportunities. Front. Plant Sci. 2018, 9, 393. [Google Scholar] [CrossRef]
- Abreha, K.B.; Enyew, M.; Carlsson, A.S.; Vetukuri, R.R.; Feyissa, T.; Motlhaodi, T.; Ng’uni, D.; Geleta, M. Sorghum in dryland: Morphological, physiological, and molecular responses of sorghum under drought stress. Planta 2021, 255, 20. [Google Scholar] [CrossRef] [PubMed]
- Dichio, B.; Montanaro, G.; Sofo, A.; Xiloyannis, C. Stem and whole-plant hydraulics in olive (Olea europaea) and kiwifruit (Actinidia deliciosa). Trees 2013, 27, 183–191. [Google Scholar] [CrossRef]
- Hahne, G.; Tomlinson, L.; Nogué, F. Precision genetic engineering tools for next-generation plant breeding. Plant Cell Rep. 2019, 38, 435–436. [Google Scholar] [CrossRef]
- Lockhart, D.J.; Winzeler, E.A. Genomics, gene expression and DNA arrays. Nature 2000, 405, 827–836. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Chen, J.; Wang, T.; Gao, C.; Li, Z.; Guo, L.; Xu, J.; Cheng, Y. Linking plant secondary metabolites and plant microbiomes: A Review. Front. Plant Sci. 2021, 12, 621276. [Google Scholar] [CrossRef]
- Guijas, C.; Montenegro-Burke, J.R.; Warth, B.; Spilker, M.E.; Siuzdak, G. Metabolomics activity screening for identifying metabolites that modulate phenotype. Nat. Biotechnol. 2018, 36, 316–320. [Google Scholar] [CrossRef]
- Fiehn, O. Metabolomics by Gas Chromatography-Mass Spectrometry: Combined Targeted and Untargeted Profiling. Curr. Protoc. Mol. Biol. 2016, 114, 30–34. [Google Scholar] [CrossRef] [PubMed]
- Luo, F.; Yu, Z.; Zhou, Q.; Huang, A. Multi-omics-based discovery of plant signaling molecules. Metabolites 2022, 12, 76. [Google Scholar] [CrossRef] [PubMed]
- Meena, K.K.; Sorty, A.M.; Bitla, U.M.; Choudhary, K.; Gupta, P.; Pareek, A.; Singh, D.P.; Prabha, R.; Sahu, P.K.; Gupta, V.K.; et al. Abiotic stress responses and microbe-mediated mitigation in plants: The omics strategies. Front. Plant Sci. 2017, 8, 172. [Google Scholar] [CrossRef]
- Yang, S.; Chu, N.; Zhou, H.; Li, J.; Feng, N.; Su, J.; Deng, Z.; Shen, X.; Zheng, D. Integrated analysis of transcriptome and metabolome reveals the regulation of chitooligosaccharide on drought tolerance in sugarcane (Saccharum spp. hybrid) under drought stress. Int. J. Mol. Sci. 2022, 23, 9737. [Google Scholar] [CrossRef]
- Chen, S.; Zhong, K.; Li, Y.; Bai, C.; Xue, Z.; Wu, Y. Joint transcriptomic and metabolomic analysis provides new insights into drought resistance in watermelon (Citrullus lanatus). Front. Plant Sci. 2024, 15, 1364631. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Yang, F.; Lv, G. Combined analysis of transcriptome and metabolome reveals the molecular mechanism and candidate genes of Haloxylon drought tolerance. Front. Plant Sci. 2022, 13, 1020367. [Google Scholar] [CrossRef]
- Yue, J.; Liu, J.; Tang, W.; Wu, Y.Q.; Tang, X.; Li, W.; Yang, Y.; Wang, L.; Huang, S.; Fang, C.; et al. Kiwifruit genome database (kgd): A comprehensive resource for kiwifruit genomics. Hortic. Res. 2020, 7, 117. [Google Scholar] [CrossRef]
- Jung, S.; Lee, T.; Cheng, C.H.; Buble, K.; Zheng, P.; Yu, J.; Humann, J.; Ficklin, S.P.; Gasic, K.; Scott, K.; et al. 15 years of GDR: New data and functionality in the Genome Database for Rosaceae. Nucleic Acids Res. 2019, 47, D1137–D1145. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zi, Y.; Yang, X.; Yang, X.; Zhu, L.; Cai, H.; Yin, T.; Liu, X.; Zhang, H. Genome-wide analysis of the CsAP2/ERF gene family of sweet orange (Citrus sinensis) and joint analysis of transcriptional metabolism under salt stress. Ann. Bot. 2025, mcaf006. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F. CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017, 45, D200–D203. [Google Scholar] [CrossRef]
- Zi, Y.; Zhang, Z.; Zhao, K.; Yang, X.; Zhu, L.; Yin, T.; Chen, C.; Wen, K.; Li, X.; Zhang, H.; et al. Genome-wide identification of kiwifruit K+ channel Shaker family members and their response to low-K+ stress. BMC Plant Biol. 2024, 24, 833. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein identification and analysis tools in the ExPASy server. In 2-D Proteome Analysis Protocols; Humana Press: Totowa, NJ, USA, 1999; Volume 112, pp. 531–552. [Google Scholar] [CrossRef]
- Chou, K.C.; Shen, H.B. Plant-mPLoc: A top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS ONE 2010, 5, e11335. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Xie, J.; Chen, Y.; Cai, G.; Cai, R.; Hu, Z.; Wang, H. Tree Visualization By One Table (tvBOT): A web application for visualizing, modifying and annotating phylogenetic trees. Nucleic Acids Res. 2023, 51, W587–W592. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Zi, Y.; Zhang, M.; Yang, X.; Zhao, K.; Yin, T.; Wen, K.; Li, X.; Liu, X.; Zhang, H. Identification of the sweet orange (Citrus sinensis) bHLH gene family and the role of CsbHLH55 and CsbHLH87 in regulating salt stress. Plant Genome. 2024, 17, e20502. [Google Scholar] [CrossRef]
- Cantalapiedra, C.P.; Hernández-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. eggNOG-mapper v2: Functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol. Biol. Evol. 2021, 38, 5825–5829. [Google Scholar] [CrossRef]
- Valmonte-Cortes, G.R.; Higgins, C.M.; MacDiarmid, R.M. Arabidopsis calcium dependent protein kinase 3, and its orthologues OsCPK1, OsCPK15, and AcCPK16, are involved in biotic and abiotic stresses. Plants 2025, 14, 294. [Google Scholar] [CrossRef] [PubMed]
- Tan, Z.; Lu, D.; Yu, Y.; Li, L.; Dong, W.; Xu, L.; Yang, Q.; Wan, X.; Liang, H. Genome-wide identification and characterization of the bHLH gene family and its response to abiotic stresses in Carthamus tinctorius. Plants 2023, 12, 3764. [Google Scholar] [CrossRef]
- Mahmood, T.; Khalid, S.; Abdullah, M.; Ahmed, Z.; Shah, M.K.N.; Ghafoor, A.; Du, X. Insights into drought stress signaling in plants and the molecular genetic basis of cotton drought tolerance. Cells 2019, 9, 105. [Google Scholar] [CrossRef] [PubMed]
- Ullah, A.; Manghwar, H.; Shaban, M.; Khan, A.H.; Akbar, A.; Ali, U.; Ali, E.; Fahad, S. Phytohormones enhanced drought tolerance in plants: A coping strategy. Environ. Sci. Pollut. Res. Int. 2018, 25, 33103–33118. [Google Scholar] [CrossRef] [PubMed]
- Jogawat, A.; Yadav, B.; Chhaya Lakra, N.; Singh, A.K.; Narayan, O.P. Crosstalk between phytohormones and secondary metabolites in the drought stress tolerance of crop plants: A review. Physiol. Plant. 2021, 172, 1106–1132. [Google Scholar] [CrossRef]
- Wang, S.; Qiu, Y.; Zhu, F. Kiwifruit (Actinidia spp.): A review of chemical diversity and biological activities. Food Chem. 2021, 350, 128469. [Google Scholar] [CrossRef]
- Mukaka, M.M. Statistics corner: A guide to appropriate use of correlation coefficient in medical research. Malawi Med. J. 2012, 24, 69–71. [Google Scholar]
- Cradock-Henry, N.A. New Zealand kiwifruit growers’ vulnerability to climate and other stressors. Reg. Environ. Change 2017, 17, 245–259. [Google Scholar] [CrossRef]
- Bardi, L. Early kiwifruit decline: A soil-borne disease syndrome or a climate change effect on plant–soil relations? Front. Agron. 2020, 2, 3. [Google Scholar] [CrossRef]
- Gupta, A.; Sinha, R.; Fernandes, J.L.; Abdelrahman, M.; Burritt, D.J.; Tran, L.P. Phytohormones regulate convergent and divergent responses between individual and combined drought and pathogen infection. Crit. Rev. Biotechnol. 2020, 40, 320–340. [Google Scholar] [CrossRef]
- Mubarik, M.S.; Khan, S.H.; Sajjad, M.; Raza, A.; Hafeez, M.B.; Yasmeen, T.; Rizwan, M.; Ali, S.; Arif, M.S. A manipulative interplay between positive and negative regulators of phytohormones: A way forward for improving drought tolerance in plants. Physiol. Plant. 2021, 172, 1269–1290. [Google Scholar] [CrossRef]
- Zuo, Z.F.; Lee, H.Y.; Kang, H.G. Basic helix-loop-helix transcription factors: Regulators for plant growth development and abiotic stress responses. Int. J. Mol. Sci. 2023, 24, 1419. [Google Scholar] [CrossRef] [PubMed]
- Bailey, P.C.; Martin, C.; Toledo-Ortiz, G.; Quail, P.H.; Huq, E.; Heim, M.A.; Jakoby, M.; Werber, M.; Weisshaar, B. Update on the basic helix-loop-helix transcription factor gene family in Arabidopsis thaliana. Plant Cell 2003, 15, 2497–2502. [Google Scholar] [CrossRef]
- Wang, B.; Yang, X.; Jia, Y.; Xu, Y.; Jia, P.; Dang, N.; Wang, S.; Xu, T.; Zhao, X.; Gao, S.; et al. High-quality Arabidopsis thaliana Genome Assembly with Nanopore and HiFi Long Reads. Genom. Proteom. Bioinform. 2022, 20, 4–13. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Chen, Y.; Zhou, Y.; Zhang, Y.; Li, M.; Ouyang, Y.; Chebotarov, D.; Mauleon, R.; Zhao, H.; Xie, W.; et al. Rice Gene Index: A comprehensive pan-genome database for comparative and functional genomics of Asian rice. Mol. Plant 2023, 16, 798–801. [Google Scholar] [CrossRef]
- Mao, K.; Dong, Q.; Li, C.; Liu, C.; Ma, F. Genome wide identification and characterization of apple bHLH transcription factors and expression analysis in response to drought and salt stress. Front. Plant Sci. 2017, 8, 480. [Google Scholar] [CrossRef]
- Chen, X.; Yao, C.; Liu, J.; Liu, J.; Fang, J.; Deng, H.; Yao, Q.; Kang, T.; Guo, X. Basic helix-loop-helix (bHLH) gene family in rye (Secale cereale L.): Genome-wide identification, phylogeny, evolutionary expansion and expression analyses. BMC Genom. 2024, 25, 67. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Chen, W.; Liu, R.; Shi, B.; Shu, Y.; Zhang, H. Genome-wide characterization and expression analysis of bHLH gene family in physic nut (Jatropha curcas L.). PeerJ 2022, 10, e13786. [Google Scholar] [CrossRef]
- Yu, L.; Fei, C.; Wang, D.; Huang, R.; Xuan, W.; Guo, C.; Jing, L.; Meng, W.; Yi, L.; Zhang, H.; et al. Genome-wide identification, evolution and expression profiles analysis of bHLH gene family in Castanea mollissima. Front. Genet. 2023, 14, 1193953. [Google Scholar] [CrossRef]
- Seo, J.S.; Joo, J.; Kim, M.J.; Kim, Y.K.; Nahm, B.H.; Song, S.I.; Cheong, J.J.; Lee, J.S.; Kim, J.K.; Choi, Y.D. OsbHLH148, a basic helix-loop-helix protein, interacts with OsJAZ proteins in a jasmonate signaling pathway leading to drought tolerance in rice. Plant J. 2011, 65, 907–921. [Google Scholar] [CrossRef]
- Le Hir, R.; Castelain, M.; Chakraborti, D.; Moritz, T.; Dinant, S.; Bellini, C. AtbHLH68 transcription factor contributes to the regulation of ABA homeostasis and drought stress tolerance in Arabidopsis thaliana. Physiol. Plant. 2017, 160, 312–327. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Liu, B.; Zhang, X.; Wang, M.; Liang, H. Phytohormone response of drought-acclimated Illicium difengpi (Schisandraceae). Int. J. Mol. Sci. 2023, 24, 16443. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Yan, J.; Su, Z.; Chang, J.; Yang, J.; Wei, C.; Zhang, Y.; Ma, J.; Zhang, X.; Li, H. Abscisic acid mediates grafting-induced cold tolerance of watermelon via interaction with melatonin and methyl jasmonate. Front. Plant Sci. 2021, 12, 785317. [Google Scholar] [CrossRef]
- Fu, J.; Wu, H.; Ma, S.; Xiang, D.; Liu, R.; Xiong, L. OsJAZ1 Attenuates drought resistance by regulating ja and ABA signaling in rice. Front. Plant Sci. 2017, 8, 2108. [Google Scholar] [CrossRef]
- Liao, Z.; Zhang, Y.; Yu, Q.; Fang, W.; Chen, M.; Li, T.; Liu, Y.; Liu, Z.; Chen, L.; Yu, S.; et al. Coordination of growth and drought responses by GA-ABA signaling in rice. New Phytol. 2023, 240, 1149–1161. [Google Scholar] [CrossRef] [PubMed]
- Dong, Q.; Duan, D.; Zheng, W.; Huang, D.; Wang, Q.; Yang, J.; Liu, C.; Li, C.; Gong, X.; Li, C.; et al. Overexpression of MdVQ37 reduces drought tolerance by altering leaf anatomy and SA homeostasis in transgenic apple. Tree Physiol. 2022, 42, 160–174. [Google Scholar] [CrossRef]
- Zhang, H.; Cao, Y.; Shang, C.; Li, J.; Wang, J.; Wu, Z.; Ma, L.; Qi, T.; Fu, C.; Bai, Z.; et al. Genome-wide characterization of GRAS family genes in Medicago truncatula reveals their evolutionary dynamics and functional diversification. PLoS ONE 2017, 12, e0185439. [Google Scholar] [CrossRef]
- Singh, B.; Norvell, E.; Wijewardana, C.; Wallace, T.; Chastain, D.; Reddy, K.R. Assessing morphological characteristics of elite cotton lines from different breeding programmes for low temperature and drought tolerance. J. Agron. Crop. Sci. 2018, 204, 467–476. [Google Scholar] [CrossRef]
- Fan, G.; Xia, X.; Yao, W.; Cheng, Z.; Zhang, X.; Jiang, J.; Zhou, B.; Jiang, T. Genome-wide identification and expression patterns of the F-box family in poplar under salt stress. Int. J. Mol. Sci. 2022, 23, 10934. [Google Scholar] [CrossRef]
- Yu, Z.; Duan, X.; Luo, L.; Dai, S.; Ding, Z.; Xia, G. How plant hormones mediate salt stress responses. Trends Plant Sci. 2020, 25, 1117–1130. [Google Scholar] [CrossRef]
- Zhu, C.; Li, X.; Zhang, M.; Wang, S.; Jing, B.; Hu, C.; Thomas, H.R.; Zhou, Y.; Yu, J.; Hu, Z. ERF.D2 negatively controls drought tolerance through synergistic regulation of abscisic acid and jasmonic acid in tomato. Plant Biotechnol. J. 2025. [Google Scholar] [CrossRef] [PubMed]
- Chai, M.; Yang, F.; Cai, S.; Liu, T.; Xu, X.; Huang, Y.; Xi, X.; Yang, J.; Cao, Z.; Sun, L.; et al. Overexpression of the Transcription Factor GmbZIP60 Increases Salt and Drought Tolerance in Soybean (Glycine max). Int. J. Mol. Sci. 2025, 26, 3455. [Google Scholar] [CrossRef] [PubMed]
- Lymperopoulos, P.; Msanne, J.; Rabara, R. Phytochrome and phytohormones: Working in tandem for plant growth and development. Front. Plant Sci. 2018, 9, 1037. [Google Scholar] [CrossRef] [PubMed]
- Wasternack, C.; Hause, B. Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 2013, 111, 1021–1058. [Google Scholar] [CrossRef]
- Vick, B.A.; Zimmerman, D.C. The biosynthesis of jasmonic acid: A physiological role for plant lipoxygenase. Biochem. Biophys. Res. Commun. 1983, 111, 470–477. [Google Scholar] [CrossRef] [PubMed]
- Staswick, P.E.; Tiryaki, I. The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. Plant Cell 2004, 16, 2117–2127. [Google Scholar] [CrossRef]
- Fonseca, S.; Chini, A.; Hamberg, M.; Adie, B.; Porzel, A.; Kramell, R.; Miersch, O.; Wasternack, C.; Solano, R. (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat. Chem. Biol. 2009, 5, 344–350. [Google Scholar] [CrossRef]
- Liu, Y.; Du, M.; Deng, L.; Shen, J.; Fang, M.; Chen, Q.; Lu, Y.; Wang, Q.; Li, C.; Zhai, Q. MYC2 Regulates the termination of jasmonate signaling via an autoregulatory negative feedback loop. Plant Cell 2019, 31, 106–127. [Google Scholar] [CrossRef] [PubMed]
- Lorenzo, O.; Chico, J.M.; Sánchez-Serrano, J.J.; Solano, R. JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 2004, 16, 1938–1950. [Google Scholar] [CrossRef]
- Luo, L.; Wang, Y.; Qiu, L.; Han, X.; Zhu, Y.; Liu, L.; Man, M.; Li, F.; Ren, M.; Xing, Y. MYC2: A master switch for plant physiological processes and specialized metabolite synthesis. Int. J. Mol. Sci. 2023, 24, 3511. [Google Scholar] [CrossRef]
- Wang, R.; Yu, M.; Xia, J.; Xing, J.; Fan, X.; Xu, Q.; Cang, J.; Zhang, D. Overexpression of TaMYC2 confers freeze tolerance by ICE-CBF-COR module in Arabidopsis thaliana. Front. Plant Sci. 2022, 13, 1042889. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Movahedi, A.; Xu, C.; Sun, W.; Li, L.; Wang, P.; Li, D.; Zhuge, Q. Overexpression of PtHMGR enhances drought and salt tolerance of poplar. Ann. Bot. 2020, 125, 785–803. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, K.; Xu, R.; Yin, T.; Chen, X.; Ding, R.; Liu, X.; Zhang, H. Genome-Wide Identification of the bHLH Gene Family in Kiwifruit (Actinidia chinensis) and the Responses of AcbHLH84 and AcbHLH97 Under Drought Stress. Agronomy 2025, 15, 1598. https://doi.org/10.3390/agronomy15071598
Zhao K, Xu R, Yin T, Chen X, Ding R, Liu X, Zhang H. Genome-Wide Identification of the bHLH Gene Family in Kiwifruit (Actinidia chinensis) and the Responses of AcbHLH84 and AcbHLH97 Under Drought Stress. Agronomy. 2025; 15(7):1598. https://doi.org/10.3390/agronomy15071598
Chicago/Turabian StyleZhao, Ke, Rong Xu, Tuo Yin, Xia Chen, Renzhan Ding, Xiaozhen Liu, and Hanyao Zhang. 2025. "Genome-Wide Identification of the bHLH Gene Family in Kiwifruit (Actinidia chinensis) and the Responses of AcbHLH84 and AcbHLH97 Under Drought Stress" Agronomy 15, no. 7: 1598. https://doi.org/10.3390/agronomy15071598
APA StyleZhao, K., Xu, R., Yin, T., Chen, X., Ding, R., Liu, X., & Zhang, H. (2025). Genome-Wide Identification of the bHLH Gene Family in Kiwifruit (Actinidia chinensis) and the Responses of AcbHLH84 and AcbHLH97 Under Drought Stress. Agronomy, 15(7), 1598. https://doi.org/10.3390/agronomy15071598






