Identification and Genetic Diversity Analysis of Cucurbita Varieties Based on SSR Markers
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Source of SSR Primer
2.3. Field Planting and Traits Investigation
2.4. DNA Extraction
2.5. PCR Amplification
2.6. Polyacrylamide Gel Electrophoresis (PAGE)
2.6.1. Gel Making
2.6.2. Electrophoresis
2.6.3. Silver Staining
2.7. Fluorescent Capillary Electrophoresis
2.8. Data Analysis
3. Results
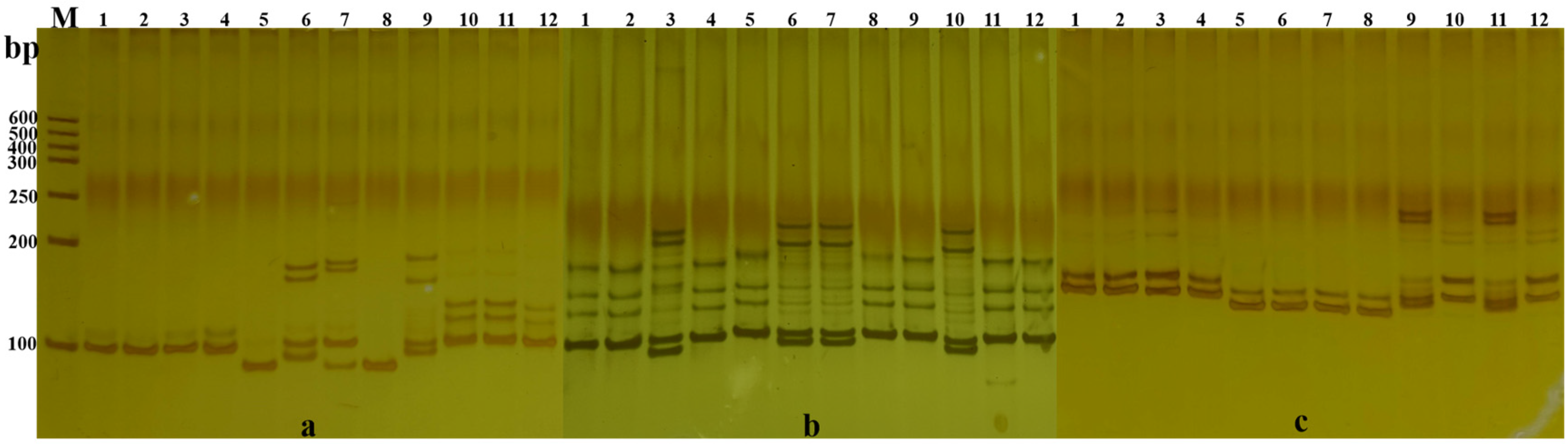
3.1. Primer Screening
3.2. Analysis of Core Primers
3.3. Construction of DNA Fingerprint Database
3.4. Selection of Reference Varieties
3.5. Analysis of Genetic Diversity of Cucurbita Varieties
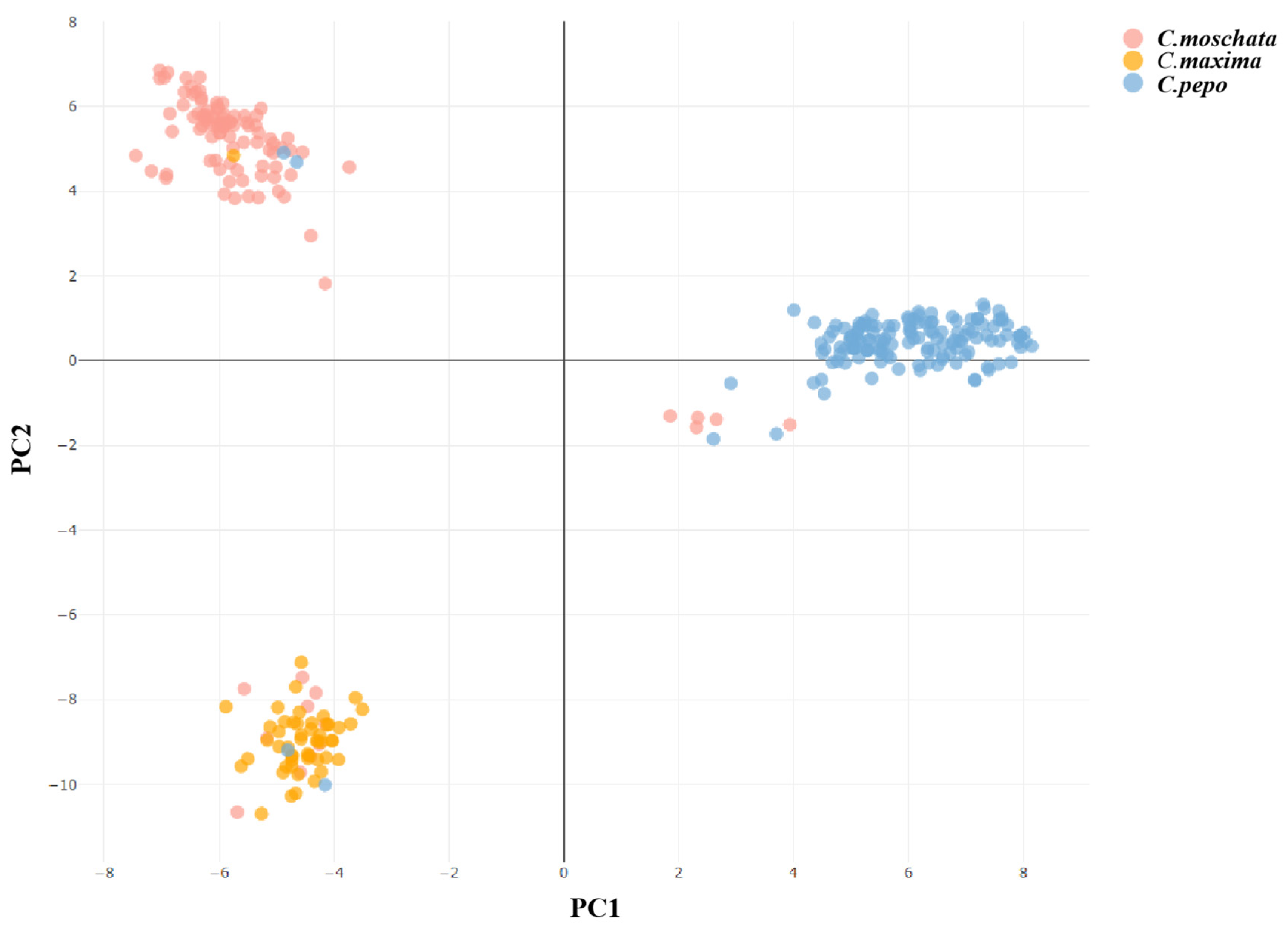
3.6. Principal Component Analysis
3.7. Analysis of Population Genetic Structure
3.8. Analysis of Diversity of Cucurbita Varieties in Different Regions
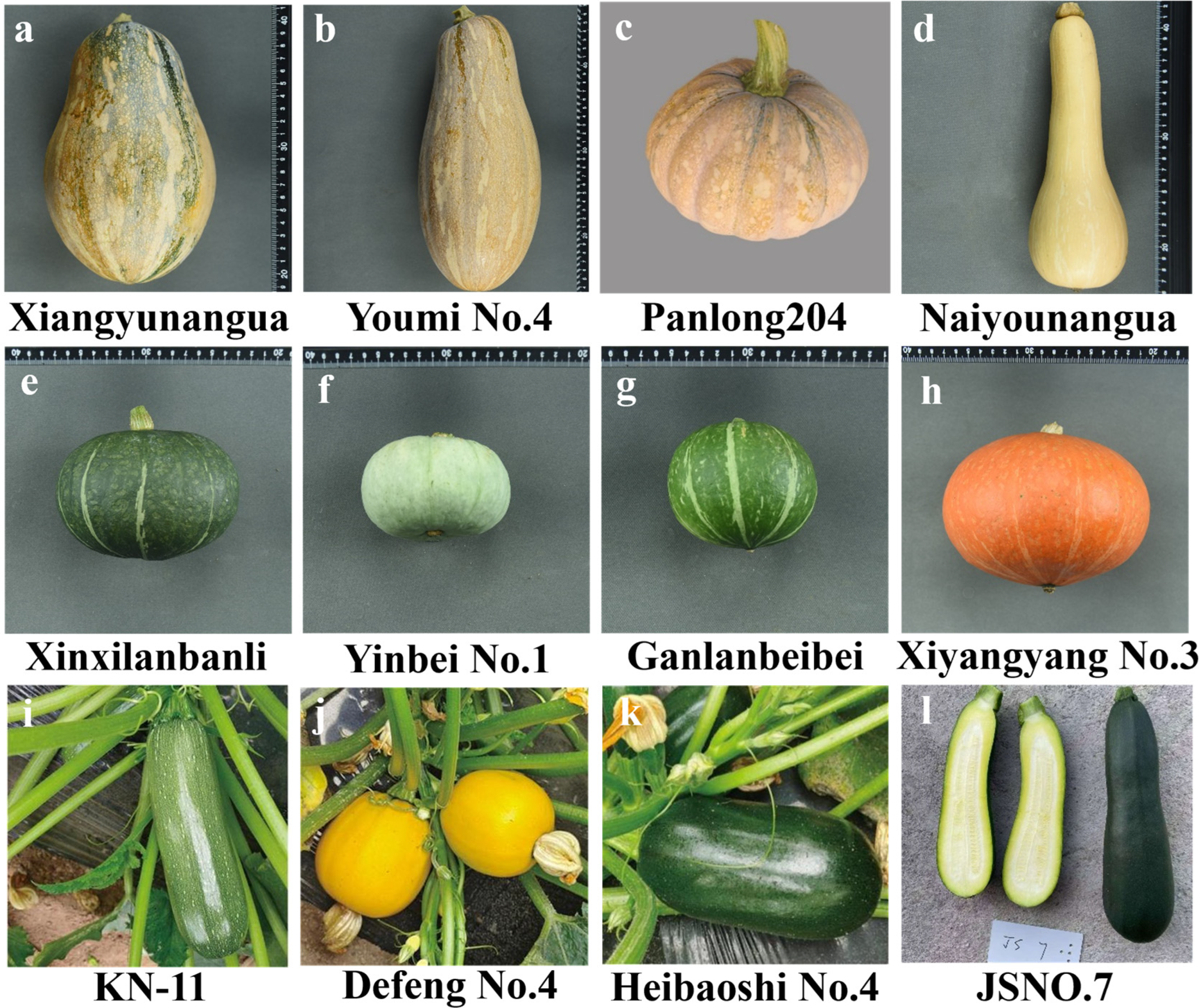
3.9. Phenotypic Identification of Molecular-Undifferentiated Varieties
4. Discussion
4.1. Evaluation of SSR Core Primer Polymorphism and Variety Identification Ability
4.2. Identification of Germplasm Resources and Varieties Combined with Molecular Markers and Phenotypes
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Appendix A
| NO. | Variety Name | Variety Type | Region | NO. | Variety Name | Variety Type | Region | NO. | Variety Name | Variety Type | Region |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Panlong204 A | C. moschata | Anhui | 103 | Xuelong A | C. maxima | Xinjiang | 205 | ZT17429 B | C. pepo | Gansu |
| 2 | Xinjiangmi2hao A | C. moschata | Anhui | 104 | Meiyaqiantu A | C. maxima | Anhui | 206 | ZT5816 B | C. pepo | Gansu |
| 3 | Jiangyimopan16005 A | C. moschata | Anhui | 105 | RCH2521 B | C. moschata | Shandong | 207 | ZT5818 B | C. pepo | Gansu |
| 4 | RWP93102 B | C. moschata | Shandong | 106 | RTCU9267 B | C. moschata | Shandong | 208 | ZT5886 B | C. pepo | Gansu |
| 5 | Jinyoumi1hao A | C. moschata | Hunan | 107 | Weiermixiaobei A | C. maxima | Shandong | 209 | Dingyan2202 A | C. pepo | Henan |
| 6 | Heliqihao A | C. moschata | Shandong | 108 | RCA2203 B | C. moschata | Shandong | 210 | Xinlü4306 A | C. pepo | Shandong |
| 7 | Youmi1hao A | C. moschata | Guangdong | 109 | Weiermihuangfei A | C. moschata | Shandong | 211 | Zhongtai16 A | C. pepo | Gansu |
| 8 | Tianmi A | C. moschata | Guangdong | 110 | Weiermijinling A | C. maxima | Shandong | 212 | Xiuyu916 A | C. pepo | Shandong |
| 9 | C128 B | C. moschata | Guangdong | 111 | Weiermiyihao A | C. maxima | Shandong | 213 | TBB9083 B | C. pepo | Neimenggu |
| 10 | Wawamiyihao A | C. moschata | Hunan | 112 | Jinpingguo33 A | C. maxima | Gansu | 214 | Dingyan2201 A | C. pepo | Henan |
| 11 | Xiaohei A | C. moschata | Beijing | 113 | Jinpingguo77 A | C. pepo | Gansu | 215 | Yuhu816 A | C. pepo | Hainan |
| 12 | Xingshudaguomiben A | C. moschata | Hunan | 114 | RCP21423 B | C. moschata | Shandong | 216 | Luhu3hao A | C. pepo | Shandong |
| 13 | Heliyihao A | C. moschata | Shandong | 115 | RCP21442 B | C. moschata | Shandong | 217 | Zifeng119 A | C. pepo | Neimenggu |
| 14 | Helisanhao A | C. moschata | Shandong | 116 | RCP1767 B | C. moschata | Shandong | 218 | ZT9092 B | C. pepo | Gansu |
| 15 | Tianli A | C. moschata | Sichuan | 117 | PK1582 B | C. maxima | Jiangsu | 219 | Zifeng49 A | C. pepo | Neimenggu |
| 16 | Jinbaoliyihao A | C. moschata | Shandong | 118 | Chunyu60 A | C. pepo | Shandong | 220 | Yuyan16 A | C. pepo | Gansu |
| 17 | Fulu333 A | C. moschata | Guangdong | 119 | Boshou410 A | C. pepo | Beijing | 221 | JH15835 B | C. pepo | Beijing |
| 18 | Xingshudaguo A | C. moschata | Hunan | 120 | Boshou607 A | C. pepo | Beijing | 222 | JH865 B | C. pepo | Beijing |
| 19 | Yunnan2hao-1-1 A | C. moschata | yunnan | 121 | Weikeduo A | C. pepo | Beijing | 223 | Bofeng A | C. pepo | Xinjiang |
| 20 | Panlong203 A | C. moschata | Guangdong | 122 | Jinghu36 A | C. pepo | Beijing | 224 | Baijiale A | C. pepo | Xinjiang |
| 21 | Youmi1hao A | C. moschata | Guangdong | 123 | Jinlierhao | C. pepo | Shanxi | 225 | Haojie9hao A | C. pepo | Xinjiang |
| 22 | RCP1730 B | C. moschata | Shandong | 124 | Kuaihulu A | C. pepo | Beijing | 226 | Suchengyihao A | C. pepo | Shandong |
| 23 | Guangzhoujinling A | C. moschata | Guangdong | 125 | 9794Xihulu A | C. pepo | Beijing | 227 | HS513 B | C. pepo | Xinjiang |
| 24 | Mitianxing A | C. moschata | Guangdong | 126 | Jinghu36 A | C. pepo | Beijing | 228 | Kairuite A | C. pepo | Xinjiang |
| 25 | Jinchuanmixiang A | C. moschata | Guangdong | 127 | Jiuyuanjinfeng68 A | C. pepo | Neimenggu | 229 | HS524 B | C. pepo | Xinjiang |
| 26 | Jinchuangaoming A | C. moschata | Guangdong | 128 | Jinfengerhao A | C. pepo | Neimenggu | 230 | HS512 B | C. pepo | Xinjiang |
| 27 | Jinchuanjinxiang A | C. moschata | Guangdong | 129 | Zhongzhongrekang5hao A | C. pepo | Beijing | 231 | Yuyan17 A | C. pepo | Gansu |
| 28 | Panlong211 A | C. moschata | Shandong | 130 | Zhongzhongrekang1hao A | C. pepo | Beijing | 232 | Yuyan112 A | C. pepo | Gansu |
| 29 | Nanmiyihao A | C. moschata | Tianjin | 131 | Zhongzhongzi2hao A | C. pepo | Beijing | 233 | HCA2 B | C. pepo | Neimenggu |
| 30 | Jibei A | C. moschata | Tianjin | 132 | Zhongzhongzi1hao A | C. pepo | Beijing | 234 | Farui A | C. pepo | Xinjiang |
| 31 | Runbei A | C. moschata | Tianjin | 133 | Cuiying108 A | C. pepo | Beijing | 235 | JP2102 B | C. pepo | Gansu |
| 32 | Fubei A | C. moschata | Tianjin | 134 | Zhongzhongz10 A | C. pepo | Beijing | 236 | JP2203 B | C. pepo | Gansu |
| 33 | Yinjueerhao A | C. moschata | Tianjin | 135 | P7 A | C. pepo | Beijing | 237 | TEQ3 B | C. pepo | Gansu |
| 34 | Yinzhu A | C. moschata | Tianjin | 136 | Lüfu95 A | C. pepo | Jiangsu | 238 | X066 B | C. pepo | Gansu |
| 35 | Guifeimi A | C. moschata | Hunan | 137 | Jinhui5hao A | C. pepo | Heilongjiang | 239 | Zhangfengerhao A | C. pepo | Gansu |
| 36 | Jinyuan A | C. moschata | Hunan | 138 | Younite928 A | C. pepo | Henan | 240 | HC0024 B | C. pepo | Neimenggu |
| 37 | Jiuyuanjinfeng108 A | C. moschata | Neimenggu | 139 | 8M03 B | C. pepo | Gansu | 241 | HCE674 B | C. pepo | Neimenggu |
| 38 | Panlong212 A | C. moschata | Anhui | 140 | Yuncui A | C. pepo | Shandong | 242 | Nongren22 | C. pepo | Shandong |
| 39 | PJB0012 B | C. moschata | Fujian | 141 | S736 B | C. pepo | Henan | 243 | Yuyan109 A | C. pepo | Gansu |
| 40 | Sizhuang17 A | C. moschata | Zhejiang | 142 | Shengrun817 A | C. pepo | Henan | 244 | HCA5 B | C. pepo | Neimenggu |
| 41 | Beiluli2hao A | C. moschata | Shandong | 143 | HC208 B | C. pepo | Neimenggu | 245 | Hongchangzhangfeng A | C. pepo | Neimenggu |
| 42 | Beiluli1hao A | C. moschata | Shandong | 144 | Hongchang212 A | C. pepo | Neimenggu | 246 | HCE671 B | C. pepo | Neimenggu |
| 43 | Madun2hao A | C. moschata | Shandong | 145 | Chuanqihongchang211 A | C. pepo | Neimenggu | 247 | HCE682 B | C. pepo | Neimenggu |
| 44 | Xiaojinbei A | C. moschata | Xinjiang | 146 | HC203 B | C. pepo | Neimenggu | 248 | Hongchangyihao A | C. pepo | Neimenggu |
| 45 | Zhengyuan3hao A | C. moschata | Guangdong | 147 | Zhongtai268 A | C. pepo | Gansu | 249 | Nongren91 B | C. pepo | Shandong |
| 46 | Sizhuang21 A | C. moschata | Zhejiang | 148 | Nongren518 A | C. pepo | Hebei | 250 | Nongren2230 B | C. pepo | Shandong |
| 47 | Zhengyuan1hao A | C. moschata | Guangdong | 149 | H020 B | C. pepo | Gansu | 251 | Donghu19hao A | C. pepo | Shanxi |
| 48 | PJB0092 B | C. moschata | Fujian | 150 | Donghu32hao A | C. pepo | Shanxi | 252 | Dingyan2205 A | C. pepo | Henan |
| 49 | Zhengyuan2hao A | C. moschata | Guangdong | 151 | Donghu No.16 A | C. pepo | Shanxi | 253 | Linong1haoxiangyunangua A | C. moschata | Guangdong |
| 50 | Sizhuang19 A | C. moschata | Zhejiang | 152 | PK No.2 B | C. pepo | Gansu | 254 | Zhengyuan1haomibennangua A | C. moschata | Guangdong |
| 51 | Deli327 A | C. moschata | Shandong | 153 | Jiarui600 A | C. pepo | Gansu | 255 | Chengxingmibennangua A | C. moschata | Guangdong |
| 52 | Dinglibahao A | C. moschata | Shandong | 154 | Jiarui570 A | C. pepo | Gansu | 256 | Shanhaimibennangua A | C. moschata | Guangdong |
| 53 | Deli902 A | C. moschata | Shandong | 155 | Huangbeibei A | C. pepo | Shanghai | 257 | Xiangxiangnangua A | C. moschata | Guangdong |
| 54 | Lüxiuer A | C. moschata | Xinjiang | 156 | Baibeibei A | C. pepo | Shanghai | 258 | Linonghuashengnangua A | C. moschata | Guangdong |
| 55 | Xinhuali A | C. moschata | Zhejiang | 157 | Nongren968 A | C. pepo | Shandong | 259 | Zajiaofaxinangua A | C. moschata | Guangdong |
| 56 | Fangfang A | C. moschata | Guangdong | 158 | Meihuicheng216 A | C. pepo | Gansu | 260 | V201 B | C. moschata | Guangdong |
| 57 | Meimei A | C. moschata | Guangdong | 159 | Bulanding A | C. pepo | Shandong | 261 | Linongmopannangua A | C. moschata | Guangdong |
| 58 | Shilengmi1hao A | C. moschata | Anhui | 160 | Meixiu A | C. pepo | Xinjiang | 262 | Xinxilanbanli A | C. maxima | Guangdong |
| 59 | Jinpingguo203 A | C. maxima | Gansu | 161 | Yuanbaosanhao A | C. pepo | Xinjiang | 263 | Linongnennangua A | C. moschata | Guangdong |
| 60 | Shengshengmi2hao A | C. moschata | Anhui | 162 | Qiusheng A | C. pepo | Xinjiang | 264 | Linonglüguizunangua A | C. maxima | Guangdong |
| 61 | Jifeng77 A | C. moschata | Guangxi | 163 | Jialing A | C. pepo | Xinjiang | 265 | Ganlanguizunangua A | C. maxima | Guangdong |
| 62 | Hongjianguizu A | C. maxima | Guangdong | 164 | Badao A | C. pepo | Xinjiang | 266 | Linonghuipiduancuguizu2haonangua A | C. maxima | Guangdong |
| 63 | Lüyuan28hao A | C. maxima | Zhejiang | 165 | JR003CM B | C. pepo | Gansu | 267 | Linong3haobanlinangua A | C. maxima | Guangdong |
| 64 | Chengzhang42hao A | C. maxima | Zhejiang | 166 | JR19D B | C. pepo | Gansu | 268 | Linong9haobanlinangua A | C. maxima | Guangdong |
| 65 | Jifei A | C. moschata | Guangdong | 167 | ZTPK04 B | C. pepo | Gansu | 269 | Aibisibanlinangua A | C. maxima | Guangdong |
| 66 | Jixianfeng A | C. moschata | Hebei | 168 | ZT2H B | C. pepo | Gansu | 270 | Yinlinangua A | C. maxima | Guangdong |
| 67 | Fuwaxianfeng A | C. pepo | Gansu | 169 | Xiuyu160 A | C. pepo | Shandong | 271 | Beilishihaonangua A | C. maxima | Guangdong |
| 68 | Hongfenjiaren A | C. maxima | Guangdong | 170 | Jiuyuanjinfeng118 A | C. pepo | Neimenggu | 272 | Dabeinangua A | C. maxima | Guangdong |
| 69 | RCP1257 B | C. moschata | Shandong | 171 | Zhenhu9hao A | C. pepo | Shandong | 273 | Huiyou1haobeibeinangua A | C. maxima | Guangdong |
| 70 | RWP9402 B | C. moschata | Shandong | 172 | Shenghu11 A | C. pepo | Shandong | 274 | Huiyou2haobeibeinangua A | C. maxima | Guangdong |
| 71 | RCP6321 B | C. moschata | Shandong | 173 | Luhu2hao A | C. pepo | Shandong | 275 | Huiyou3haobeibeinangua A | C. maxima | Guangdong |
| 72 | RWP281 B | C. moschata | Shandong | 174 | Gangqinuo A | C. pepo | Shandong | 276 | Linong3haobeibeinangua A | C. maxima | Guangdong |
| 73 | RWP297 B | C. moschata | Shandong | 175 | Dongdiou A | C. pepo | Shandong | 277 | Linong23haobeibeinangua A | C. maxima | Guangdong |
| 74 | RCB2263 B | C. moschata | Shandong | 176 | Donghu688 A | C. pepo | Henan | 278 | G163 B | C. maxima | Guangdong |
| 75 | RCB8N25 B | C. moschata | Shandong | 177 | Xiaboke A | C. pepo | Shandong | 279 | Q1174 B | C. moschata | Guangdong |
| 76 | RWP286 B | C. moschata | Shandong | 178 | Maodun A | C. pepo | Shandong | 280 | Q1177 B | C. moschata | Guangdong |
| 77 | Sizhuang11 A | C. moschata | Zhejiang | 179 | Jinyu1hao A | C. pepo | Gansu | 281 | V222 B | C. moschata | Guangdong |
| 78 | Weiermiheizhenzhu A | C. maxima | Shandong | 180 | 9M13 B | C. pepo | Gansu | 282 | V224 B | C. moschata | Guangdong |
| 79 | Linongjinxiang A | C. moschata | Guangdong | 181 | Zhangfengyihao A | C. pepo | Gansu | 283 | V264 B | C. maxima | Guangdong |
| 80 | RTSY671 B | C. moschata | Shandong | 182 | Ziguan12hao A | C. pepo | Shanxi | 284 | V379 B | C. maxima | Guangdong |
| 81 | Beili4Hao A | C. maxima | Anhui | 183 | Ziguan No.8 A | C. pepo | Shanxi | 285 | Xiangyunangua A | C. moschata | Anhui |
| 82 | Fuwa808 A | C. maxima | Gansu | 184 | Sanruishuoguo A | C. pepo | Xinjiang | 286 | Youmi No.4 A | C. moschata | Anhui |
| 83 | RCP1734 B | C. moschata | Shandong | 185 | Haojie5hao A | C. pepo | Xinjiang | 287 | Naiyounangua A | C. moschata | Anhui |
| 84 | RTCU2534 B | C. moschata | Shandong | 186 | Sanruiyuguo A | C. pepo | Xinjiang | 288 | Shengsheng1hao A | C. moschata | Anhui |
| 85 | RTCU2530 B | C. moschata | Shandong | 187 | Gongxi518 A | C. pepo | Shandong | 289 | Shengsheng4hao A | C. moschata | Anhui |
| 86 | RTDL6802 B | C. moschata | Shandong | 188 | Gongxi568 A | C. pepo | Shandong | 290 | Shilengmi3hao A | C. moschata | Anhui |
| 87 | PK201 B | C. maxima | Neimenggu | 189 | SF2080D2 B | C. pepo | Xinjiang | 291 | Youmi18hao A | C. moschata | Anhui |
| 88 | PK529 B | C. moschata | Neimenggu | 190 | Haojie4hao A | C. pepo | Xinjiang | 292 | Yinbei No.1 A | C. maxima | Anhui |
| 89 | Sizhuang33 A | C. maxima | Zhejiang | 191 | Haojie1hao A | C. pepo | Xinjiang | 293 | Ganlanbeibei A | C. maxima | Anhui |
| 90 | Sizhuang31 A | C. moschata | Zhejiang | 192 | SF2021XT B | C. pepo | Xinjiang | 294 | Xiyangyang No.3 A | C. maxima | Anhui |
| 91 | Tianmi1hao A | C. moschata | Beijing | 193 | SF2025dapian B | C. pepo | Xinjiang | 295 | Beili4hao A | C. maxima | Anhui |
| 92 | Jinpinyinbei2hao A | C. maxima | Fujian | 194 | 9X218 B | C. pepo | Gansu | 296 | Jiabei6hao A | C. maxima | Anhui |
| 93 | RTCU8401 B | C. moschata | Shandong | 195 | 9M34 B | C. pepo | Gansu | 297 | Jinbei No.5 A | C. maxima | Anhui |
| 94 | Z10 B | C. moschata | Beijing | 196 | ZT5133 B | C. pepo | Gansu | 298 | Yinbei2hao A | C. maxima | Anhui |
| 95 | RTCU4767 B | C. moschata | Shandong | 197 | Jindianjiuhao A | C. pepo | Tianjin | 299 | Jiangli5hao A | C. maxima | Anhui |
| 96 | RTCU6730 B | C. moschata | Shandong | 198 | Zhongtai266 A | C. pepo | Gansu | 300 | Yinli6hao A | C. maxima | Anhui |
| 97 | RTCU6457 B | C. moschata | Shandong | 199 | Zhongtai13 A | C. pepo | Gansu | 301 | JSNO.7 A | C. pepo | Anhui |
| 98 | RWP9604 B | C. moschata | Shandong | 200 | ZT3H B | C. pepo | Gansu | 302 | Mingzhu1hao A | C. pepo | Anhui |
| 99 | RCH3063 B | C. moschata | Shandong | 201 | Nongren818 A | C. pepo | Shandong | 303 | Mingzhu6hao A | C. pepo | Anhui |
| 100 | Meiyasihao A | C. maxima | Anhui | 202 | Nongren778 A | C. pepo | Shandong | 304 | KN-11 A | C. pepo | Beijing |
| 101 | Jindian A | C. moschata | Xinjiang | 203 | Youhulü058 A | C. pepo | Shandong | 305 | Defeng No.4 A | C. pepo | Beijing |
| 102 | Lüfeicui A | C. moschata | Xinjiang | 204 | ZT17428 B | C. pepo | Gansu | 306 | Heibaoshi No.4 A | C. pepo | Beijing |
| Primer No. | Forward Primer Sequence (5′→3′) | Reverse Primer Sequence (5′→3′) | Primer Source |
|---|---|---|---|
| NG1 | CACCAAAATGGCCGCTCAAA | AGCTGCGACGAATGTGAAGA | Independent development |
| NG2 | CAGCTTCTTCAATCTCGCGC | CCTCCACAACAACAAGCAGC | Independent development |
| NG3 | GTGACCCAACTGACAGAGGG | TCGACTTCGAACGCAACAGA | Independent development |
| NG4 | GTGCCTTGGTTCTGTCGGTA | GCGCGCGTAAATATGTGTGT | Independent development |
| NG5 | CTCGTCGGGTCTTCGATCAG | AACAGTTCGCGTGCAACTTC | Independent development |
| NG6 | TGTGAAACGCCTCCAGTACC | GAGATCCGGTAAGCCACGAG | Independent development |
| NG7 | ACCTACCCCAGCCATACCAT | GATCGGCTGACGTCAATTGC | Independent development |
| NG8 | CGCGGTTGATCATTGTCGTC | CCATGCTGTGTGTGTGTGTG | Independent development |
| NG9 | GAATGAGCTTCGTCGAACGC | GCTCTGATCGGGGCTAGTTC | Independent development |
| NG10 | TGCATAGGTATCGGCAGCTG | AGTGGGTTCCAAGTGCAACA | Independent development |
| NG11 | GGCTGAGCCCCTCTTCAATT | CATCGTCGCCATTGCTTCTG | Independent development |
| NG12 | CAGAGGCTTGTTGTGTTGGC | CCACACTCGGAATTCCACGA | Independent development |
| NG13 | CCCTGGTTTTGGCTCCCTAG | CATGATGCGCACGAACTGAG | Independent development |
| NG14 | GATCAACGCAATACACCGGC | CTAGGTCGTCTTCGTCGTCG | Independent development |
| NG15 | CGGAATCGCCTGCAATTCAG | GGGAATTGGCGCGAAGAATC | Independent development |
| NG16 | TCATCCCAGGCATTTGGACC | GAATTTGACTCCGCCGCATC | Independent development |
| NG17 | GAGGCTAACGACGATGCGTA | CGTTACGTATCGCCGTCAGA | Independent development |
| NG18 | CGTTTATCTGTTGCTCGGCG | GAGGGATTCCAGGAGAGGGT | Independent development |
| NG19 | AGACCAAGCTCTATCCGGGT | GTTCGACGCTGCATCCATTC | Independent development |
| NG20 | GCCGACGGATCTTTTTACGC | CTGCAAACACGTCGTTTCGT | Independent development |
| NG21 | CGCCGAGTTTGAGTAATGCG | CCAAACTCCTCGACAACGGA | Independent development |
| NG22 | GAAGACGCCGATTTCGTTCG | CCATCATACGAGTGAGCGCT | Independent development |
| NG23 | CGTAACGTGCGAATCGTTCC | GACCGTCCAACCACCATGAT | Independent development |
| NG24 | GCCATAACTTGCTCTTCGCG | TGTTCCATAGCCGCCATTGT | Independent development |
| NG25 | CGGACTCTAGCGTCGAGTTC | CAACTTCCGAGAACGTGGGA | Independent development |
| NG26 | CGGCGTATGGCTCAGTACAT | ACCACAGCGTTGAAATTGGC | Independent development |
| NG27 | GATTTGCATCGGAACTCGGC | TCTGTTTCTCCAGCGGACAC | Independent development |
| NG28 | CATAGTCGCCTGATTCCGCT | CTTCGATTTCGCAGTCGCAG | Independent development |
| NG29 | GTTGTGCCATGACGGTCCTA | GAAAGCGGCGTTGATAGCAG | Independent development |
| NG30 | GTGGCCTCTGTTTGCACTTG | GACGTGTGATGAACGATGCG | Independent development |
| NG31 | GGAGGAAGAGAAGATCGCCG | GAGGCGAAAATGGCGGATTC | Independent development |
| NG32 | CTAAGCAGTAGTCGGCCTCG | CGTGAACTTTTGCAGCGTCA | Independent development |
| NG33 | GCCGTGTGAAAGGTTGTGAC | CCGCGTTTCGTTTGACTTGT | Independent development |
| NG34 | TTCGCGACTCCTAATCGACG | CGCACTTGTCGCTAATCGTG | Independent development |
| NG35 | GCCGCAGAAAAAGCACTTCA | CCCTAGCCCCTCTTCTTCCT | Independent development |
| NG36 | CCAATGGCGTTGCTAATCCG | CGTTAATCGGTTCGACGCAC | Independent development |
| NG37 | TCGCTGTTGGAGCTGTGAAT | GGCAACGTGTCGATCGATTG | Independent development |
| NG38 | ATGCCGCGTCATTCAAATCG | CTCATTGCTCGATGGCGTTG | Independent development |
| NG39 | CGCTTTCTCGTGCTCAATCG | GATTTGAACCGTATGCGCCC | Independent development |
| NG40 | GAAGCGACGGATTCAATGGC | TCACGTCGTGTTTTCAACGC | Independent development |
| NG41 | CCATCAGCAATGTCGAAGCG | GTGTTGTCGGCACAGTTGAC | Independent development |
| NG42 | TGGGATAAGGGCAGCCAAAG | GCGACTAGCGTTCGGGAATA | Independent development |
| NG43 | ACGTCGAACAAGCAAACGTG | CAATTGCCGATCAAGCCTCG | Independent development |
| NG44 | CCTCTTGATTTGCATCGCCG | CTACCAAGAAAATCGCCGCG | Independent development |
| NG45 | CACAGCCCAACTCAACAACG | TTCATGTGTTTGCTGCAGGC | Independent development |
| NG46 | GCCGCGTTTTGATGAGATCC | GCTCCATATGCCAGCAAACG | Independent development |
| NG47 | CACCAAAATGGCCGCTCAAA | CCTCTGCAACTTCGTCTCGT | Independent development |
| NG48 | GCAGCTTCTTCAATCTCGCG | CCTCCACAACAACAAGCAGC | Independent development |
| NG49 | GGTGACCCAACTGACAGAGG | TCGACTTCGAACGCAACAGA | Independent development |
| NG50 | ACCAACTGGAGCTCGAAAGG | GCTCTCGCAAATACCGCTTG | Independent development |
| NW1 | GCTTGAACAGAGATGGAGGG | AAAGTCGCTGAGAGCTGGAG | [29] |
| NW2 | GGTGCATTGTCCAAACACAA | CCGCATCCATGAAAGAAAGT | [29] |
| NW3 | ATTTGCTTACCAAACAGCCG | GTTCAGAGGAGCTGGGTACG | [29] |
| NW4 | GGACTTGAGATGGAGGTGGA | TTTGTACGTTGTTCGTTGCC | [29] |
| NW5 | TCGCTTCACCGGTAATTTTC | GCGCTGAAGAATCCATGTTT | [29] |
| NW6 | ACAACGAAGCCTCAAAGGAA | GATGCAAAGGATGGAAAGGA | [29] |
| NW7 | CTCAGTGGAGGGACAAGCTC | CCGACTCCACCATGTCCTAT | [29] |
| NW8 | ACCTCTGCATTTCAACCCAC | ATACCCACCAAGCCCTTTCT | [29] |
| NW9 | CTGTTGCTGTTGTTGCTGGT | GCGCTTCTCTCAATGCTTCT | [29] |
| NW10 | AGCTACGCATGCCTGAATCT | TGCACCTGCTGTCATAGCTC | [29] |
| NW11 | CATACCCACCGTCGACTCCT | GGGCGAAGTGGAGGTTATGA | [30] |
| NW12 | GTTGCTCCAACTCGATCTTCA | TTTCAAACGAGCACAAGCAC | [30] |
| NW13 | TAGTCGAGAAGGCCGAGAAG | AAATTCGACGACCGCTTG | [30] |
| NW14 | AAATTCGACGACCGCTTG | TTTTTAAAGGGCTGAAAATAATTG | [30] |
| NW15 | AAAATTGCTAGGCTGTAGTGGTG | AAAATTGCTAGGCTGTAGTGGTG | [30] |
| NW16 | TGTCAGCTTCCTCAGTAGGG | TGAACTGGGAGAGAGGTTTG | [30] |
| NW17 | TCCCATCCTCTACTGTTGCAC | TAAGTTGTGGGTGGGGAAAC | [30] |
| NW18 | ACCCCACCAAATTAATGCAG | AGAGCCCACTGTGATGACCT | [30] |
| NW19 | TGATTTGCGCACAAACAAAC | GCCAAAGGTTCCAAATGACA | [30] |
| NW20 | GCCAAAGGTTCCAAATGACA | TGATCGAATTGTGGCTGGT | [30] |
| NW21 | TGATCGAATTGTGGCTGGT | GTGGCCGTAGGTTTGTCAGT | [30] |
| NW22 | CACCTGGCTGTTTTGTCTGA | ACATGGGCATACCTCGAATC | [30] |
| NW23 | TGAAATGAACGCAGAATTGC | ACTTGCGGACTTTCACACCT | [30] |
| NW24 | TTTTTGAAATTCTTTGCATCACT | AAGCCAAAGCCCCTTATCT | [30] |
| NW25 | GCCAAAGGTTCCAAATGAC | GCAACAAATTGTAGTTGCAAAG | [30] |
| NW26 | CACGAAGATTTGATGGCCTTA | GGATTGGGATGGTGAAGATG | [31] |
| NW27 | GCAGAGGAGAAGTGGGTTTG | CTTTATCCGACCAAGCGTTC | [31] |
| NW28 | AGCCTTTCAGAAGAACCAAG | GGCTTCAAACAAATACTAACCA | [31] |
| NW29 | ATTAAATGCTCCTCCCCACC | GGAGAGAGAGGGAAAAACGG | [32] |
| NW30 | AGTCCGACGAAGCTCAGGTA | TACATGTCTCTGCGAGCGTC | [32] |
| NW31 | TCAATGGATCTGCCTTTTCC | AGGGAAGGATGCTAAGGAGC | [32] |
| NW32 | GGTGGGTATGGAGGAGGTG | ACCGCCACGTGGATAACTAA | [32] |
| NW33 | CCTTTCAAAATGGCTTCCAA | TCTTCTTCCCAAGCTGCCTA | [32] |
| NW34 | CAGACGGCTTTTGAAGGAAG | TCGAAGAGCTCTGTTGGTGA | [33] |
| NW35 | TTCTCAGGTGCTGTTGATGC | TCCTTTCCTTCGCTTCCTCT | [33] |
| NW36 | GCTTTTGAAGATGAGGCGAG | CACTCAAGCAGATTGCCAAA | [33] |
| NW37 | CGGTCGTGAATACATCATGG | GCTCCACCAATGGGAAACTA | [33] |
| NW38 | CGATATGATGGAGCTGCTGA | CCAGCTCCCGAGCTTCTAAT | [33] |
| NW39 | GAAGAGGAAGAAGCAGCACG | TGTCCACGATCTCTGCTTTG | [33] |
| NW40 | AGAGACGAGAATGGGGGAGT | GCGAAATCGGTGCAATAAAT | [33] |
| NW41 | TTTCTTTTCTCCTCTGCCCA | AATCACACCTTGGGCACTTC | [33] |
| NW42 | ATCATAGTCGTCGTCGGGTC | GCCGATTCTTGAGGAACAGA | [33] |
| NW43 | TTCAAAGCTTCTCTGGTAAGGC | ACATCGCCCAAGAGAAGTTG | [33] |
| NW44 | GAAGGCGAGGTTTTGAGTTG | GGCGGAACCCTAAGAAATGT | [34] |
| NW45 | TCGGACCAAAGTACCCTCCA | TCATCGCCGGTTGTGATCT | [34] |
| NW46 | GCACTTGAATCTTCGTCAAC | CGAGAAAGAATTAACGAGCA | [34] |
| NW47 | ATGGCTTCCAAGCTCCTCTT | GTCGGCCATGAGCTTGAG | [34] |
| NW48 | GAAGGACCGTGAGTGAAAGG | ATCTTGTGCCAAAGCTCCAT | [34] |
| NW49 | CAGGCTATTCGCACCCTCTA | CCTCATGCATTTTGCGTGTA | [34] |
| NW50 | CGTGAACATTCGTTTGTTGG | TCATCCGTTTCCTTTTCAGC | [34] |
| NW51 | TCACTTTACAACCAGAAGCTGA | CACTTTGCTGCTCATCCAC | [34] |
| NW52 | GGCTGGCTCATAAAGAAAGC | GGGGTTTTTGAAGATGCTTG | [34] |
| NW53 | ATGATGGAGTCCCAGTCGAG | ACCCACACACCCTCTCCTC | [34] |
| NW54 | ATCCACAAACAAGGCACCTC | GTAGTGGAGGCTCGGGTGTA | [34] |
| NW55 | CACAAGCCATCACACAAACC | AGGTGGAGCTGACCGTAGTG | [34] |
| NW56 | TCTCACTCATCCACCACACC | CGTTTCGAGTCATTTGTTCG | [34] |
| NW57 | CCTCTCATTCTTCCCCATCTC | TGATCCGGTAGGGGTCTACTC | [34] |
| NW58 | GCCCAGAAGACAAAAGTTCG | TTTTTGTGTGCGTGTGTGG | [34] |
| NW59 | ATTGGTGCCGAAGCTATCAC | CCCACGTTATGGAGCAGAAT | [34] |
| NW60 | ATGCTCAGACATCCATGCAC | GCGAAAGATTACCGATGCTC | [34] |
| NW61 | AACACTCGGCCACAACATC | CTCCTTGTAAAACGGGTTGC | [34] |
| NW62 | CTCCATTCCCATGGCTTC | CCATGAGCTTGAGAGAGGTG | [34] |
| NW63 | CAAATTCAGACGCTTCTTTTGG | AGAATTGAGCAAAAAGGAGATGG | [34] |
| NW64 | TTAAGATAGTTTCAGGATTCCATGT | AGGAGTTTGAAACAAATGAAGG | [34] |
| NW65 | GAGTGATGTTTTGAGTAAACAG | CTTGTTCATCATCATCTGTG | [34] |
| NW66 | AGGTGGCATCTGTACACTGAG | TGAACAAACTCCACACCAATAG | [34] |
| NW67 | GACAGGACAGGTCAACACCTC | AACCCAATTGCACAGCTTCT | [34] |
| NW68 | TGTTCAGTAGCCATTGATCTATCC | TGGATGACTTCTGGGTTGGT | [34] |
| NW69 | TTATAAGAATGATGTTACTCGAT | CATCATCATACATCTTACATTG | [34] |
| NW70 | GAACTCTAATCCAGCCGTTG | TTAAAATAAATGGAGCAAATAATGAG | [34] |
| NW71 | GGTCAAATTCAAGGGCTTACC | AGGAGCATCCTTGTCGTTGT | [34] |
| NW72 | CTGGTTTTTCCACACAGAATCA | CCCAGAAGGGAATTGAAACA | [34] |
| NW73 | CCGCCACCACTGAACAACT | GGCTGTGGCCCTCCATAGT | [34] |
| NW74 | TCACAAAGTCAAAACAGACAAACA | TGTTTCAGGGAAATGAAGAGG | [34] |
| NW75 | GGCGAAAAGGAAGAACGAAT | TTTTTCTCCCCCTTCCACAT | [34] |
| NW76 | ACCTACCGTCACACCCACAT | CCACCTGAAAACAGGGCTAA | [34] |
| NW77 | GAACTTCGTGTGTGCGTGTC | TTGCTGGAACTTCCTCTCGT | [34] |
| NW78 | CTCTCCCCCTCCTCCACTC | GCGTTCTGCATTGTGGAAGT | [34] |
| NW79 | CAAATTTTCTTCTACAATTTGGT | GTGAATGAACATGCGTCTC | [34] |
| NW80 | ATGGAGACGCGCAAGTAGAT | ACCTCGAGGAAGCAAAAATG | [34] |
| NW81 | TATGTAGCGCTCTGCACAAT | CCTCAATGTAATGTTTTGAATCC | [34] |
| NW82 | TGAAACTACACTACATGACCTTGG | TGGGTTGGTAGACTTGTAGTTGA | [34] |
| NW83 | CATCAGGTTATTGAGTTTTACTCAGAC | TCGTCTGCCCCAATAATTC | [34] |
| NW84 | CGTGTTTGTGTTGGAAAAGC | AAGCAAACCCACAGAAGGAG | [34] |
| NW85 | ACGTTCTTTCTGCCTCCAT | ATGCCCATTATGCAATTCTC | [34] |
| NW86 | ACAATGGTTCTTTTGATCCTTGA | TGCAAACAATGTGTGTGTGTG | [34] |
| NW87 | TGTGGGGTTTTGCTTTTAGG | ATCCAAAATGGTGGTGCATT | [35] |
| NW88 | GCAAGCCCTAGCTGATTTTT | GGGCGAAAACAGAGTGAGAG | [35] |
| NW89 | AACAGAGAATCTGGTGCTGGA | GCCAATTCCTCCTTTTCTCC | [35] |
| NW90 | GGATTGCCTTGCTGGAGAG | CAAATCCAGGTGGAAGGCTA | [35] |
| NW91 | TCACCAACTTGCCATAGACG | CGCGCAACTGATAAGGATTT | [35] |
| NW92 | TGATCTGACAGCAACCGAAG | CCATTCCCTTAGTTTCTAAACCACT | [35] |
| NW93 | TGATGACAAAGAAGCCATCG | ATCTTATGCCGGAGCAGATG | [35] |
| NW94 | TTTCGTTGCAGAGAATGGTG | CCCATTTCTTTCTCGCTCAA | [35] |
| NW95 | AATTCTAACCGTTGGGGGTT | CCCAGAAACGAAAGAAGCAG | [35] |
| NW96 | ATGGCATAATCAGCCCTCAC | TTTGAAGAAGGGAAGAGGGG | [35] |
| NW97 | TCAAACTCTCAACTGGGTCG | TCCGATTGAGAGCTGGAGTT | [35] |
| NW98 | TCCTCGTCGGAACAGAACTT | TCTAACTTGGAGCCTGGAGC | [35] |
| NW99 | GAGCTTCTGGATGATAGCGG | GCTTCGAACTTTCGTTTGCT | [35] |
| NW100 | TGTAATTTGAAGATGAGATTATGAAGA | GGGTCTTGTTTCTCGAGCTG | [35] |
| NW101 | TTTCAAATTCGCTCGTTTCC | GAGTCGTCGATGTCGTCAAA | [35] |
| NW102 | TCTCGAATCCGAAGAAATCG | TGCGTTGCAGAATATCAAGC | [35] |
| NW103 | GGGTGAAGCTGTGGATTCAT | TCTCTAGTGCCACACTCAAGTACA | [35] |
| NW104 | GTGGGCTAAGTTCAAATCGTTC | GAACCCAATCTTCTCATTTCCA | [36] |
| NW105 | CAAGCCTTTACCAAAACCAAAC | ACGATCTTCCTCCTCCTCTTCT | [36] |
| NW106 | GAACCTCGTTGCTGGTTCTTCT | TCTAGCATCTTACGCACGCTTC | [36] |
| NW107 | TGTCAAATTCGCTTCCATCATC | ACGACTGTGAGAACGGTGAAAA | [36] |
| NW108 | TGGAATCCAGAGACTGATGAAG | CACGATTCCGATAACAACAAGA | [36] |
| NW109 | TGGAGGTATCCTTCCGTATGTT | CATACGGGAGTTTCGTTTTTCT | [36] |
| NW110 | ATCCCAGGTCCCAATTTTCTTC | CCATACCTGAGGGACCTGAAAC | [36] |
| NW111 | CGAATTTCTCCGAAAAGAAACC | CCTCGATCAATCCTCCTACACC | [36] |
| NW112 | CGACTACAGCTCAACAAAGACC | GAAGTCTATTACGCCCAATTCC | [36] |
| NW113 | CGAGTACGTTTACAACCATCCA | CAACTCTCAACAAGAACCCAAG | [36] |
| NW114 | TCTTCTCGGAGTTTGATGTCCA | TTTACCCTCATCGGAACCTCAT | [36] |
| NW115 | GACACTCACGAGCAAAATGACC | CGTGTGGAGCTTTCTCACAACT | [36] |
| NW116 | GCAGCACACTCGATTCCTCTTT | TAGGAAAAATGGCGGTGAAGAA | [36] |
| NW117 | AGACTTTGGGTTTGTAGGAAGG | GCTAAGCATTGTTAGGGCTTCT | [36] |
| NW118 | GACGAATACTTTCGCAATCAAG | CATGTGGAAGTTGTTGTTGTTG | [36] |
| NW119 | CTAATCAGGCTGGCCAAGAAGA | CGACGTTACCGACAAGATTTCC | [36] |
| NW120 | AGCACTGGGTTAACAGAGGAAA | ATGATACAGTAATCGGCGTCCT | [36] |
| NW121 | TGTGATTTGTTGGGCTCTCTGT | ACATGGAAATCCACCTCTTCGT | [36] |
| NW122 | TCCACACCAGTGGTTCTTTCGT | TTTCACCATCCGGACAATCATC | [36] |
| NW123 | GCCGGTGATTCTGGATGAAGTA | CAGGTAGAAGGTGGGGAGATGG | [36] |
| NW124 | TTGGACAATCTGAGGAAGTTGG | AAGGTTTGTCCTTCGACAAGGT | [36] |
| NW125 | CCCTCGGTAGCAATTTTGTAGT | TTTGGTGGGAGTGATACTGATG | [36] |
| No. | Trait Name | Observation Method | Trait Type |
|---|---|---|---|
| 1 | Cotyledon Shape | VG | PQ |
| 2 | Main Vine Length | MS | QN |
| 3 | Leaf Size | MS | QN |
| 4 | Degree of Leaf Margin Incision | VG | QN |
| 5 | Greenness of the Front Leaf Surface | VG | QN |
| 6 | Presence or Absence of White Spots on the Front Leaf Surface | VG | QL |
| 7 | Petiole Length | MS | QN |
| 8 | Petiole Thickness | MS | QN |
| 9 | Greenness of the Melon Rind | VG | QN |
| 10 | Fruit Longitudinal Diameter | MS/VG | QN |
| 11 | Fruit Transverse Diameter | MS/VG | QN |
| 12 | Ratio of Fruit Longitudinal Diameter to Transverse Diameter | MS/VG | QN |
| 13 | Position of the Maximum Transverse Diameter of the Fruit | VG | QN |
| 14 | Fruit Shape | VG | PQ |
| 15 | Prominence of the Melon Neck | VG | QN |
| 16 | Melon Neck Length | VG | QN |
| 17 | Degree of Fruit Curvature | VG | QN |
| 18 | Shape of the Fruit Stalk | VG | PQ |
| 19 | Shape of the Fruit Navel | VG | QN |
| 20 | Presence or Absence of Fruit Furrows | VG | QL |
| 21 | Spacing of Fruit Furrows | VG | QN |
| 22 | Depth of Fruit Furrows | VG | QN |
| 23 | Patterns on the Fruit Surface | VG | QN |
| 24 | Maturity Period | VG | QN |
| 25 | Main Color of the Fruit Peel | VG | PQ |
| 26 | Depth of the Main Color of the Fruit Peel | VG | QN |
| 27 | Presence or Absence of Wax Powder on the Fruit Surface | VG | QL |
| 28 | Presence or Absence of Fruit Nodules | VG | QL |
| 29 | Main Color of the Fruit Pulp | VG | PQ |
| 30 | Thickness of the Fruit Pulp | VG | QN |
| 31 | Diameter of the Fruit Navel | VG | QN |
| 32 | Seed Length | VG | QN |
| 33 | Ratio of Seed Width to Length | VG | QN |
| 34 | Color of the Outer Seed Coat | VG | PQ |
| 35 | Presence or Absence of the Outer Seed Coat | VG | QL |
| No. | Trait Name | Observation Method | Trait Type |
|---|---|---|---|
| 1 | Cotyledon Shape | VG | PQ |
| 2 | Cotyledon Cross-Section Shape | VG | PQ |
| 3 | Presence of Inner Corolla Wreath in Female Flowers | VG | QL |
| 4 | Color of Inner Corolla Wreath in Female Flowers | VG | PQ |
| 5 | Color Intensity of Inner Corolla Wreath in Female Flowers | VG | QN |
| 6 | Presence of Inner Corolla Wreath in Male Flowers | VG | QL |
| 7 | Color of Inner Corolla Wreath in Male Flowers | VG | PQ |
| 8 | Color Intensity of Inner Corolla Wreath in Male Flowers | VG | QN |
| 9 | Color of Male Flower Corolla | VG | PQ |
| 10 | Shape of Male Flower Tube | VG | PQ |
| 11 | Apex Shape of Male Flower Petals | VG | PQ |
| 12 | Shape of Male Flower Buds | VG | PQ |
| 13 | Length of Male Flower Sepals | VG | QN |
| 14 | Plant Growth Habit | VG | PQ |
| 15 | Presence of Plant Branches | VG | QL |
| 16 | Number of Plant Branches | MS | QN |
| 17 | Stem Color | VG | PQ |
| 18 | Greenness Intensity of Stem | VG | QN |
| 19 | Presence of Stem Patterns | VG | QL |
| 20 | Presence of Stem Tendrils | VG | QL |
| 21 | Leaf Shape | VG | PQ |
| 22 | Leaf Margin Shape | VG | PQ |
| 23 | Leaf Size | MS | QN |
| 24 | Degree of Leaf Sinus | VG | QN |
| 25 | Greenness Intensity of Leaf Adaxial Surface | VG | QN |
| 26 | Presence of White Spots on Leaf Adaxial Surface | VG | QL |
| 27 | Area of White Spots on Leaves | VG | QN |
| 28 | Size of Commercial Fruit | MS | QN |
| 29 | Presence of Fruit Neck in Commercial Fruit | VG | QL |
| 30 | Curvature of Fruit Neck in Commercial Fruit | VG | QL |
| 31 | Number of Fruit Surface Colors in Commercial Fruit | VG | QL |
| 32 | Primary Color of Commercial Fruit Surface | VG | PQ |
| 33 | Color Intensity of Primary Fruit Surface Color | VG | QN |
| 34 | Glossiness of Commercial Fruit Surface | VG | QN |
| 35 | Type of Fruit Surface Patterns | VG | PQ |
| 36 | Color of Primary Fruit Surface Patterns | VG | PQ |
| 37 | Size of Flower Scar on Commercial Fruit | VG | QN |
| 38 | Morphology of Flower Scar End | VG | QN |
| 39 | Length of Fruit Stalk | MS | QN |
| 40 | Color of Fruit Stalk | VG | PQ |
| 41 | Overall Shape of Mature Fruit | VG | PQ |
| 42 | Length of Mature Fruit | MS | QN |
| 43 | Maximum Diameter of Mature Fruit | MS | QN |
| 44 | Fruit Shape Index (Length/Diameter Ratio) | MS | QN |
| 45 | Primary Color of Mature Fruit Surface | VG | PQ |
| 46 | Color Intensity of Primary Mature Fruit Surface Color | VG | QN |
| 47 | Secondary Color of Mature Fruit Surface | VG | PQ |
| 48 | Type of Mature Fruit Surface Patterns | VG | PQ |
| 49 | Presence of Fruit Furrows | VG | QL |
| 50 | Depth of Fruit Furrows | VG | QN |
| 51 | Presence of Fruit Ridges | VG | QL |
| 52 | Color of Fruit Ridges | VG | QL |
| 53 | Presence of Fruit Nodules | VG | QL |
| 54 | Number of Fruit Nodules | VG | QN |
| 55 | Color of Fruit Pulp | VG | PQ |
| 56 | Structure of Fruit Pulp | VG | PQ |
| 57 | Seed Size | VG | QN |
| 58 | Seed Shape | VG | PQ |
| 59 | Presence of Outer Seed Coat | VG | QL |
| 60 | Color of Outer Seed Coat | VG | PQ |
| 61 | Color of Inner Seed Coat | VG | PQ |
| No. | Trait Name | Observation Method | Trait Type |
|---|---|---|---|
| 1 | Cotyledon Shape | VG | PQ |
| 2 | Main Vine Length | MS | QN |
| 3 | Main Vine Color | VG | PQ |
| 4 | Number of Lateral Branches | MS | QN |
| 5 | Leaf Size | MS | QN |
| 6 | Degree of Leaf Margin Incision | VG | QN |
| 7 | Greenness of Adaxial Leaf Surface | VG | QN |
| 8 | Density of White Spots on Adaxial Leaf Surface | VG | QN |
| 9 | Initial Flowering Stage | VG | QN |
| 10 | Yellowness of Female Flower Corolla | VG | QN |
| 11 | Length of Female Flower Sepals | MS | QN |
| 12 | Length of Male Flower Sepals | MS | QN |
| 13 | Fruit Longitudinal Diameter | MS/VG | QN |
| 14 | Fruit Transverse Diameter | MS/VG | QN |
| 15 | Ratio of Fruit Longitudinal to Transverse Diameter | MS/VG | QN |
| 16 | Fruit Shape | VG | PQ |
| 17 | Position of Maximum Fruit Transverse Diameter | VG | QN |
| 18 | Outline of Fruit Stalk End | VG | PQ |
| 19 | Outline of Flower Stalk End | VG | PQ |
| 20 | Presence/Absence of Fruit Furrows | VG | QL |
| 21 | Spacing of Fruit Furrows | VG | QN |
| 22 | Depth of Fruit Furrows | VG | QN |
| 23 | Number of Fruit Peel Colors | VG | QN |
| 24 | Primary Color of Fruit Peel | VG | PQ |
| 25 | Intensity of Primary Fruit Peel Color | VG | QN |
| 26 | Secondary Color of Fruit Peel | VG | PQ |
| 27 | Intensity of Secondary Fruit Peel Color | VG | QN |
| 28 | Distribution Pattern of Secondary Fruit Color | VG | PQ |
| 29 | Lignification Status of Fruit Peel | VG | PQ |
| 30 | Diameter of Flower Stalk Base | VG | QN |
| 31 | Primary Color of Fruit Pulp | VG | PQ |
| 32 | Seed Size | VG | QN |
| 33 | Seed Shape | VG | PQ |
| 34 | Seed Coat Color | VG | PQ |
| 35 | Surface Texture of Seed Coat | VG | PQ |
| Marker | Missing Rate | Null Alleles Detection |
|---|---|---|
| NG2 | 0.00% | Existence |
| NW1 | 1.31% | Existence |
| NW112 | 1.31% | Existence |
| NW121 | 0.00% | Existence |
| NW124 | 0.00% | Existence |
| NW125 | 0.00% | Existence |
| NW18 | 0.00% | Existence |
| NW22 | 0.00% | Existence |
| NW23 | 0.33% | Existence |
| NW26 | 0.33% | Existence |
| NW37 | 0.00% | Existence |
| NW42 | 0.00% | Existence |
| NW44 | 0.00% | Existence |
| NW5 | 0.00% | Existence |
| NW51 | 0.00% | Existence |
| NW56 | 0.00% | Existence |
| NW57 | 0.98% | Existence |
| NW59 | 0.00% | Existence |
| NW63 | 0.00% | Existence |
| NW66 | 0.33% | Existence |
| NW84 | 0.98% | Existence |
| NW85 | 0.98% | Existence |
| NW87 | 0.33% | Existence |
| NW98 | 0.33% | Existence |
| Marker Pairs | r² Value | Linkage Strength |
|---|---|---|
| NW1 & NW121 | 0.46 | Weak linkage |
| NW5 & NW51 | 0.44 | Weak linkage |
| NW18 & NW22 | 0.42 | Weak linkage |
| NW37 & NW42 | 0.41 | Weak linkage |
References
- Shen, C.W.; Yuan, J.P.; Li, S.; Xu, Y.; Sun, B.; Zhang, Y.Y.; Khan, N.; Guo, X.L. Genome-wide characterization and expression analysis of yabby gene family in three species of cucurbita and their response of salt stress in Cucurbita moschata. In Horticulture, Environment, and Biotechnology; Springer: Berlin/Heidelberg, Germany, 2025. [Google Scholar]
- Zhang, M.J.; Zhou, C.L.; Ma, L.; Su, W.; Jiang, J.; Hu, X.Y. Influence of ultrasound on the microbiological, physicochemical properties, and sensory quality of different varieties of pumpkin juice. Heliyon 2024, 10, e27927. [Google Scholar] [CrossRef] [PubMed]
- Li, J.-X.; Liu, X.-Q.; Zhao, G.-J.; Luo, J.-N.; Gong, H.; Zheng, X.-M.; Wu, H.-B. Research Progress of Cucurbita (Cucurbita moschata) Breeding. Guangdong Agric. Sci. 2021, 48, 12–21. [Google Scholar]
- FAOSTAT. Food and Agriculture Organization of the United Nations; FAO: Rome, Italy, 2022; Available online: https://www.fao.org/faostat/en/#data/QCL/visualize (accessed on 13 December 2024).
- Protection List of agricultural plant varieties of the People’s Republic of China (10th batch); Gazette of the State Council of the People’s Republic of China: Beijing, China, 2016; pp. 57–60.
- Ministry of Agriculture and Rural Affairs of the People’s Republic of China. 2024. Available online: https://www.moa.gov.cn/ (accessed on 13 December 2024).
- Development Center of Science and Technology, Ministry of Agriculture and Rural Affairs. 2024. Available online: http://www.nybkjfzzx.cn/ (accessed on 5 August 2024).
- Liu, K.-Y.; Yu, S.-S.; Wen, Z.-Y.; Duan, H.; Lu, Y.; Peng, C.-C.; Feng, J.-Y.; Li, Y.-X.; Zheng, H. Test Current Situation and Cultivation Techniques of the Protection of New Variety of Cucurbita moschata Duch. J. Anhui Agric. Sci. 2019, 50, 57–59. [Google Scholar]
- Zheng, Y.-H.; Zhang, H.; Wang, D.-J.; Sun, J.-M.; Wang, X.-M.; Duan, L.-L.; Li, H.; Wang, W.; Li, R.-Y. Development of a Wheat Variety Identification System Based on Fluorescently Labeled SSR Markers. Sci. Agric. Sin. 2014, 47, 3725–3735. [Google Scholar]
- Wang, F.-G.; Yang, Y.; Yi, H.-M.; Zhao, J.-R.; Ren, J.; Wang, L.; Ge, J.-R.; Jiang, B.; Zhang, X.-C.; Tian, H.-L.; et al. Construction of an SSR-Based Standard Fingerprint Database for Corn Variety Authorized in China. Sci. Agric. Sin. 2017, 50, 1–14. [Google Scholar]
- GB/T 39917-2021; Variety Genuineness and Purity Testing of Main Crops with SSR Markers Rice. Ministry of Agriculture and Rural Affairs, People’s Republic of China: Beijing, China, 2021.
- Li, Y.; Ma, X.-F.; Tang, H.; Li, N.; Jiang, D.; Long, G.-Y.; Li, D.-Z.; Niu, Y.; Han, R.-X.; Deng, Z.-N.; et al. SSR Markers Screening for Identification of Citrus Cultivar and Construction of DNA Fingerprinting Library. Sci. Agric. Sin. 2018, 51, 149–159. [Google Scholar]
- Ling, C.; Zhao, H.; Yan, J.; Ma, Y.-X.; Feng, Y.-F.; Zhang, J.-L.; Li, B.; Deng, C.-H.; Xu, Z.-J. Establishment and Application of Variety Identification System Based on SSR Markers for Lettuce. Acta Agric. Boreali-Sin. 2022, 37, 34–44. [Google Scholar]
- Yin, J.; Zhao, H.; Wu, X.; Ma, Y.; Zhang, J.; Li, Y.; Shao, G.; Chen, H.; Han, R.; Xu, Z. SSR marker based analysis for identification and of genetic diversity of non-heading Chinese cabbage varieties. Front. Plant Sci. 2023, 14, 1112748. [Google Scholar] [CrossRef]
- Su, G.-Z.; Li, A.-A.; Liu, Z.-H.; Chen, Y.-H.; Zhang, X.-J.; Ma, Y.-X.; Yang, X.-H.; Deng, C.; Xu, Z.-J. Construction and Application of SSR Marker Identification System for Bitter Gourd Varieties. Sci. Agric. Sin. 2019, 57, 2227–2242. [Google Scholar]
- Sun, H.; Wu, S.; Zhang, G.; Jiao, C.; Guo, S.; Ren, Y.; Zhang, J.; Zhang, H.; Gong, G.; Jia, Z.; et al. Karyotype stability and unbiased fractionation in the paleo-allotetraploid Cucurbita genomes. Mol. Plant. 2017, 10, 1293–1306. [Google Scholar] [CrossRef]
- Montero-Pau, J.; Blanca, J.; Bombarely, A.; Ziarsolo, P.; Esteras, C.; Marti-Gomez, C.; Ferriol, M.; Gomez, P.; Jamilena, M.; Mueller, L.; et al. De novo assembly of the zucchini genome reveals a whole—Genome duplication associated with the origin of the Cucurbita genus. Plant Biotechnol. J. 2018, 16, 1161–1171. [Google Scholar] [CrossRef] [PubMed]
- Yun, T.-H.; Zheng, D.-J.; Xie, L.-S.; Deng, C.-Z. Analysis of Genetic Specificity and DNA Fingerprinting Establishment for the Hainan Island Landraces of Cucurbita moschata. J. Plant Genet. Resour. 2013, 14, 679–685. [Google Scholar]
- Sim, S.; Hong, J.; Kwon, Y. DNA profiling of commercial pumpkin cultivars using simple sequence repeat polymorphisms. Hortic. Environ. Biotechnol. 2015, 56, 811–820. [Google Scholar] [CrossRef]
- Nguyen, N.N.; Kim, M.; Jung, J.K.; Shim, E.J.; Chung, S.M.; Park, Y.; Lee, G.P.; Sim, S.C. Genome-wide SNP discovery and core marker sets for assessment of genetic variations in cultivated pumpkin (Cucurbita spp.). Hortic. Res. 2020, 7, 10. [Google Scholar] [CrossRef]
- Tao, A.-F.; Wei, J.-J.; Liu, X.; Xu, J.-T.; Zhu, Z.-N.; Qi, J.-M. Construction of Molecular Fingerprinting Map for 88 Accessions of Cucurbita by SRAP Markers. J. Plant Genet. Resour. 2017, 18, 225–232. [Google Scholar]
- Liu, Z.F. Pumpkin (C. maxima. L.) Varieties Fingerprint Construction and Molecular Markers Application in Hybrid Purity Detection. Master’s Thesis, Hunan Agricultural University, Changsha, China, 2009. [Google Scholar]
- Zhang, Y.; Wang, P.; Li, E.-N.; Tian, X.-C.; Chen, P. Genetic diversity analysis and SSR fingerprinting of seed-use pumpkin(Cucurbita pepo L.) germplasm. China Cucurbits Veg. 2002, 35, 9–15. [Google Scholar]
- Xiang, C.-G.; Wang, Y.-L.; Zhang, X.-M.; Wang, C.-L.; Yin, L.; Wang, Y.-J. Application of SSR Markers in Genetic Relationships Analysis of Cucurbita. North. Hortic. 2013, 15, 104–108. [Google Scholar]
- NT/Y 2594-2016; General Rules for DNA Molecular Markers for Identification of Plant Varieties. Ministry of Agriculture and Rural Affairs of the People’s Republic of China: Beijing, China, 2016; p. 2.
- Song, W.-L. Establishment and Application of a Technology Platform for DNA Fingerprint Identification of Rapeseed Cultivars Basedon Capillary Electrophoresis with SSR Fluorescence Makers. Master’s Thesis, Chinese Academy of Agricultural Sciences, Beijing, China, 2013. [Google Scholar]
- Zhang, D.; Ren, L.-F.; Zhang, L.-L.; Zhang, W.-H.; Wang, T.-Z. Construction of SSR Fingerprint of Alfalfa Based on Fluorescent Capillary Electrophoresis. Chin. J. Grassl. 2019, 42, 10–14. [Google Scholar]
- Wei, M.-M.; Huang, X.; Li, W.-G.; Huang, H.-S. Using the SSR Fluorescent Labeling to Establish SSR Fingerprints for 37 Cultivars in rubber tree (Hevea brasiliensis). Chin. J. Trop. Crops 2019, 43, 1565–1576. [Google Scholar]
- Zhu, H.-S.; Wang, B.; Ye, X.-R.; Liu, J.-T.; Li, Y.-P.; Chen, M.-D.; Lin, H.; Wen, Q.-F. Analysis on SSR Information in Transcriptome and Development of Molecular Markers in Cucurbita moschata Duch. Chin. J. Cell Biol. 2019, 41, 468–475. [Google Scholar]
- Wang, Y.-E.; Xing, N.-L.; Ying, Q.-S.; Zhang, H.-B.; Huang, Y.-P.; Wang, Y.-H. SSR analysis of genetic diversity of Cucurbita germplasm resources. J. Zhejiang Agric. Sci. 2017, 58, 1161–1165. [Google Scholar]
- Huang, K.-M.; Zou, Y.-J.; Ying, Y.-N.; Yan, S.-B.; Bao, J.-S. SSR molecular marker analysis and agronomic character diversity of Cucurbita cultivars. J. Nucl. Agric. Sci. 2019, 35, 2746–2755. [Google Scholar]
- Zou, J.F. Analysis of Genetic Diversity of Cucurbita moschata Germplasm Resources by SSR and InDel Markers. Master’s Thesis, Foshan University of Science and Technology, Foshan, China, 2020. [Google Scholar]
- Wang, R.; Wu, T.-Q.; Zhong, Y.-Q.; Huang, H.-X. Genetic Relationship Analysis of 95 Accessions of Squash Germplasms by SSR Markers. Chin. Agric. Sci. Bull. 2016, 32, 135–142. [Google Scholar]
- Gong, L.; Stift, G.; Kofler, R.; Pachner, M.; Lelley, T. Microsatellites for the genus cucurbita and an SSR-based genetic linkage map of Cucurbita pepo L. Theor. Appl. Genet. 2008, 117, 37–48. [Google Scholar] [CrossRef]
- Wu, T.; Luo, S.; Wang, R.; Zhong, Y.; Xu, X.; Lin, Y.; He, X.; Sun, B.; Huang, H. The first Illumina-based de novo transcriptome sequencing and analysis of Cucurbita (Cucurbita moschata Duch.) and SSR marker development. Mol. Breed. 2014, 34, 1437–1447. [Google Scholar] [CrossRef]
- Blanca, J.; Canizares, J.; Roig, C.; Ziarsolo, P.; Nuez, F.; Pico, B. Transcriptome characterization and high throughput SSRs and SNPs discovery in Cucurbita pepo (Cucurbitaceae). BMC Genom. 2011, 12, 104. [Google Scholar] [CrossRef]
- Xie, Q.; Zhang, S.-Y.; Jiang, L.; Ding, M.-Y.; Liu, L.-L.; Wu, R.-J.; Chen, Q.-X. Analysis of Canarium album Transcriptome SSR Information and Molecular Marker Development and Application. Acta Hortic. Sin. 2019, 50, 2350–2364. [Google Scholar]
- Wang, H.; Nie, Z.-X.; Yang, S.-H.; Shao, Z.-Y.; Wang, T.-L.; Zheng, J.-R. Analysis of Simple Sequence Repeat (SSR) Loci and Marker Development in Eggplant (Solanum melongena L.) Pericarp Transcriptome. North. Hortic. 2024, 21, 1–8. [Google Scholar]
- NY/T 2762-2015; Guidelines for Testing Distinctness, Uniformity and Stability of New Plant Varieties Cucurbitas (Cucurbita moschata). Ministry of Agriculture and Rural Affairs, People’s Republic of China: Beijing, China, 2015.
- NY/T 2343-2013; Guidelines for Testing Distinctness, Uniformity and Stability of New Plant Varieties Zucchini. Ministry of Agriculture and Rural Affairs, People’s Republic of China: Beijing, China, 2013.
- Xiong, F.-Q.; Liu, J.; Liu, J.; He, L.-Q.; Jiang, J.; Tang, X.-M.; Huang, Z.-P.; Wu, H.-N.; Zhong, R.-C.; Han, J.-Q.; et al. Comparative Analysis and Application of Five Improved CTAB Extraction Methods for Peanut DNA. Mol. Plant Breed. 2019, 17, 2207–2216. [Google Scholar]
- Schenk, J.J.; Becklund, L.E.; Carey, S.J.; Fabre, P.P. What is the “modified” ctab protocol? Characterizing modifications to the ctab dna extraction protocol. Appl. Plant Sci. 2023, 11, e11517. [Google Scholar] [CrossRef]
- Maloo, S.R.; Sharma, R.; Soan, H. SSR based genetic diversity analysis in fenugreek (trigonella foenum-graecum l.) Genotypes. Legume Res. 2023, 46, 307–311. [Google Scholar] [CrossRef]
- Liu, X.N.; Xing, H.Y.; Kong, F.Q.; Zhang, K.F.; Cao, Y.; Guo, X.Y.; Li, Q.; Wang, J.X.; Jing, T.Z.; Zhan, Y.G.; et al. Molecular identification of f1 hybrids of fraxinus mandshurica x fraxinus chinensis using SSR markers. Gene 2025, 959, 149507. [Google Scholar] [CrossRef]
- Hu, Q.; Yao, Y.H.; Cui, Y.M.; Li, X.; An, L.K.; Bai, Y.X.; Ding, B.J.; Yao, X.H.; Wu, K.L. Genetic diversity analysis and DNA fingerprinting of primary qingke (hordeum vulgare l. Var. Nudum hook. F.) Cultivars. Genet. Resour. Crop Evol. 2025, 72, 1803–1818. [Google Scholar] [CrossRef]
- Nagpal, S.; Sirari, A.; Sharma, P.; Singh, S.; Mandahal, K.S.; Singh, H.; Singh, S. Marker trait association for biological nitrogen fixation traits in an interspecific cross of chickpea (cicer arietinum x cicer reticulatum). Physiol. Mol. Biol. Plants 2023, 29, 1005–1018. [Google Scholar] [CrossRef] [PubMed]
- Chiou, C.Y.; Shih, H.C.; Tsai, C.C.; Jin, X.L.; Ko, Y.Z.; Mantiquilla, J.A.; Weng, I.S.; Chiang, Y.C. The genetic relationships of indian jujube (ziziphus mauritiana lam.) Cultivars using SSR markers. Heliyon 2020, 6, e05078. [Google Scholar] [CrossRef]
- Bai, X.Q.; Zhang, S.J.; Wang, W.; Chen, Y.; Zhao, Y.Q.; Shi, F.H.; Zhu, C.C. Genetic relationships of 118 castanea specific germplasms and construction of their molecular ID based on morphological characteristics and SSR markers. Plants 2023, 12, 1438. [Google Scholar] [CrossRef]
- Basile, B.; Rao, R.; Corrado, G. Genotypic diversity and population structure of the apricot landraces of the campania region (southern italy) based on fluorescent SSRs. Genet. Resour. Crop Evol. 2023, 70, 125–134. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenALEx 6.5, Genetic analysis in Excel. Population genetic software for teaching and reserach, an update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef]
- Liu, K.J.; Muse, S.V. PowerMarker, An integrated analysis environment for genetic marker analysis. Bioinformatics 2005, 21, 2128–2129. [Google Scholar] [CrossRef]
- Okan, K.; Sevindik, E.; Paksoy, M.Y. Phylogenetic analysis of the endemic bornmuellera hausskn. Spp. (Brassicaceae) in turkiye based on nuclear its and chloroplast trnl intron, trnl-f, rbcl and trnq-rps16 DNA sequences. Genet. Resour. Crop Evol. 2024, 71, 1529–1539. [Google Scholar] [CrossRef]
- Dong, B.N.; Li, Q.Y.; Zhang, T.T.; Liang, X.; Jia, M.S.; Fu, Y.S.; Bai, J.; Fu, S.B. Population genetic polymorphism of skeletal muscle strength related genes in five ethnic minorities in north China. Front. Genet. 2021, 12, 756802. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.H. Data analysis with RStudio: An easygoing introduction. Biometrics 2021, 77, 1502–1503. [Google Scholar] [CrossRef]
- Li, Y.L.; Liu, J.X. Structureselector: A web-based software to select and visualize the optimal number of clusters using multiple methods. Mol. Ecol. Resour. 2018, 18, 176–177. [Google Scholar] [CrossRef] [PubMed]
- Van Oosterhout, C.; Hutchinson, W.F.; Wills, D.; Shipley, P. Micro-checker: Software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 2004, 4, 535–538. [Google Scholar] [CrossRef]
- Rousset, F. Genepop′007:: A complete re-implementation of the genepop software for windows and linux. Mol. Ecol. Resour. 2008, 8, 103–106. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE, A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Wu, C.-L.; Zhang, Q.-Q.; Dong, B.-X.; Li, S.-H.; Zhang, C.-H. Analysis of Genetic Structure and Genetic Relationships of Partial Maize Inbred Lines in China. Acta Agron. Sin. 2010, 36, 1820–1831. [Google Scholar]
- Wang, Y.Y. Studies on Genetic Diversty of Cucurbita Pepo Germplasm Resources. Master’s Thesis, Northeast Agricultural University, Harbin, China, 2016. [Google Scholar]
- Liu, C. Studies On Genentic Diversity of Seed-Used Pumpkin Germplasm Resources. Master’s Thesis, Northeast Agricultural University, Harbin, China, 2012. [Google Scholar]
- Liu, H.; Xu, Z.-J.; Rao, D.-H.; Lu, Q.; Li, S.-X.; Liu, H.-Y.; Chen, X.-P.; Liang, X.-Q.; Hong, Y.-B. Genetic diversity analysis and distinctness identification of peanut cultivars based on morphological traits and SSR markers. Acta Agron. Sin. 2019, 45, 26–36. [Google Scholar] [CrossRef]






| Primer No. | Primer Name | Chromosome | Forward Primer Sequence (5′→3′) | Reverse Primer Sequence (5′→3′) | Primer Source |
|---|---|---|---|---|---|
| NW56 | CMTp252 | 1 | TCTCACTCATCCACCACACC | CGTTTCGAGTCATTTGTTCG | [34] |
| NG2 | NG2 | 2 | CAGCTTCTTCAATCTCGCGC | CCTCCACAACAACAAGCAGC | Independent development |
| NW18 | S231 | 3 | ACCCCACCAAATTAATGCAG | AGAGCCCACTGTGATGACCT | [30] |
| NW1 | ZN1 | 4 | GCTTGAACAGAGATGGAGGG | AAAGTCGCTGAGAGCTGGAG | [29] |
| NW121 | CUTC017708 | 4 | TGTGATTTGTTGGGCTCTCTGT | ACATGGAAATCCACCTCTTCGT | [36] |
| NW5 | ZN8 | 5 | TCGCTTCACCGGTAATTTTC | GCGCTGAAGAATCCATGTTT | [29] |
| NW85 | CMTm137 | 6 | ACGTTCTTTCTGCCTCCAT | ATGCCCATTATGCAATTCTC | [34] |
| NW66 | CMTm178 | 7 | AGGTGGCATCTGTACACTGAG | TGAACAAACTCCACACCAATAG | [34] |
| NW98 | Unigene0001517 | 8 | TCCTCGTCGGAACAGAACTT | TCTAACTTGGAGCCTGGAGC | [35] |
| NW59 | CMTm42 | 9 | ATTGGTGCCGAAGCTATCAC | CCCACGTTATGGAGCAGAAT | [34] |
| NW44 | CMTp34 | 10 | GAAGGCGAGGTTTTGAGTTG | GGCGGAACCCTAAGAAATGT | [34] |
| NW84 | CMTm37 | 11 | CGTGTTTGTGTTGGAAAAGC | AAGCAAACCCACAGAAGGAG | [34] |
| NW26 | CMTp257 | 12 | CACGAAGATTTGATGGCCTTA | GGATTGGGATGGTGAAGATG | [31] |
| NW125 | CUTC046645 | 13 | CCCTCGGTAGCAATTTTGTAGT | TTTGGTGGGAGTGATACTGATG | [36] |
| NW42 | S65 | 14 | ATCATAGTCGTCGTCGGGTC | GCCGATTCTTGAGGAACAGA | [33] |
| NW22 | S254 | 15 | CACCTGGCTGTTTTGTCTGA | ACATGGGCATACCTCGAATC | [30] |
| NW87 | Unigene0000190 | 15 | TGTGGGGTTTTGCTTTTAGG | ATCCAAAATGGTGGTGCATT | [35] |
| NW23 | S263 | 16 | TGAAATGAACGCAGAATTGC | ACTTGCGGACTTTCACACCT | [30] |
| NW57 | CMTp265 | 16 | CCTCTCATTCTTCCCCATCTC | TGATCCGGTAGGGGTCTACTC | [34] |
| NW51 | CMTp209 | 17 | TCACTTTACAACCAGAAGCTGA | CACTTTGCTGCTCATCCAC | [34] |
| NW37 | S35 | 18 | CGGTCGTGAATACATCATGG | GCTCCACCAATGGGAAACTA | [33] |
| NW63 | CMTm127 | 18 | CAAATTCAGACGCTTCTTTTGG | AGAATTGAGCAAAAAGGAGATGG | [34] |
| NW112 | CUTC006703 | 19 | CGACTACAGCTCAACAAAGACC | GAAGTCTATTACGCCCAATTCC | [36] |
| NW124 | CUTC022867 | 20 | TTGGACAATCTGAGGAAGTTGG | AAGGTTTGTCCTTCGACAAGGT | [36] |
| Group | FAM | TAMRA | ROX | HEX |
|---|---|---|---|---|
| 1 | NG2 (152–175) | NW26 (105–150) | NW98 (190–225) | NW57 (76–100) |
| NW37 (235–275) | ||||
| 2 | NW18 (125–155) | NW23 (115–155) | NW56 (158–200) | NW63 (85–115) |
| NW59 (202–233) | ||||
| 3 | NW44 (116–134) | NW85 (80–105) | NW51 (105–128) | NW121 (205–235) |
| NW22 (135–175) | ||||
| 4 | NW66 (110–134) | NW84 (134–160) | NW112 (80–134) | NW125 (170–200) |
| NW5 (168–184) | ||||
| 5 | NW1 (190–235) | NW87 (105–155) | NW124 (114–137) | |
| NW42 (139–166) |
| Primer No. | MAF | GT | Na | Ne | Ho | He | PIC | I |
|---|---|---|---|---|---|---|---|---|
| NG2 | 0.35 | 19 | 7 | 4.362 | 0.268 | 0.771 | 0.737 | 1.621 |
| NW1 | 0.394 | 17 | 9 | 3.939 | 0.321 | 0.746 | 0.709 | 1.548 |
| NW112 | 0.664 | 6 | 4 | 1.99 | 0.265 | 0.497 | 0.442 | 0.907 |
| NW121 | 0.407 | 11 | 5 | 3.529 | 0.333 | 0.717 | 0.67 | 1.393 |
| NW124 | 0.609 | 8 | 4 | 2.292 | 0.137 | 0.564 | 0.514 | 1.037 |
| NW125 | 0.345 | 7 | 4 | 3.036 | 0.15 | 0.671 | 0.598 | 1.13 |
| NW18 | 0.43 | 18 | 7 | 3.824 | 0.395 | 0.739 | 0.706 | 1.565 |
| NW22 | 0.386 | 20 | 10 | 4.25 | 0.297 | 0.765 | 0.734 | 1.703 |
| NW23 | 0.307 | 16 | 8 | 4.26 | 0.105 | 0.765 | 0.728 | 1.613 |
| NW26 | 0.339 | 16 | 7 | 3.647 | 0.151 | 0.726 | 0.677 | 1.441 |
| NW37 | 0.314 | 16 | 8 | 4.47 | 0.212 | 0.776 | 0.741 | 1.63 |
| NW42 | 0.513 | 15 | 8 | 2.786 | 0.206 | 0.641 | 0.586 | 1.231 |
| NW44 | 0.786 | 4 | 3 | 1.509 | 0.016 | 0.337 | 0.281 | 0.529 |
| NW5 | 0.324 | 10 | 5 | 4.009 | 0.225 | 0.751 | 0.708 | 1.472 |
| NW51 | 0.528 | 11 | 5 | 2.589 | 0.16 | 0.614 | 0.55 | 1.098 |
| NW56 | 0.32 | 22 | 11 | 4.14 | 0.203 | 0.758 | 0.721 | 1.621 |
| NW57 | 0.389 | 13 | 5 | 3.423 | 0.182 | 0.708 | 0.658 | 1.368 |
| NW59 | 0.418 | 13 | 7 | 3.446 | 0.111 | 0.71 | 0.662 | 1.396 |
| NW63 | 0.265 | 11 | 6 | 4.625 | 0.297 | 0.784 | 0.749 | 1.6 |
| NW66 | 0.511 | 9 | 4 | 2.408 | 0.148 | 0.585 | 0.502 | 1.002 |
| NW84 | 0.396 | 5 | 3 | 2.947 | 0.086 | 0.661 | 0.587 | 1.09 |
| NW85 | 0.31 | 8 | 5 | 4.196 | 0.297 | 0.762 | 0.722 | 1.501 |
| NW87 | 0.336 | 17 | 8 | 4.448 | 0.292 | 0.775 | 0.741 | 1.614 |
| NW98 | 0.4 | 16 | 9 | 3.882 | 0.187 | 0.742 | 0.706 | 1.584 |
| Total | 10.041 | 308 | 152 | 84.007 | 5.044 | 16.565 | 15.429 | 32.694 |
| Average | 0.418 | 12.833 | 6.333 | 3.5 | 0.21 | 0.69 | 0.643 | 1.362 |
| No. | Variety Name | Sample Preservation Site | No. | Variety Name | Sample Preservation Site |
|---|---|---|---|---|---|
| 1 | Panlong204 | New Plant Variety Preservation Center, MARA | 11 | Yuanbaosanhao | New Plant Variety Preservation Center, MARA |
| 2 | Naiyounangua | Anhui Jianghuai Horticulture Co., LTD | 12 | Ziguan No.8 | New Plant Variety Preservation Center, MARA |
| 3 | Jiangyimopan16005 | New Plant Variety Preservation Center, MARA | 13 | Jindianjiuhao | New Plant Variety Preservation Center, MARA |
| 4 | Jibei | New Plant Variety Preservation Center, MARA | 14 | HS512 | New Plant Variety Preservation Center, MARA |
| 5 | Jiuyuanjinfeng108 | New Plant Variety Preservation Center, MARA | 15 | Nongren22 | New Plant Variety Preservation Center, MARA |
| 6 | Sizhuang21 | New Plant Variety Preservation Center, MARA | 16 | HCA5 | New Plant Variety Preservation Center, MARA |
| 7 | Linongjinxiang | New Plant Variety Preservation Center, MARA | 17 | Xiyangyang No.3 | Anhui Jianghuai Horticulture Co., Ltd. (Hefei, Anhui, China) |
| 8 | PK529 | New Plant Variety Preservation Center, MARA | 18 | Linonglüguizunangua | Guangdong Helinong Biological Seed Industry Co., Ltd. (Shantou, Guangdong, China) |
| 9 | Donghu No.16 | New Plant Variety Preservation Center, MARA | 19 | Huiyou3Haobeibeinangua | Guangdong Helinong Biological Seed Industry Co., Ltd. (Shantou, Guangdong, China) |
| 10 | Huangbeibei | New Plant Variety Preservation Center, MARA | 20 | Jinbei No.5 | Anhui Jianghuai Horticulture Co., Ltd. (Hefei, Anhui, China) |
| Region | Shannon’s Information Index | Expected Heterozygosity |
|---|---|---|
| East China | 1.267 | 0.648 |
| North China | 1.203 | 0.613 |
| South China | 1.031 | 0.566 |
| Northwest China | 1.338 | 0.692 |
| Trait | Group 2 | Group 3 | |||
|---|---|---|---|---|---|
| HC203 | PK No.2 | Zhongtai268 | Jiarui570 | Jiuyuanjinfeng118 | |
| Mature fruit shape | 6 (Long oval) | 6 (Long oval) | 10 (pear shape) | 12 (bar shape) | 12 (bar shape) |
| Mature fruit surface secondary color(stripe) | 1 (milky white) | 5 (orange) | 5 (orange) | 1 (milky white) | 5 (orange) |
| Mature fruit stripe type | 1 (dot) | 4 (mixed) | 1 (dot) | 1 (dot) | 1 (dot) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zou, J.; Wu, X.; Liu, X.; Peng, C.; Ma, Y.; Zhang, X.; Yang, X.; Luo, S.; Xing, W.; Hong, H.; et al. Identification and Genetic Diversity Analysis of Cucurbita Varieties Based on SSR Markers. Agronomy 2025, 15, 1420. https://doi.org/10.3390/agronomy15061420
Zou J, Wu X, Liu X, Peng C, Ma Y, Zhang X, Yang X, Luo S, Xing W, Hong H, et al. Identification and Genetic Diversity Analysis of Cucurbita Varieties Based on SSR Markers. Agronomy. 2025; 15(6):1420. https://doi.org/10.3390/agronomy15061420
Chicago/Turabian StyleZou, Jialong, Xingting Wu, Xuejing Liu, Changcheng Peng, Yingxue Ma, Xiujie Zhang, Xuhong Yang, Shuailong Luo, Weigeng Xing, Hao Hong, and et al. 2025. "Identification and Genetic Diversity Analysis of Cucurbita Varieties Based on SSR Markers" Agronomy 15, no. 6: 1420. https://doi.org/10.3390/agronomy15061420
APA StyleZou, J., Wu, X., Liu, X., Peng, C., Ma, Y., Zhang, X., Yang, X., Luo, S., Xing, W., Hong, H., Li, L., Tan, B., Jing, R., & Xu, Z. (2025). Identification and Genetic Diversity Analysis of Cucurbita Varieties Based on SSR Markers. Agronomy, 15(6), 1420. https://doi.org/10.3390/agronomy15061420







